Further Characterization of Fungal Halogenase RadH and Its Homologs
Abstract
1. Introduction
2. Materials and Methods
2.1. Halogenase Cloning, Expression, and Purification
2.2. Method for Flavin Reductase (Fre) Cloning, Expression, and Purification
2.3. Glucose Dehydrogenase from Pseudomonas sp. (Gdhi)
2.4. Crystallization and X-ray Crystallography of RadH
2.5. Molecular Docking
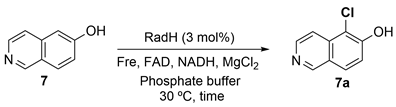
2.6. RadH-Catalyzed Halogenation
2.7. Determination of Halogenation Site
2.8. Reagents and Materials
2.9. Analytical Methods
2.9.1. Analytical HPLC Method
2.9.2. General LC-MS Method
3. Results
3.1. New Functional RadH Homologs
3.2. Reaction Optimization
3.3. Screen of Substrate Specificities of RadH and Its Homologs
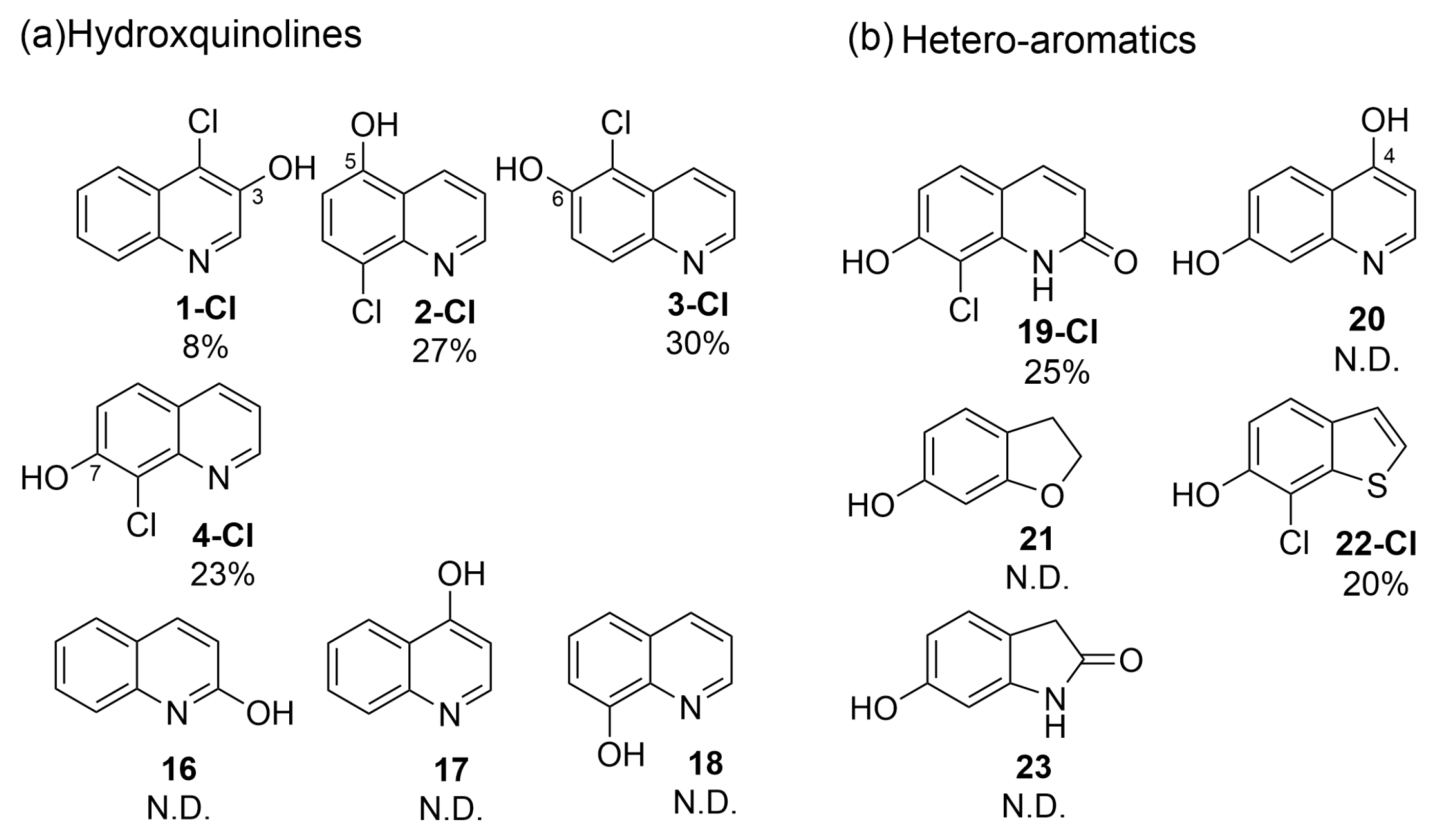
3.4. Quinoline and Heterocyclic Substrate Specificities
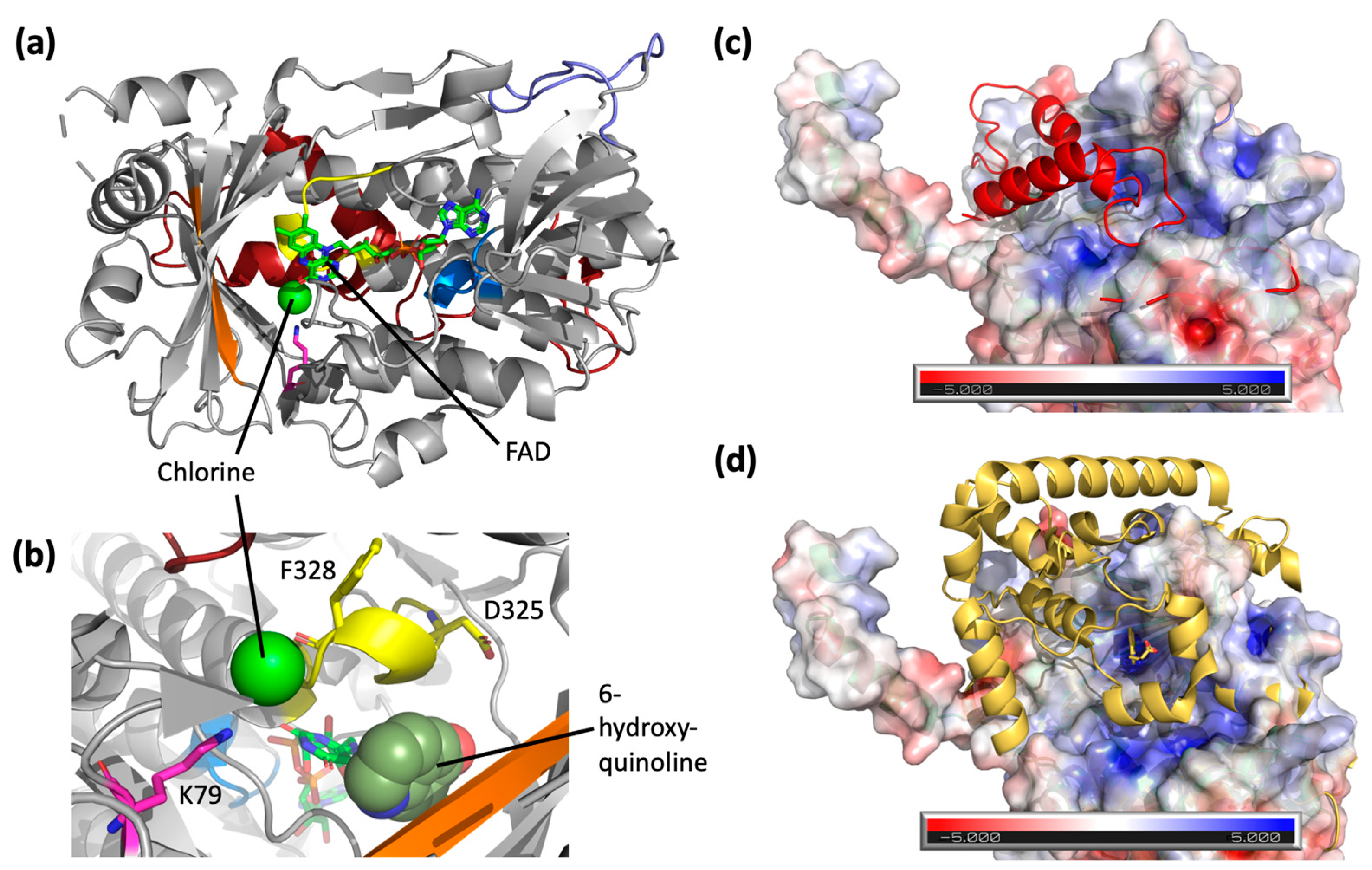
3.5. Crystal Structure of RadH
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Correction Statement
References
- Jeschke, P. Manufacturing Approaches of New Halogenated Agrochemicals. Eur. J. Org. Chem. 2022, 2022, e202101513. [Google Scholar] [CrossRef]
- Matern, J.; Bäumer, N.; Fernández, G. Unraveling Halogen Effects in Supramolecular Polymerization. J. Am. Chem. Soc. 2021, 143, 7164–7175. [Google Scholar] [CrossRef]
- Benedetto Tiz, D.; Bagnoli, L.; Rosati, O.; Marini, F.; Sancineto, L.; Santi, C. New Halogen-Containing Drugs Approved by FDA in 2021: An Overview on Their Syntheses and Pharmaceutical Use. Molecules 2022, 27, 1643. [Google Scholar] [CrossRef]
- Schnepel, C.; Sewald, N. Enzymatic Halogenation: A Timely Strategy for Regioselective C−H Activation. Chem. Eur. J. 2017, 23, 12064–12086. [Google Scholar] [CrossRef]
- Agarwal, V.; Miles, Z.D.; Winter, J.M.; Eustáquio, A.S.; El Gamal, A.A.; Moore, B.S. Enzymatic Halogenation and Dehalogenation Reactions: Pervasive and Mechanistically Diverse. Chem. Rev. 2017, 117, 5619–5674. [Google Scholar] [CrossRef] [PubMed]
- Latham, J.; Brandenburger, E.; Shepherd, S.A.; Menon, B.R.K.; Micklefield, J. Development of Halogenase Enzymes for Use in Synthesis. Chem. Rev. 2018, 118, 232–269. [Google Scholar] [CrossRef] [PubMed]
- Dong, C.; Kotzsch, A.; Dorward, M.; van Pée, K.H.; Naismith, J.H. Crystallization and X-ray diffraction of a halogenating enzyme, tryptophan 7-halogenase, from Pseudomonas fluorescens. Acta Crystallogr. D Biol. Crystallogr. 2004, 60, 1438–1440. [Google Scholar] [CrossRef]
- Yeh, E.; Blasiak, L.C.; Koglin, A.; Drennan, C.L.; Walsh, C.T. Chlorination by a Long-Lived Intermediate in the Mechanism of Flavin-Dependent Halogenases. Biochemistry 2007, 46, 1284–1292. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; De Laurentis, W.; Leang, K.; Herrmann, J.; Ihlefeld, K.; van Pée, K.H.; Naismith, J.H. Structural insights into regioselectivity in the enzymatic chlorination of tryptophan. J. Mol. Biol. 2009, 391, 74–85. [Google Scholar] [CrossRef]
- Andorfer, M.C.; Grob, J.E.; Hajdin, C.E.; Chael, J.R.; Siuti, P.; Lilly, J.; Tan, K.L.; Lewis, J.C. Understanding Flavin-Dependent Halogenase Reactivity via Substrate Activity Profiling. ACS Catal. 2017, 7, 1897–1904. [Google Scholar] [CrossRef] [PubMed]
- Chooi, Y.H.; Cacho, R.; Tang, Y. Identification of the viridicatumtoxin and griseofulvin gene clusters from Penicillium aethiopicum. Chem. Biol. 2010, 17, 483–494. [Google Scholar] [CrossRef]
- Zeng, J.; Zhan, J. A Novel Fungal Flavin-Dependent Halogenase for Natural Product Biosynthesis. ChemBioChem 2010, 11, 2119–2123. [Google Scholar] [CrossRef] [PubMed]
- Menon, B.R.K.; Brandenburger, E.; Sharif, H.H.; Klemstein, U.; Shepherd, S.A.; Greaney, M.F.; Micklefield, J. RadH: A Versatile Halogenase for Integration into Synthetic Pathways. Angew. Chem. Int. Ed. Engl. 2017, 56, 11841–11845. [Google Scholar] [CrossRef]
- Mori, S.; Pang, A.H.; Thamban Chandrika, N.; Garneau-Tsodikova, S.; Tsodikov, O.V. Unusual substrate and halide versatility of phenolic halogenase PltM. Nat. Commun. 2019, 10, 1255. [Google Scholar] [CrossRef]
- Kabsch, W. XDS. Acta Crystallogr. Sect. D 2010, 66, 125–132. [Google Scholar] [CrossRef] [PubMed]
- Vonrhein, C.; Tickle, I.J.; Flensburg, C.; Keller, P.; Paciorek, W.; Sharff, A.; Bricogne, G. Advances in automated data analysis and processing within autoPROC, combined with improved characterisation, mitigation and visualisation of the anisotropy of diffraction limits using STARANISO. Acta Crystallogr. Sect. A 2018, 74, a360. [Google Scholar] [CrossRef]
- McCoy, A.J.; Grosse-Kunstleve, R.W.; Adams, P.D.; Winn, M.D.; Storoni, L.C.; Read, R.J. Phaser crystallographic software. J. Appl. Crystallogr. 2007, 40, 658–674. [Google Scholar] [CrossRef]
- Adams, P.D.; Afonine, P.V.; Bunkóczi, G.; Chen, V.B.; Davis, I.W.; Echols, N.; Headd, J.J.; Hung, L.W.; Kapral, G.J.; Grosse-Kunstleve, R.W.; et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010, 66, 213–221. [Google Scholar] [CrossRef]
- Croll, T.I. ISOLDE: A physically realistic environment for model building into low-resolution electron-density maps. Acta Crystallogr. D Struct. Biol. 2018, 74, 519–530. [Google Scholar] [CrossRef]
- Available online: http://molprobity.biochem.duke.edu/ (accessed on 26 October 2022).
- Corso, G.; Stärk, H.; Jing, B.; Barzilay, R.; Jaakkola, T. DiffDock: Diffusion Steps, Twists, and Turns for Molecular Docking. arXiv 2022, arXiv:2210.01776v2. [Google Scholar] [CrossRef]
- Neugebauer, M.E.; Sumida, K.H.; Pelton, J.G.; McMurry, J.L.; Marchand, J.A.; Chang, M.C.Y. A family of radical halogenases for the engineering of amino-acid-based products. Nat. Chem. Biol. 2019, 15, 1009–1016. [Google Scholar] [CrossRef] [PubMed]
- Zallot, R.; Oberg, N.; Gerlt, J.A. The EFI Web Resource for Genomic Enzymology Tools: Leveraging Protein, Genome, and Metagenome Databases to Discover Novel Enzymes and Metabolic Pathways. Biochemistry 2019, 58, 4169–4182. [Google Scholar] [CrossRef] [PubMed]
- Peh, G.; Tay, T.; Tan, L.L.; Tiong, E.; Goh, Y.L.; Ye, S.; Lin, F.; Tan, C.J.X.; Tan, Y.Z.; Wong, J.; et al. Site-selective Chlorination of Pyrrolic Heterocycles by Flavin Dependent Enzyme PrnC. ChemRxiv 2022. [Google Scholar] [CrossRef]
- Crowe, C.; Molyneux, S.; Sharma, S.V.; Zhang, Y.; Gkotsi, D.S.; Connaris, H.; Goss, R.J.M. Halogenases: A palette of emerging opportunities for synthetic biology–synthetic chemistry and C–H functionalisation. Chem. Soc. Rev. 2021, 50, 9443–9481. [Google Scholar] [CrossRef] [PubMed]
- Minges, H.; Sewald, N. Recent Advances in Synthetic Application and Engineering of Halogenases. ChemCatChem 2020, 12, 4450–4470. [Google Scholar] [CrossRef]
- Buedenbender, S.; Rachid, S.; Müller, R.; Schulz, G.E. Structure and action of the myxobacterial chondrochloren halogenase CndH: A new variant of FAD-dependent halogenases. J. Mol. Biol. 2009, 385, 520–530. [Google Scholar] [CrossRef]
- Dorrestein, P.C.; Yeh, E.; Garneau-Tsodikova, S.; Kelleher, N.L.; Walsh, C.T. Dichlorination of a pyrrolyl-S-carrier protein by FADH2-dependent halogenase PltA during pyoluteorin biosynthesis. Proc. Natl. Acad. Sci. USA 2005, 102, 13843–13848. [Google Scholar] [CrossRef]
- Neubauer, P.R.; Widmann, C.; Wibberg, D.; Schröder, L.; Frese, M.; Kottke, T.; Kalinowski, J.; Niemann, H.H.; Sewald, N. A flavin-dependent halogenase from metagenomic analysis prefers bromination over chlorination. PLoS ONE 2018, 13, e0196797. [Google Scholar] [CrossRef]
- Sánchez, C.; Butovich, I.A.; Braña, A.F.; Rohr, J.; Méndez, C.; Salas, J.A. The biosynthetic gene cluster for the antitumor rebeccamycin: Characterization and generation of indolocarbazole derivatives. Chem. Biol. 2002, 9, 519–531. [Google Scholar] [CrossRef]
- Zehner, S.; Kotzsch, A.; Bister, B.; Süssmuth, R.D.; Méndez, C.; Salas, J.A.; van Pée, K.H. A regioselective tryptophan 5-halogenase is involved in pyrroindomycin biosynthesis in Streptomyces rugosporus LL-42D005. Chem. Biol. 2005, 12, 445–452. [Google Scholar] [CrossRef]
- Kirner, S.; Hammer, P.E.; Hill, D.S.; Altmann, A.; Fischer, I.; Weislo, L.J.; Lanahan, M.; van Pée, K.H.; Ligon, J.M. Functions encoded by pyrrolnitrin biosynthetic genes from Pseudomonas fluorescens. J. Bacteriol. 1998, 180, 1939–1943. [Google Scholar] [CrossRef] [PubMed]
- Keller, S.; Wage, T.; Hohaus, K.; Hölzer, M.; Eichhorn, E.; van Pée, K.H. Purification and Partial Characterization of Tryptophan 7-Halogenase (PrnA) from Pseudomonas fluorescens. Angew. Chem. Int. Ed. Engl. 2000, 39, 2300–2302. [Google Scholar] [CrossRef]
- Aly, A.A.; El-Sheref, E.M.; Mourad, A.-F.E.; Bakheet, M.E.M.; Bräse, S. 4-Hydroxy-2-quinolones: Syntheses, reactions and fused heterocycles. Mol. Divers. 2020, 24, 477–524. [Google Scholar] [CrossRef] [PubMed]
- Keri, R.S.; Chand, K.; Budagumpi, S.; Balappa Somappa, S.; Patil, S.A.; Nagaraja, B.M. An overview of benzo[b]thiophene-based medicinal chemistry. Eur. J. Med. Chem. 2017, 138, 1002–1033. [Google Scholar] [CrossRef]
- Gerry, C.J.; Schreiber, S.L. Chemical probes and drug leads from advances in synthetic planning and methodology. Nat. Rev. Drug Discov. 2018, 17, 333–352. [Google Scholar] [CrossRef]
- Prakinee, K.; Phintha, A.; Visitsatthawong, S.; Lawan, N.; Sucharitakul, J.; Kantiwiriyawanitch, C.; Damborsky, J.; Chitnumsub, P.; van Pée, K.-H.; Chaiyen, P. Mechanism-guided tunnel engineering to increase the efficiency of a flavin-dependent halogenase. Nat. Catal. 2022, 5, 534–544. [Google Scholar] [CrossRef]
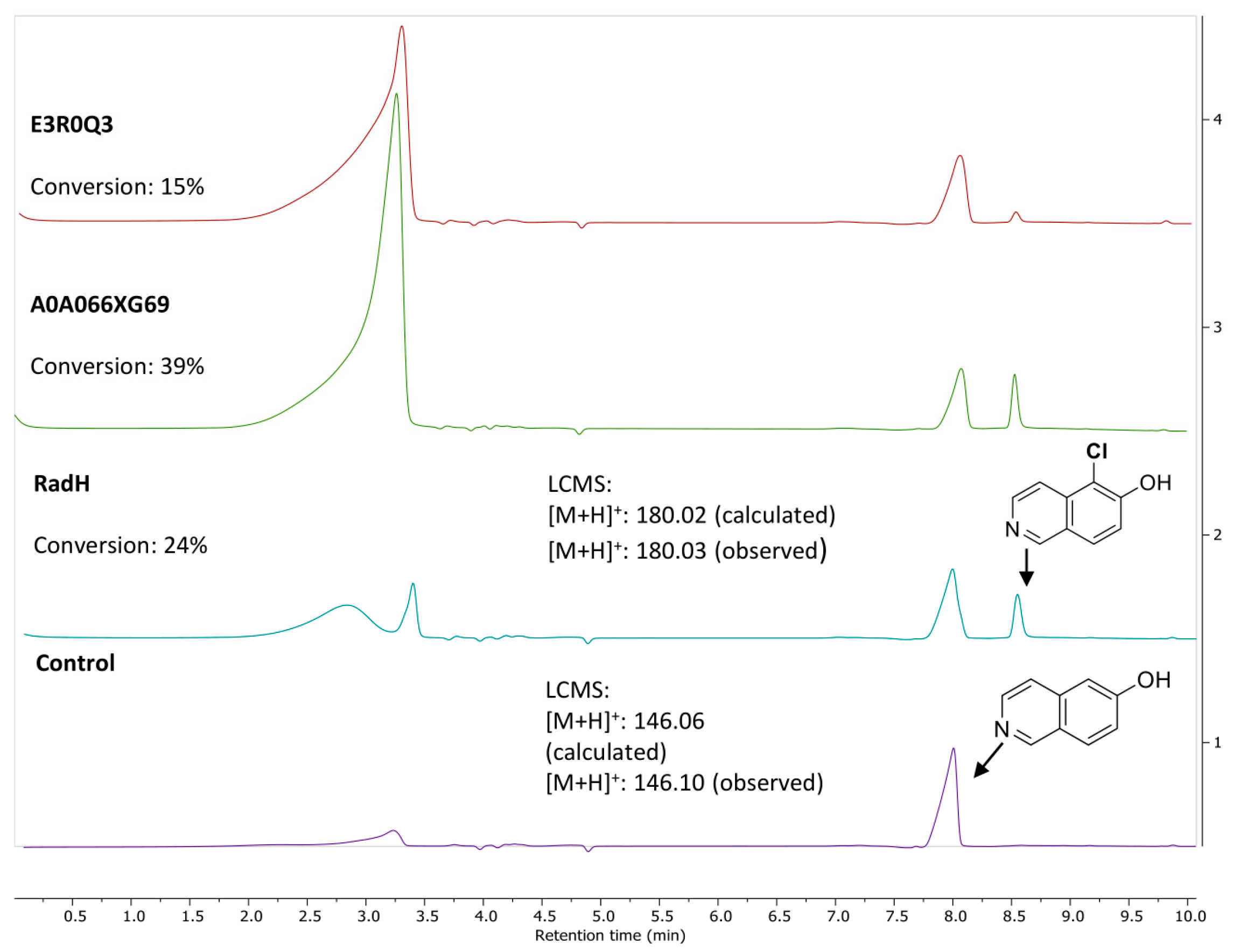

| Entry | Variation from Standard Conditions a | Time (h) | Conversion (%) b |
|---|---|---|---|
| 1 | None | 2 | 32 |
| 2 | 5 mol% RadH, 0.8 mol% of Fre | 18 | 30 |
| 3 | Addition of 0.5 mol% GdHi | 18 | 73 |
| 4 | 20 mol% of FAD, 25 µM of NADH | 18 | 31 |
| Halogenase | Variant | RMSD | TM-Score | Sequence ID (%) |
|---|---|---|---|---|
| CndH | B | 2.39 | 0.81 | 29 |
| PltA | B | 2.65 | 0.84 | 26 |
| Mpy16 | B | 2.7 | 0.84 | 27 |
| CMIS | A | 2.83 | 0.66 | 26 |
| PltM | A | 3.3 | 0.7 | 18 |
| PyrH | A | 4.05 | 0.63 | 17 |
| PrnA | A | 4.08 | 0.62 | 17 |
| RebH | A | 4.14 | 0.61 | 19 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Peh, G.; Gunawan, G.A.; Tay, T.; Tiong, E.; Tan, L.L.; Jiang, S.; Goh, Y.L.; Ye, S.; Wong, J.; Brown, C.J.; et al. Further Characterization of Fungal Halogenase RadH and Its Homologs. Biomolecules 2023, 13, 1081. https://doi.org/10.3390/biom13071081
Peh G, Gunawan GA, Tay T, Tiong E, Tan LL, Jiang S, Goh YL, Ye S, Wong J, Brown CJ, et al. Further Characterization of Fungal Halogenase RadH and Its Homologs. Biomolecules. 2023; 13(7):1081. https://doi.org/10.3390/biom13071081
Chicago/Turabian StylePeh, GuangRong, Gregory A. Gunawan, Terence Tay, Elaine Tiong, Lee Ling Tan, Shimin Jiang, Yi Ling Goh, Suming Ye, Joel Wong, Christopher J. Brown, and et al. 2023. "Further Characterization of Fungal Halogenase RadH and Its Homologs" Biomolecules 13, no. 7: 1081. https://doi.org/10.3390/biom13071081
APA StylePeh, G., Gunawan, G. A., Tay, T., Tiong, E., Tan, L. L., Jiang, S., Goh, Y. L., Ye, S., Wong, J., Brown, C. J., Zhao, H., Ang, E. L., Wong, F. T., & Lim, Y. H. (2023). Further Characterization of Fungal Halogenase RadH and Its Homologs. Biomolecules, 13(7), 1081. https://doi.org/10.3390/biom13071081







