Screening and Identification of a Prognostic Model of Ovarian Cancer by Combination of Transcriptomic and Proteomic Data
Abstract
1. Introduction
2. Materials and Methods
2.1. Extraction of Data
2.2. Construction and Assessment of a Prognosis-Related Protein Model
2.3. Verification of Clinical Relevance and Prognostic Protein Expression
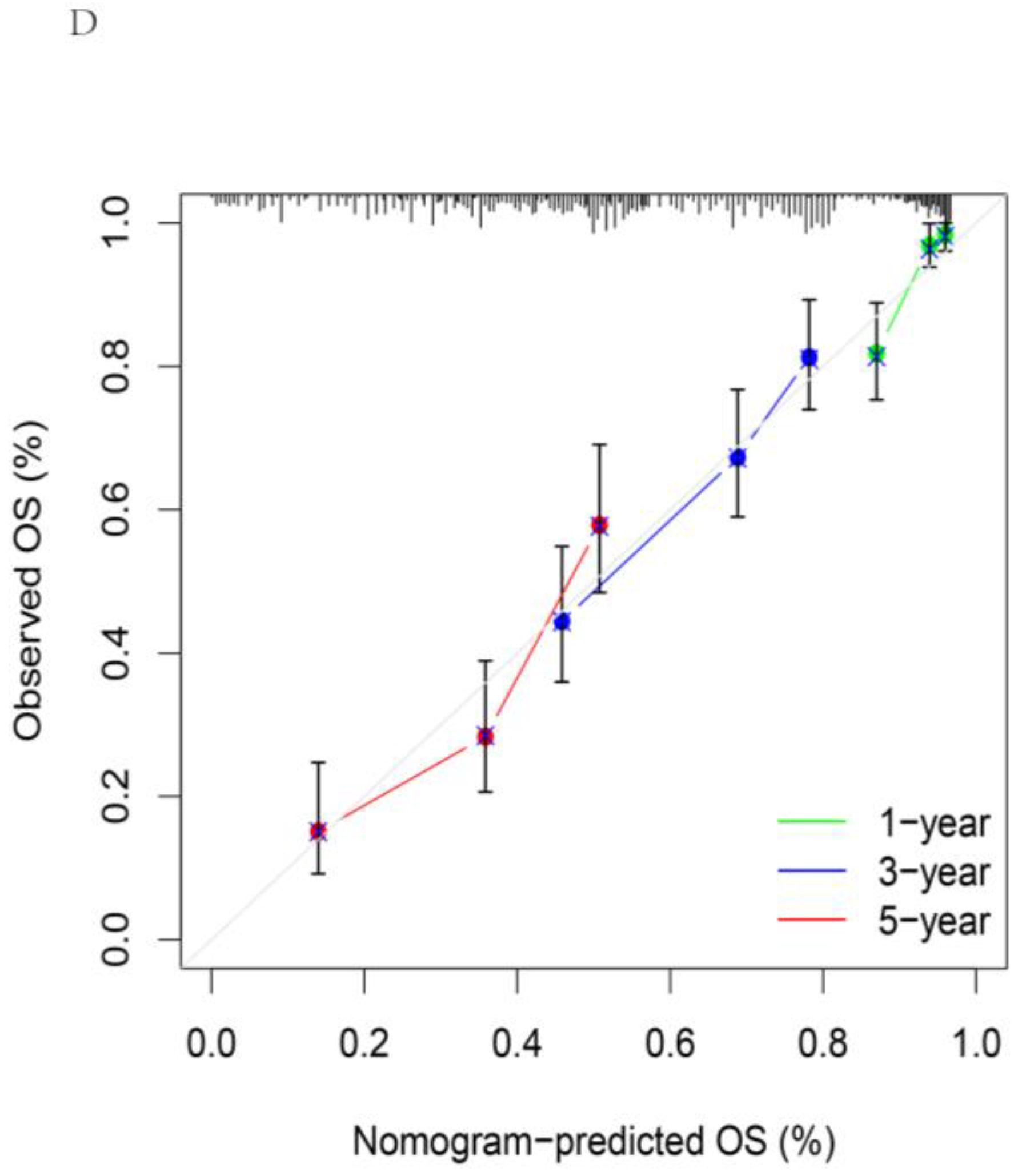
2.4. Construction of a Nomogram for Clinical Survival Prediction
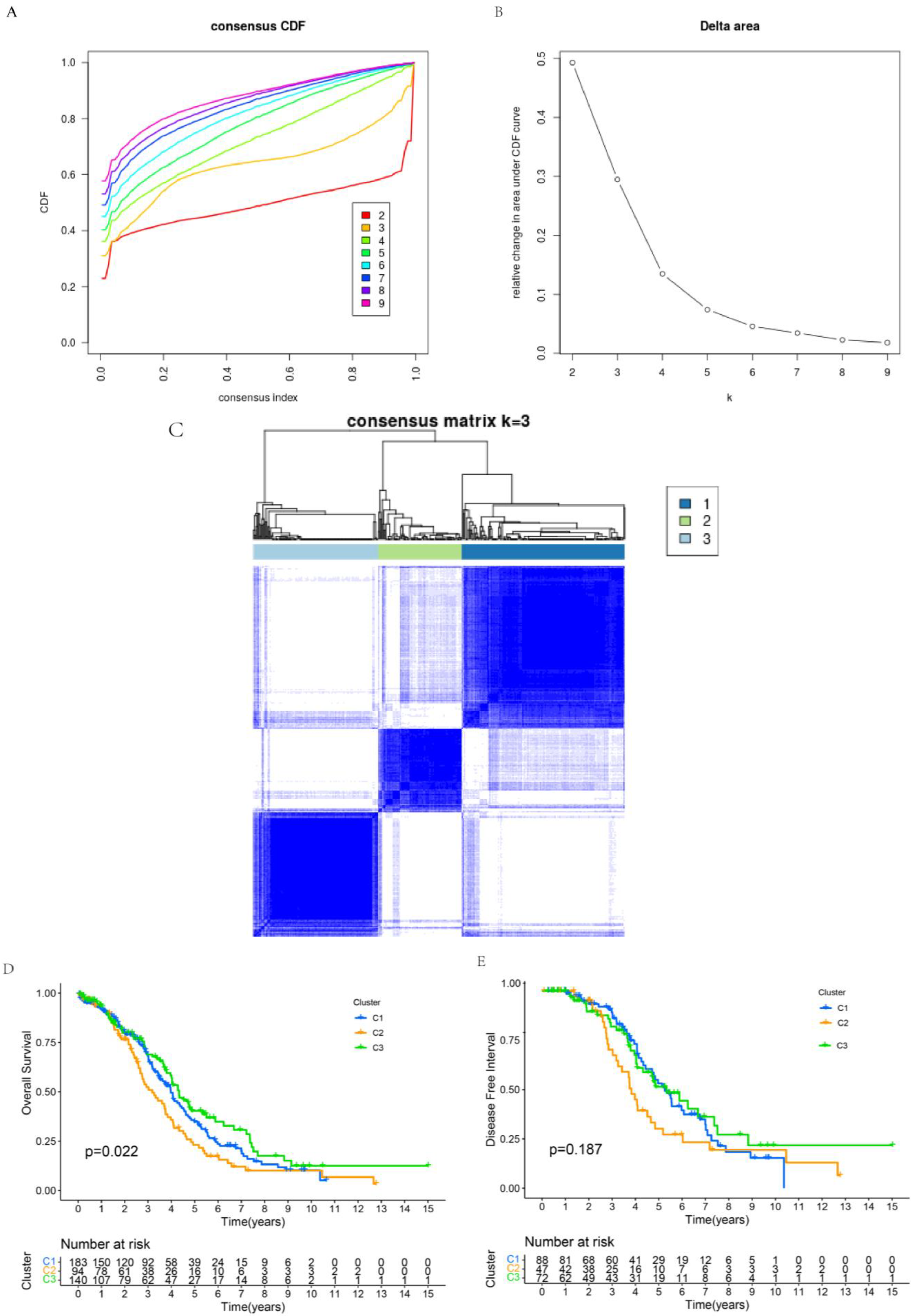
2.5. Consensus Clustering Analysis of Prognostic-Related Proteins
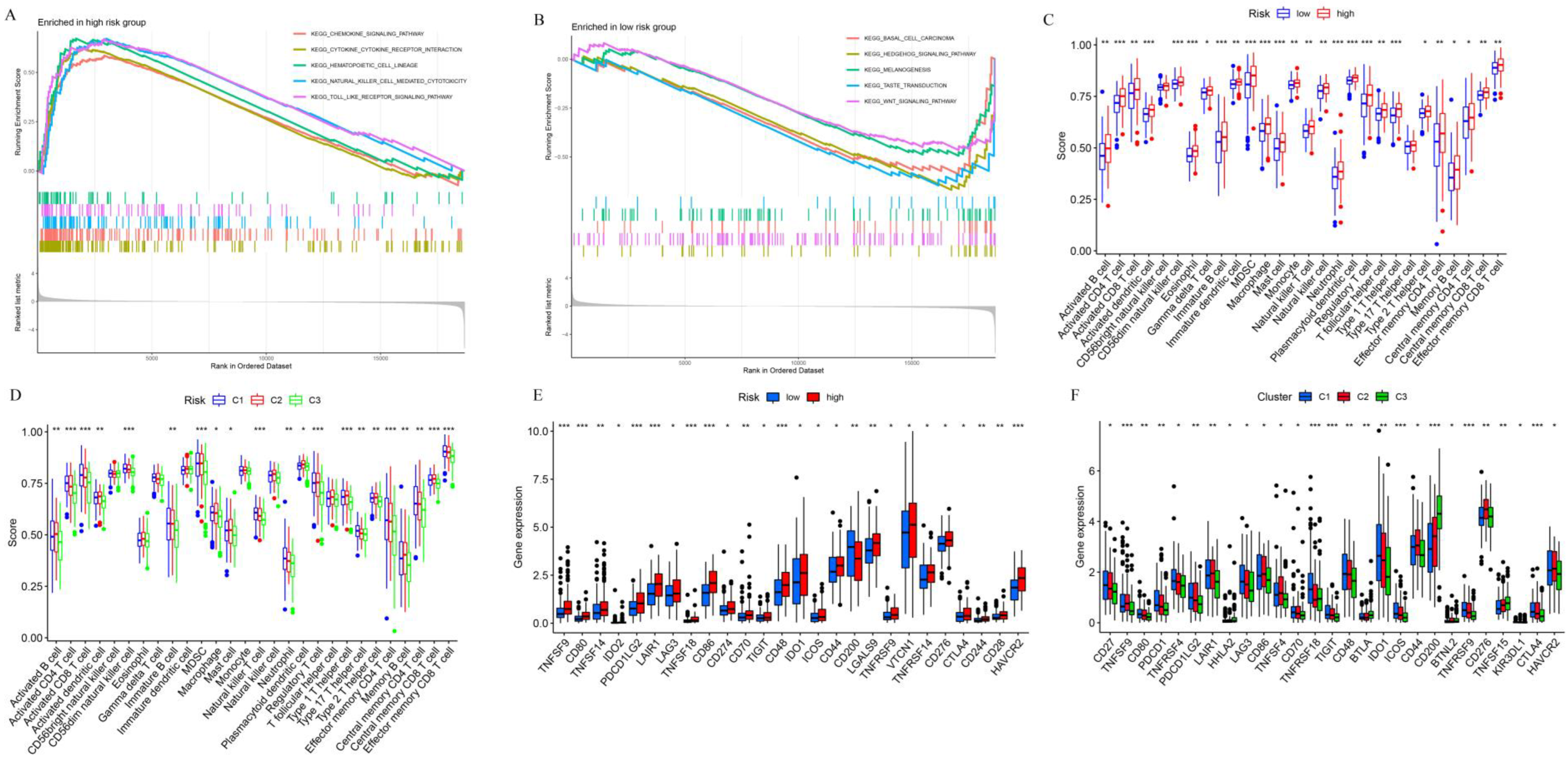
2.6. Gene Set Enrichment Analysis (GSEA) and Single-Sample GSEA (ssGSEA)
2.7. An Analysis of Proteins and Protein-Coding Genes Using Online Databases
2.8. Statistical Analysis
3. Results
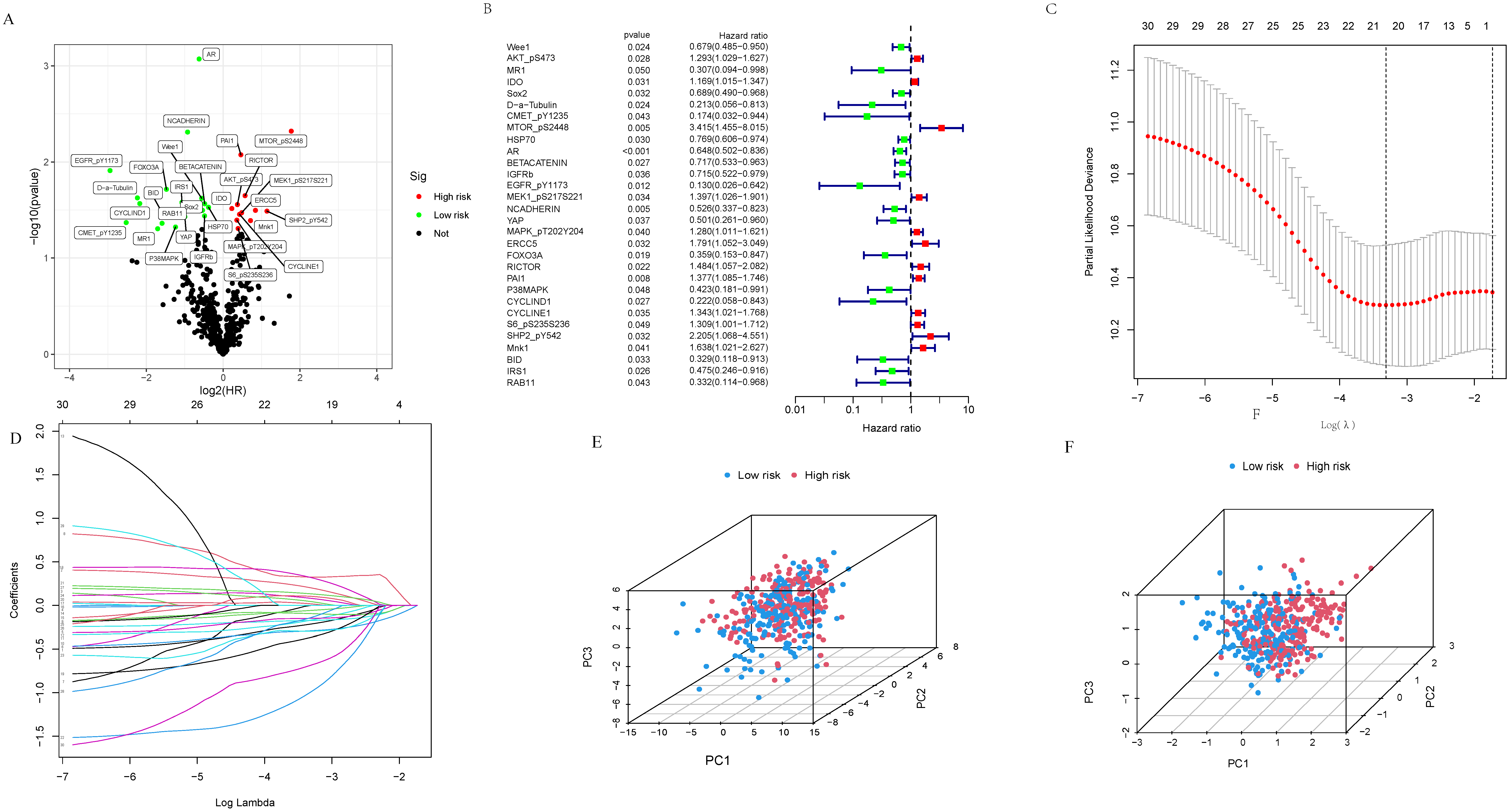
3.1. Proteins with Prognostic Relevance in Ovarian Cancer
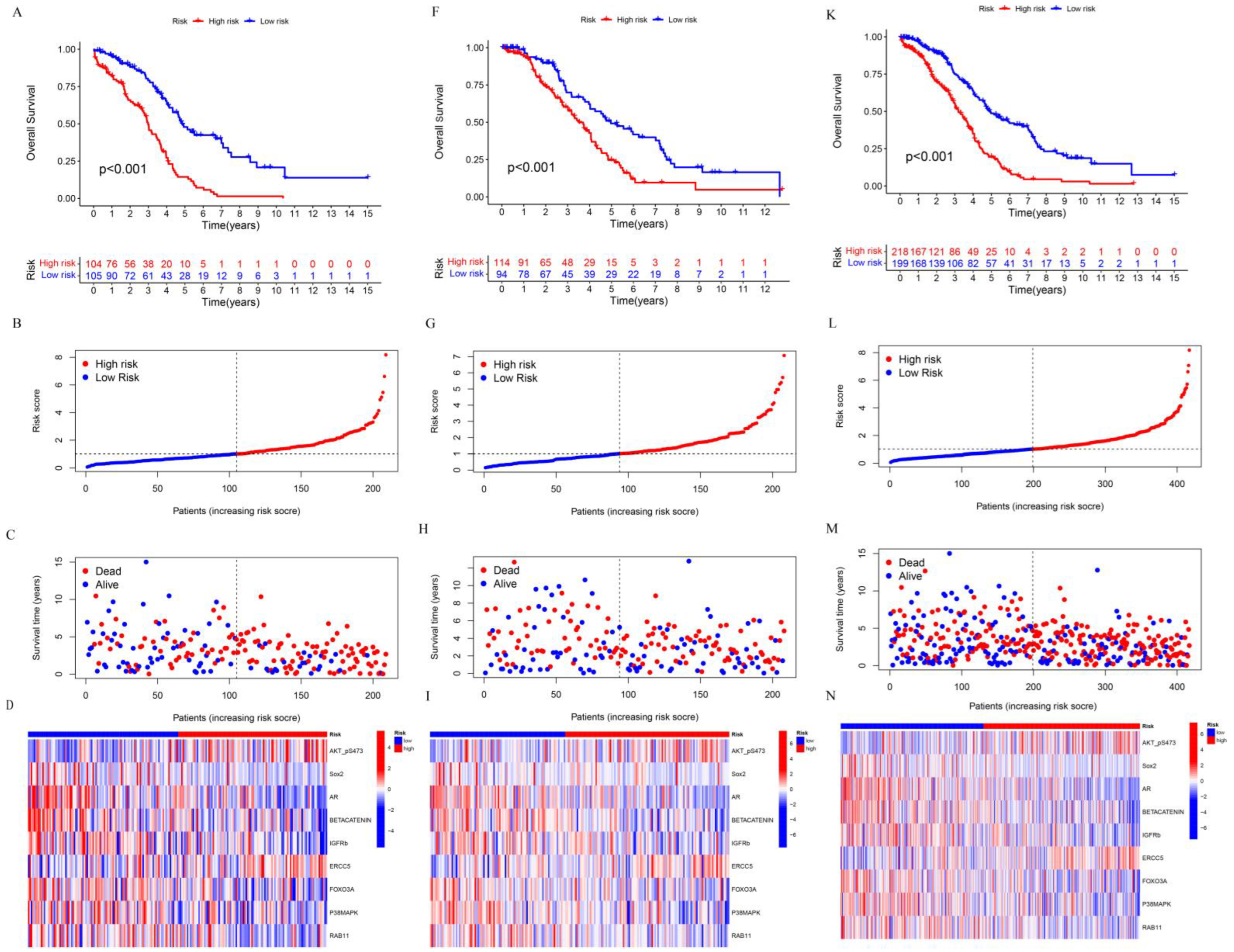
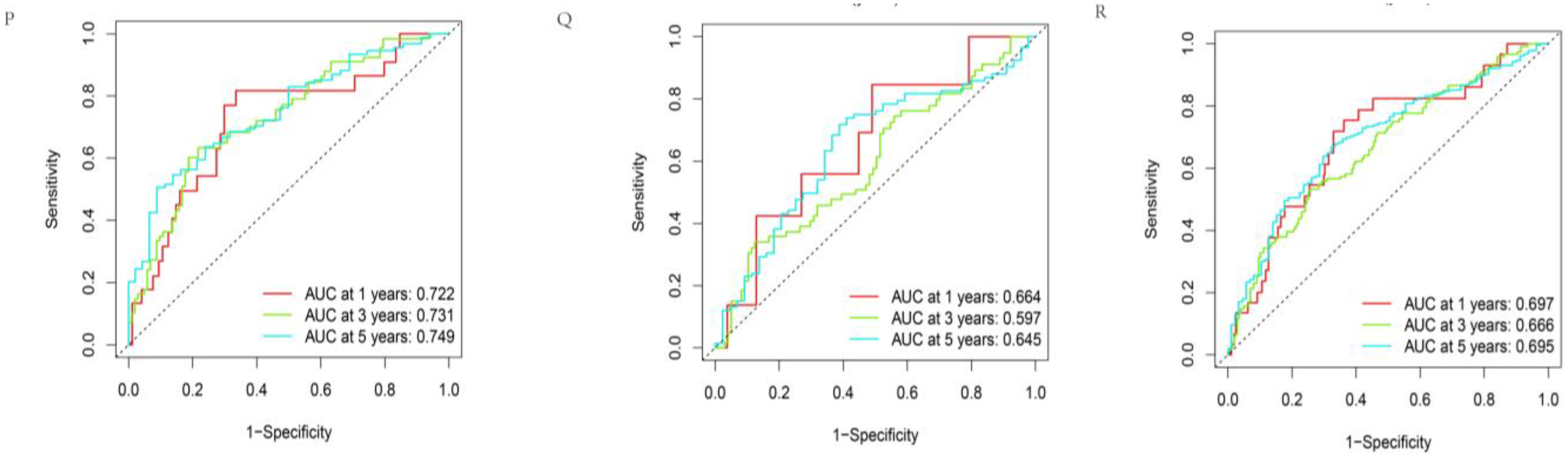
3.2. Construction and Assessment of Prognosis-Related Protein Model
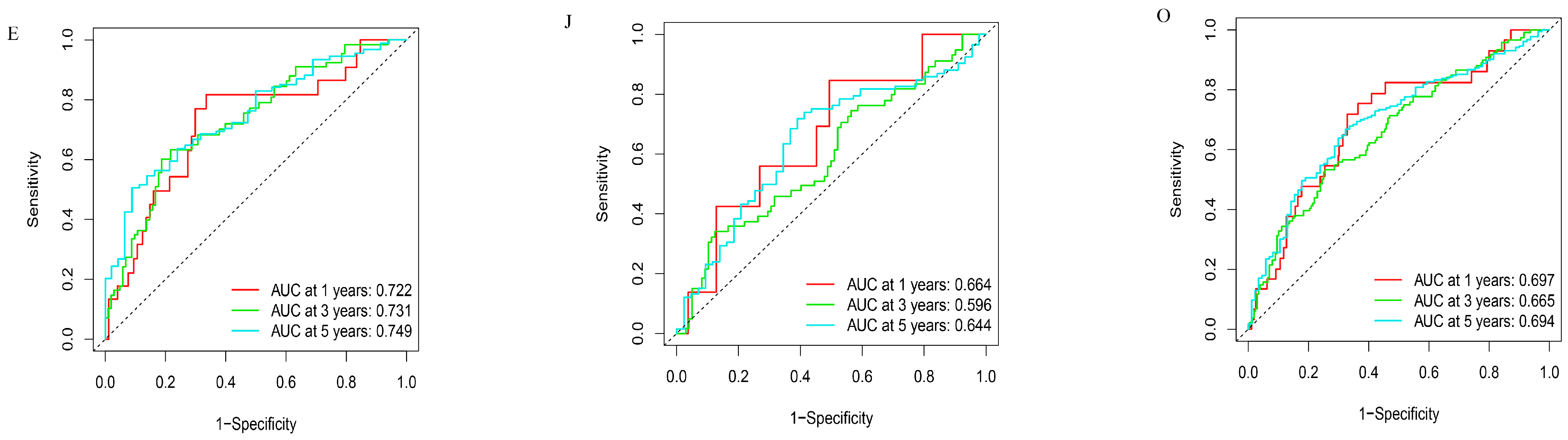
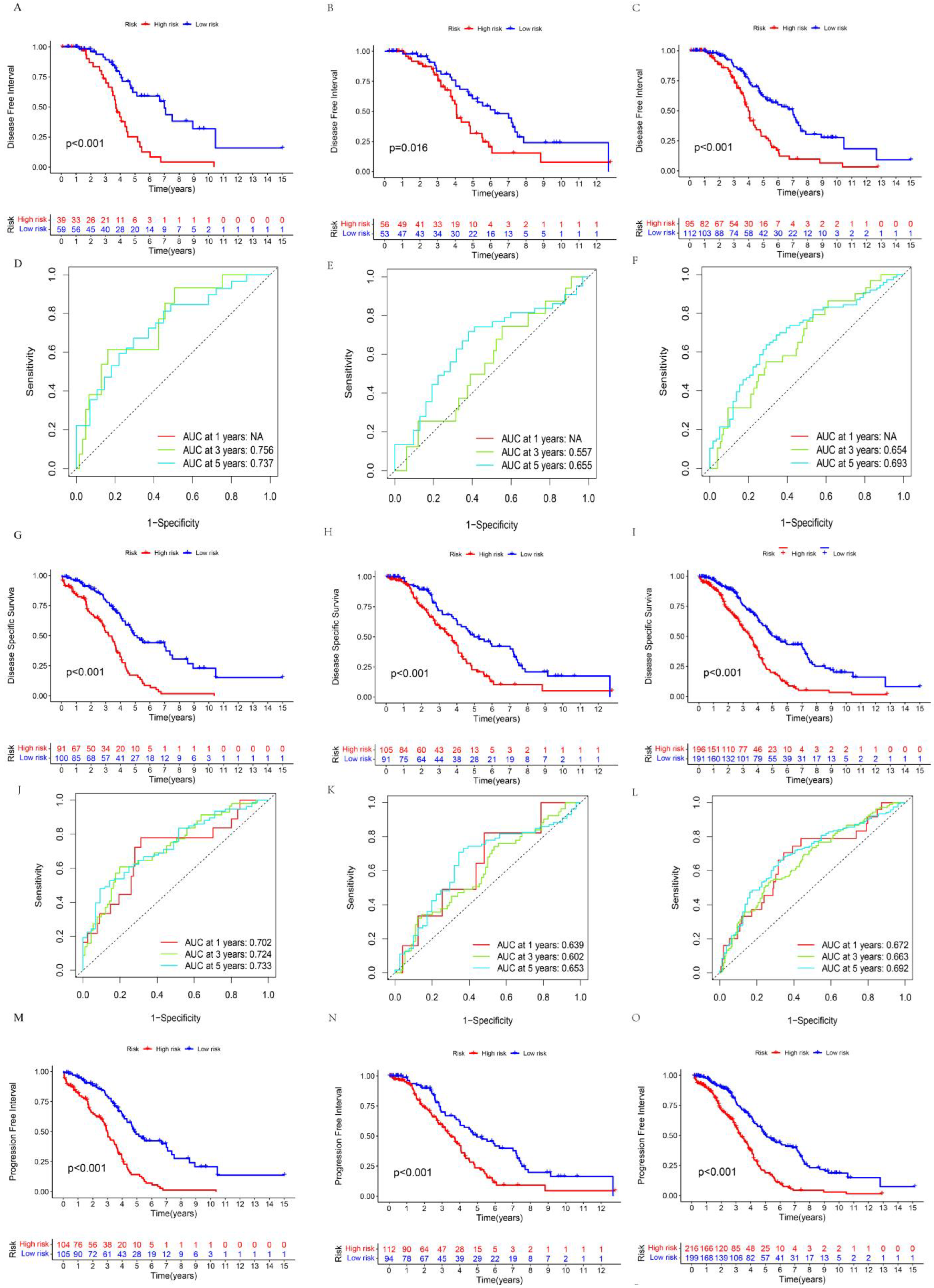
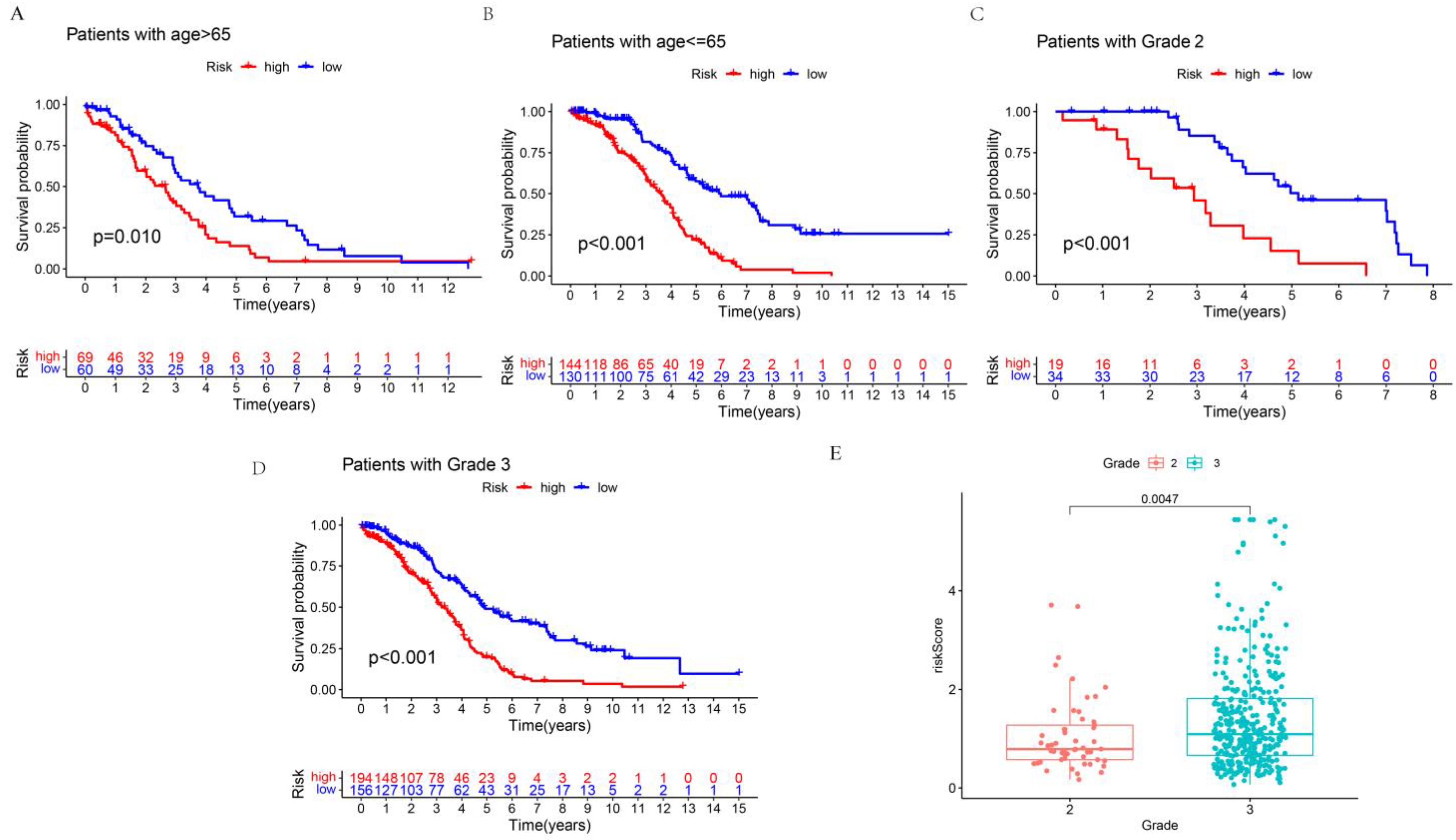
3.3. Clinical Value of Prognostic-Related Proteins
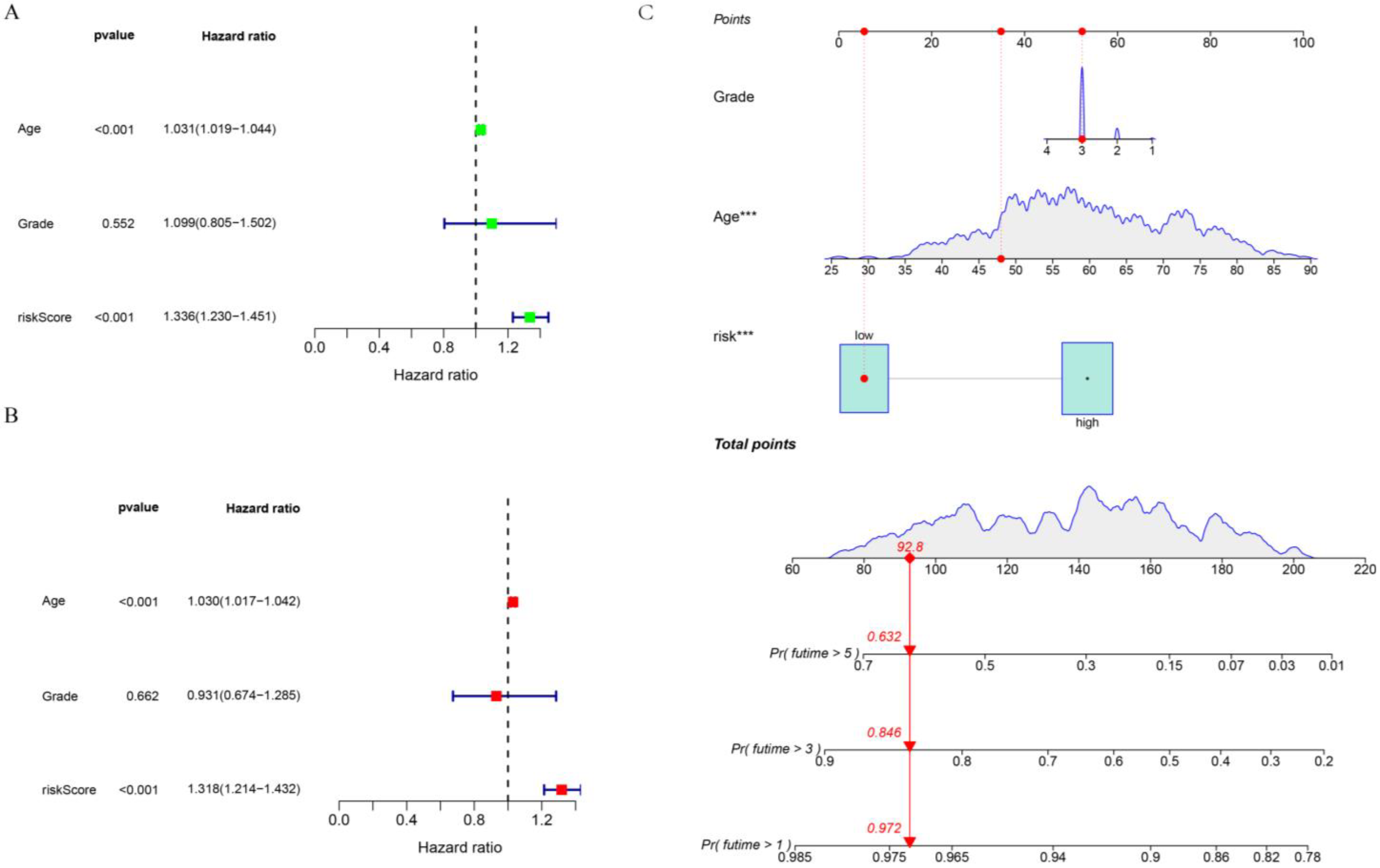
3.4. Construction of Nomogram Model
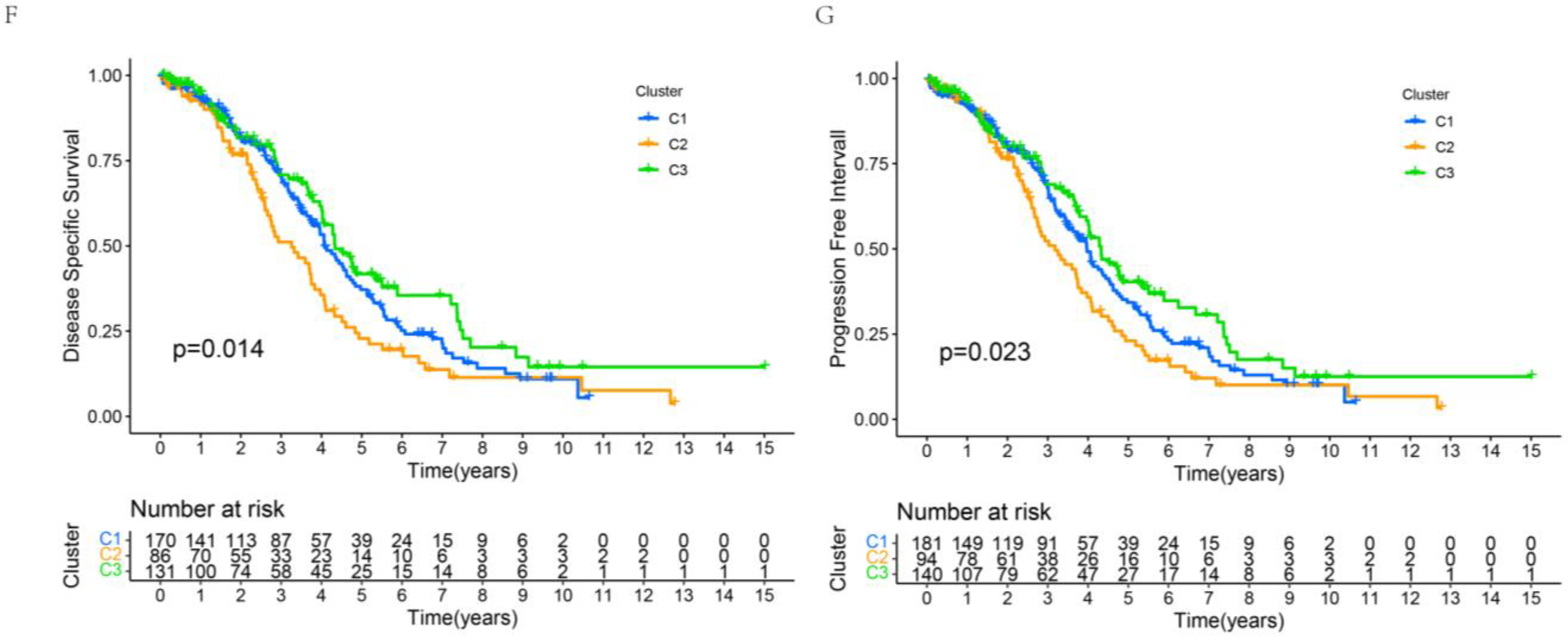
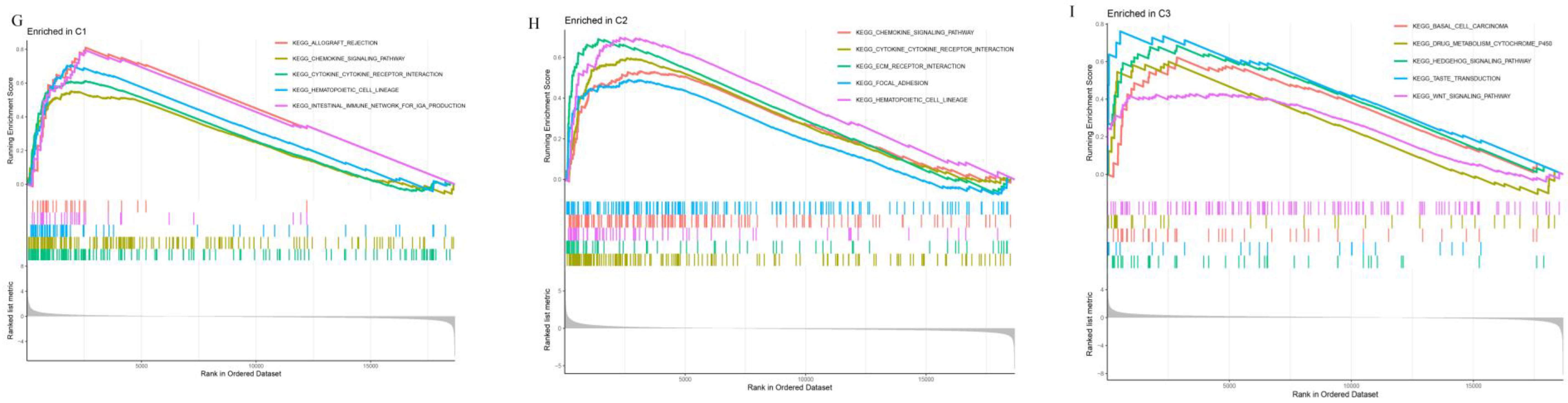
3.5. Assessment of Prognosis-Related Protein Subgroups
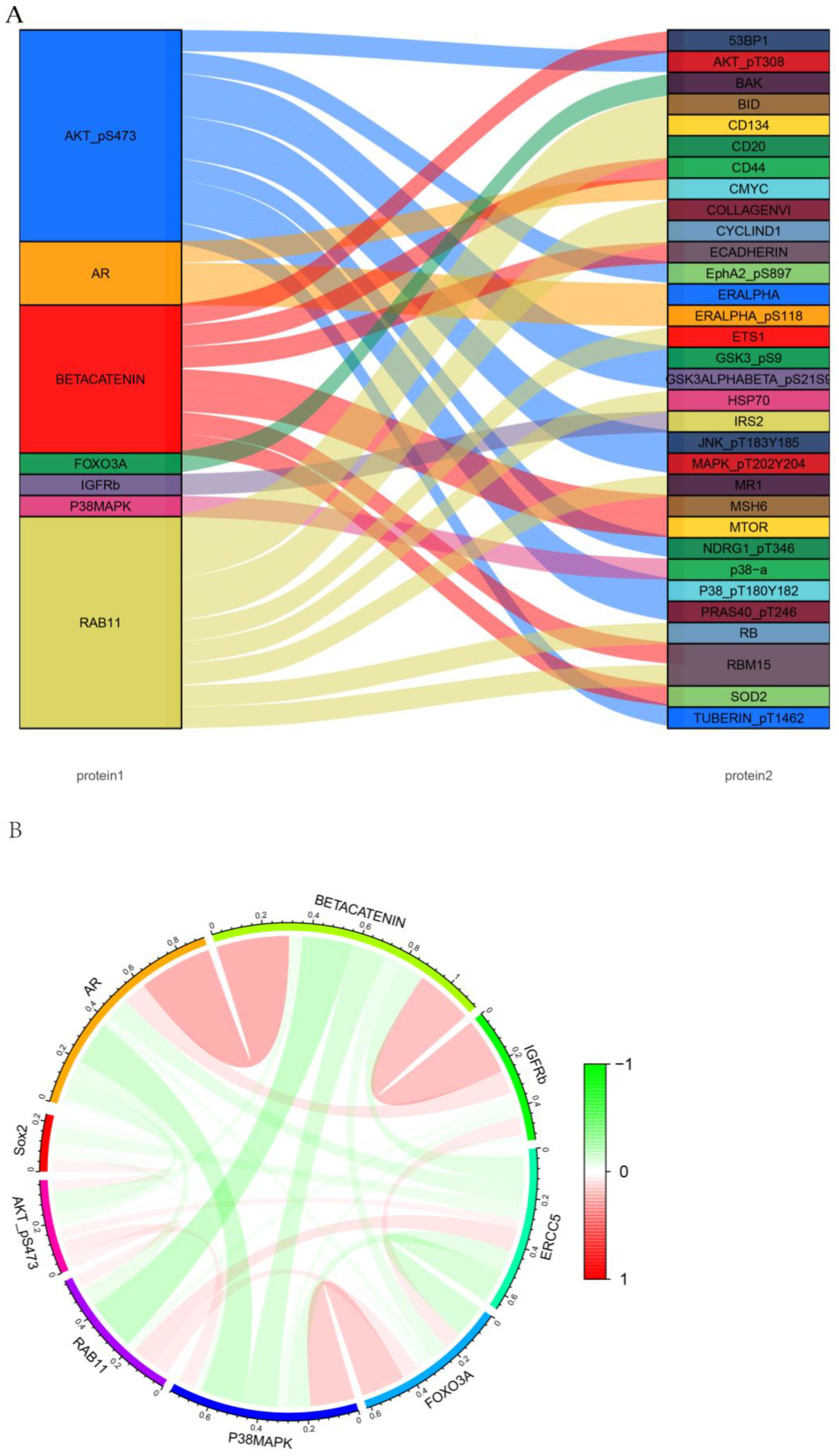
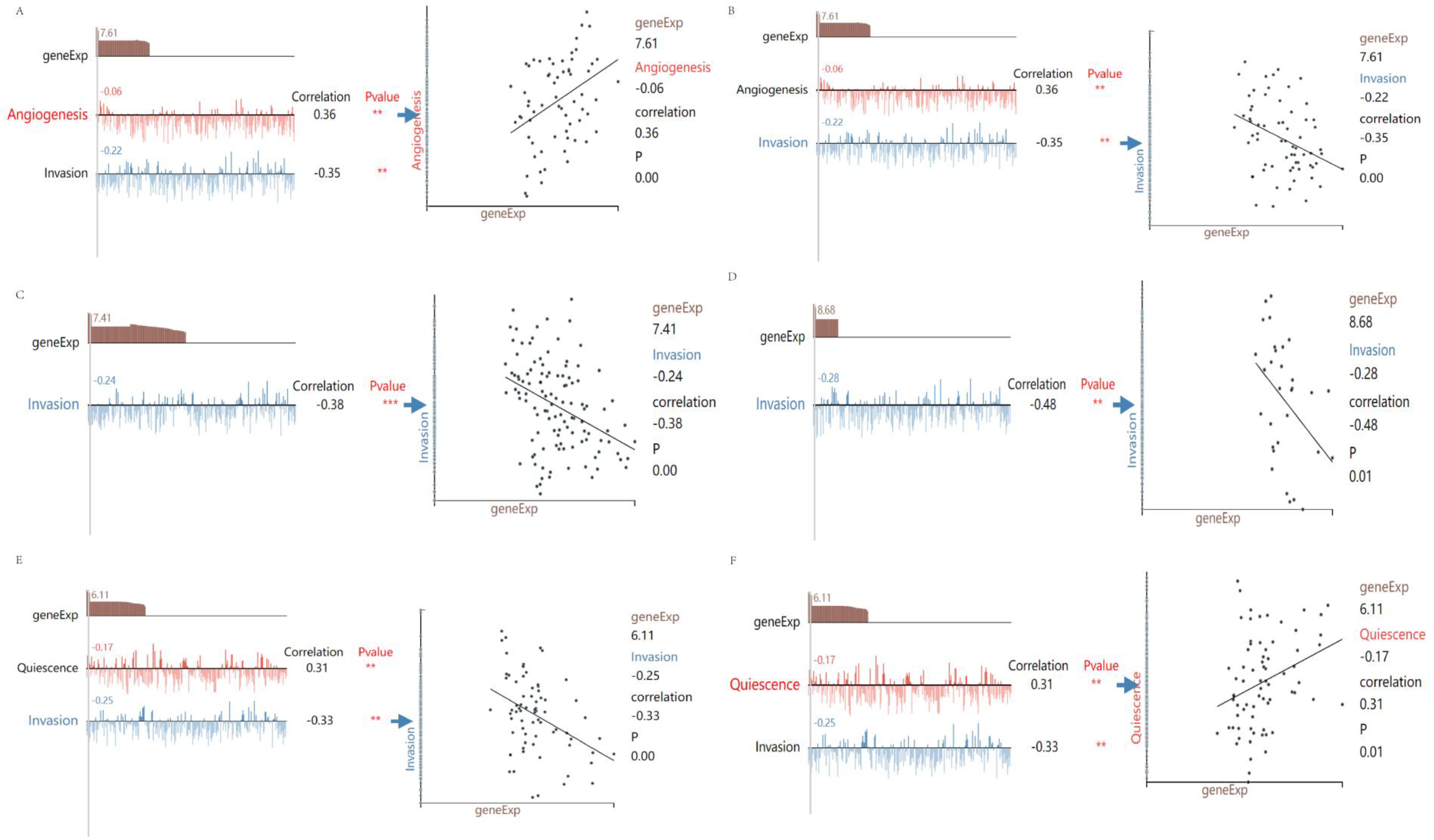
3.6. Analysis of Prognosis-Related Protein Function, Tumor-Infiltrating Immune Cells, and Immune Checkpoints
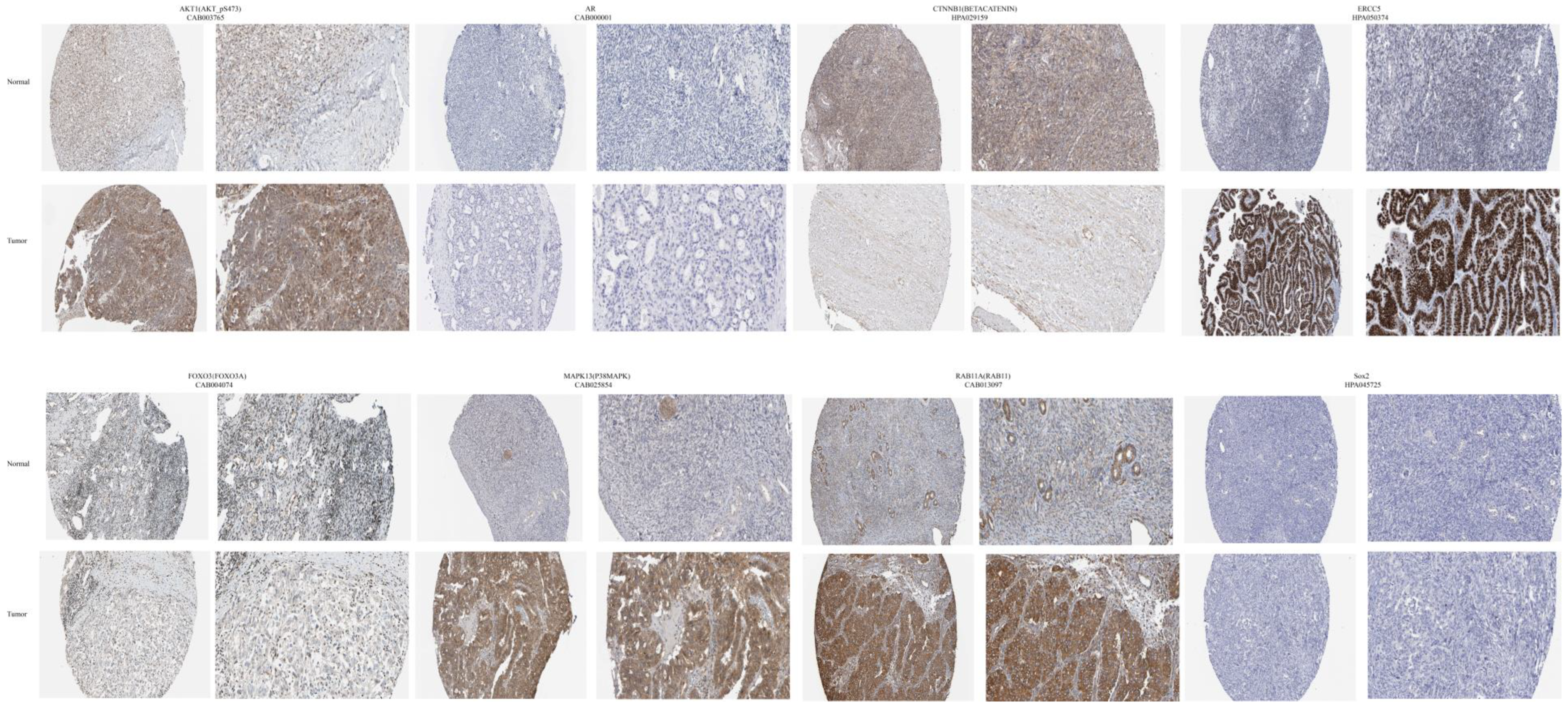
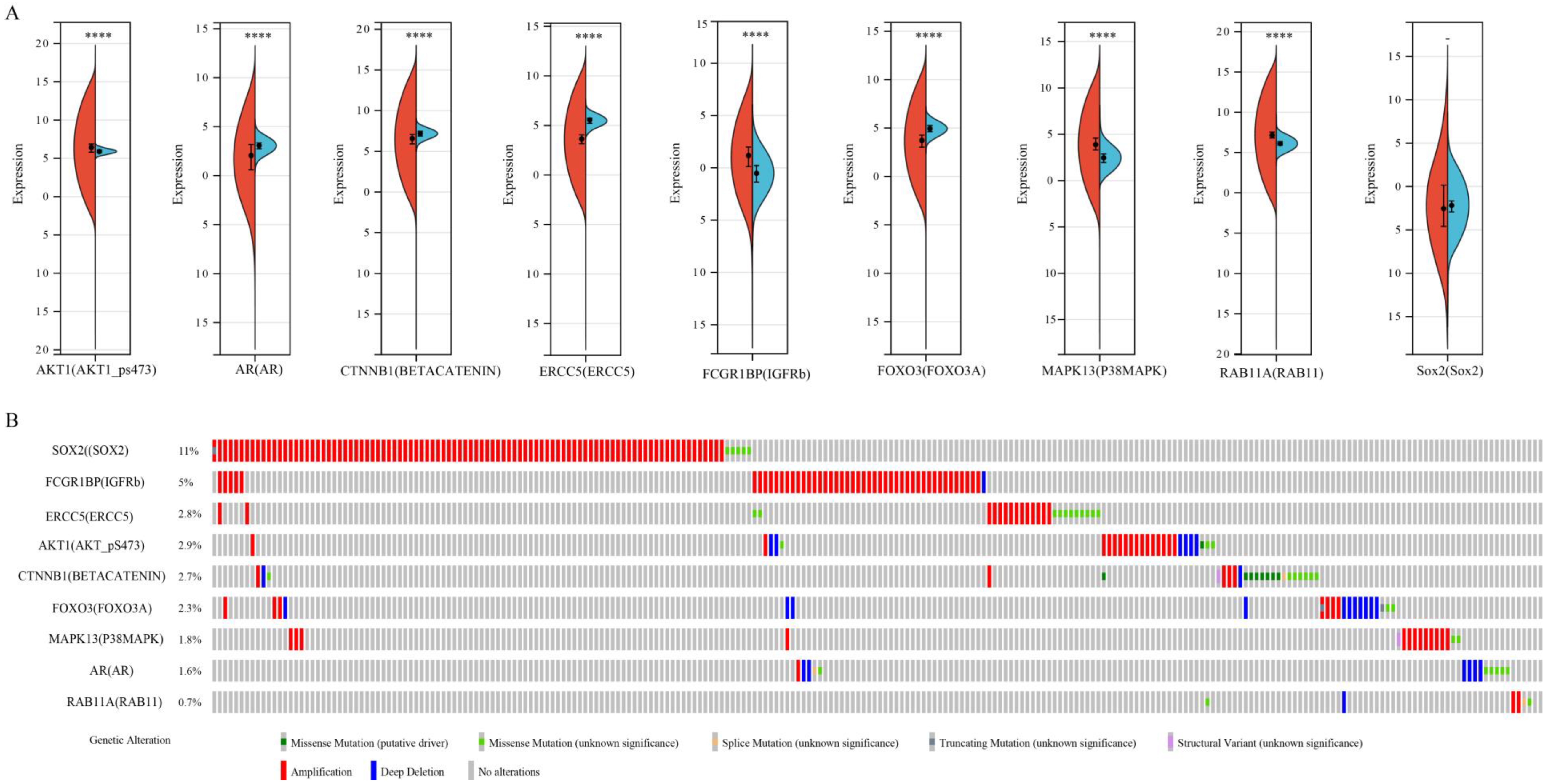
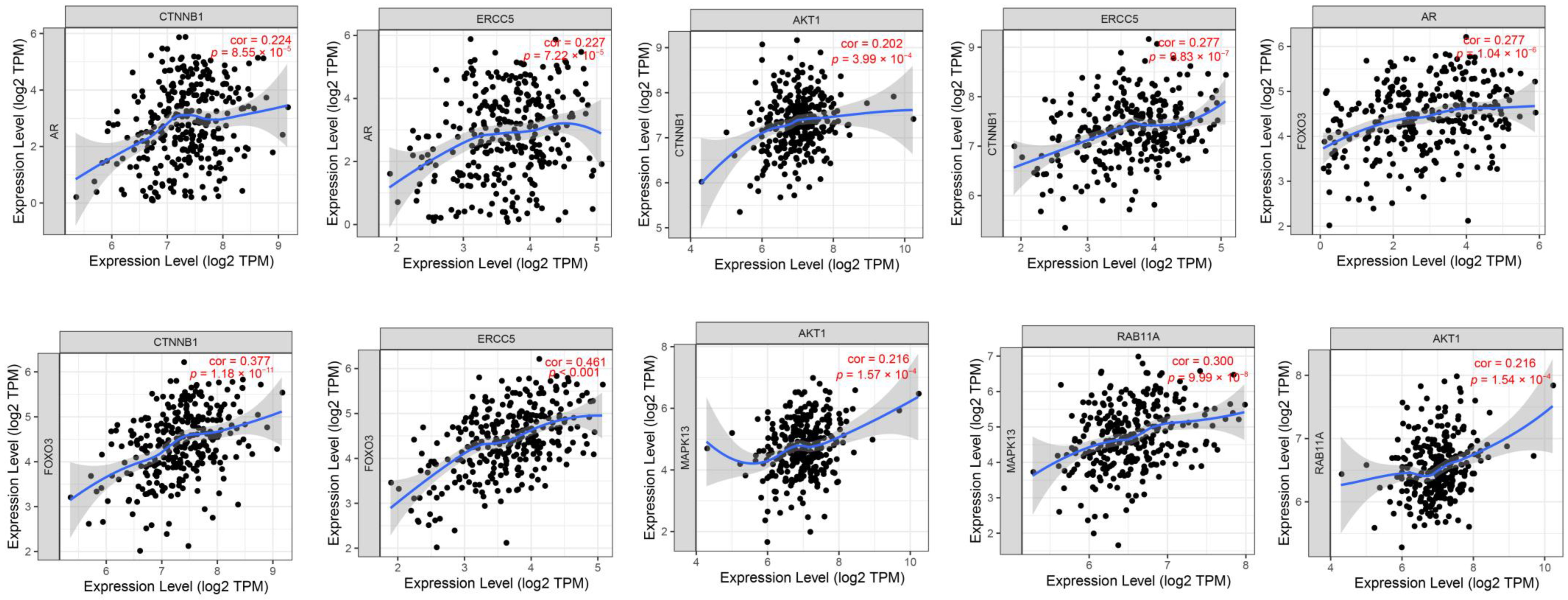
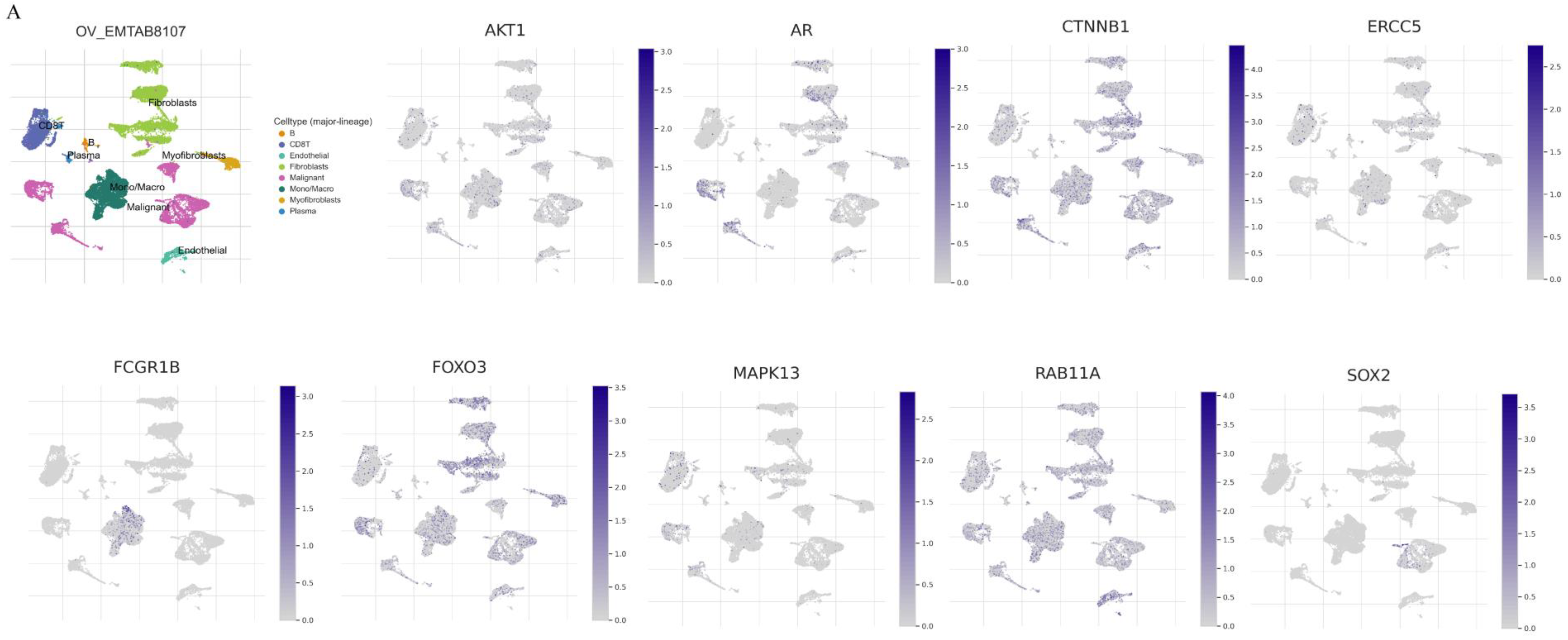
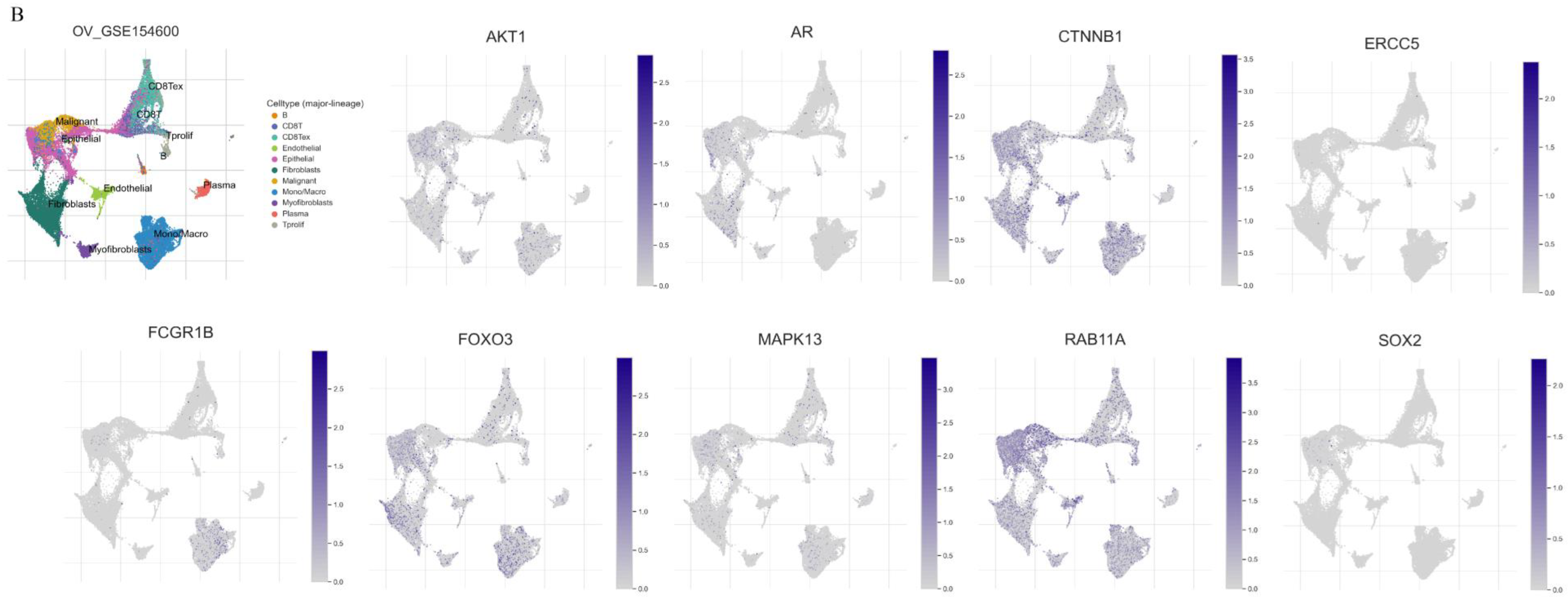
3.7. An Analysis of Proteins and Protein-Coding Genes Using Online Databases
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Gonzalez-Martin, A.; Lorusso, D.; Gourley, C.; Mirza, M.R.; Kurtz, J.; Moore, K.; Kridelka, F.; McNeish, I.; Reuss, A.; du Bois, A.; et al. Clinical research in ovarian cancer: Consensus recommendations from the Gynecologic Cancer InterGroup. Lancet Oncol. 2022, 23, e374–e384. [Google Scholar]
- Kan, T.; Zhang, S.; Zhou, S.; Zhang, Y.; Zhao, Y.; Gao, Y.; Zhang, T.; Gao, F.; Wang, X.; Zhao, L.; et al. Single-cell RNA-seq recognized the initiator of epithelial ovarian cancer recurrence. Oncogene 2022, 41, 895–906. [Google Scholar] [CrossRef] [PubMed]
- Hornburg, M.; Desbois, M.; Lu, S.; Guan, Y.; Lo, A.A.; Kaufman, S.; Elrod, A.; Lotstein, A.; DesRochers, T.M.; Munoz-Rodriguez, J.L.; et al. Single-cell dissection of cellular components and interactions shaping the tumor immune phenotypes in ovarian cancer. Cancer Cell 2021, 39, 928–944. [Google Scholar] [CrossRef] [PubMed]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global cancer statistics 2020, GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2021, 3, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Xia, C.; Dong, X.; Li, H.; Cao, M.; Sun, D.; He, S.; Yang, F.; Yan, X.; Zhang, S.; Li, N.; et al. Cancer statistics in China and United States, 2022, profiles, trends, and determinants. Chin. Med. J. 2022, 135, 584–590. [Google Scholar] [CrossRef]
- Wang, Z.; Guo, E.; Yang, B.; Xiao, R.; Lu, F.; You, L.; Chen, G. Trends and age-period-cohort effects on mortality of the three major gynecologic cancers in China from 1990 to 2019, Cervical, ovarian and uterine cancer. Gynecol. Oncol. 2021, 163, 358–363. [Google Scholar] [CrossRef]
- Olbromski, P.J.; Pawlik, P.; Bogacz, A.; Sajdak, S. Identification of New Molecular Biomarkers in Ovarian Cancer Using the Gene Expression Profile. J. Clin. Med. 2022, 11, 3888. [Google Scholar] [CrossRef]
- Arend, R.; Martinez, A.; Szul, T.; Birrer, M.J. Biomarkers in ovarian cancer: To be or not to be. Cancer-Am. Cancer Soc. 2019, 125, 4563–4572. [Google Scholar] [CrossRef]
- Olbrecht, S.; Busschaert, P.; Qian, J.; Vanderstichele, A.; Loverix, L.; Van Gorp, T.; Van Nieuwenhuysen, E.; Han, S.; Van den Broeck, A.; Coosemans, A.; et al. High-grade serous tubo-ovarian cancer refined with single-cell RNA sequencing: Specific cell subtypes influence survival and determine molecular subtype classification. Genome. Med. 2021, 13, 111. [Google Scholar] [CrossRef]
- Ji, J.X.; Cochrane, D.; Negri, G.L.; Colborne, S.; Spencer Miko, S.E.; Hoang, L.N.; Farnell, D.; Tessier Cloutier, B.; Huvila, J.; Thompson, E.; et al. The proteome of clear cell ovarian carcinoma. J. Pathol. 2022, 258, 325–338. [Google Scholar] [CrossRef]
- Mehner, C.; Hockla, A.; Coban, M.; Madden, B.; Estrada, R.; Radisky, D.C.; Radisky, E.S. Activity-based protein profiling reveals active serine proteases that drive malignancy of human ovarian clear cell carcinoma. J. Biol. Chem. 2022, 298, 102146. [Google Scholar] [CrossRef] [PubMed]
- Olalekan, S.; Xie, B.; Back, R.; Eckart, H.; Basu, A. Characterizing the tumor microenvironment of metastatic ovarian cancer by single-cell transcriptomics. Cell Rep. 2021, 35, 109165. [Google Scholar] [CrossRef] [PubMed]
- Bartoletti, M.; Musacchio, L.; Giannone, G.; Tuninetti, V.; Bergamini, A.; Scambia, G.; Lorusso, D.; Valabrega, G.; Mangili, G.; Puglisi, F.; et al. Emerging molecular alterations leading to histology-specific targeted therapies in ovarian cancer beyond PARP inhibitors. Cancer Treat. Rev. 2021, 101, 102298. [Google Scholar] [CrossRef] [PubMed]
- Zheng, G.; Zhang, G.; Zhao, Y.; Zheng, Z. Screening Protein Prognostic Biomarkers for Stomach Adenocarcinoma Based on The Cancer Proteome Atlas. Front. Oncol. 2022, 12, 901182. [Google Scholar] [CrossRef] [PubMed]
- Huang, B.; Zhang, X.; Cao, Q.; Chen, J.; Lin, C.; Xiang, T.; Zeng, P. Construction and validation of a prognostic risk model for breast cancer based on protein expression. BMC Med. Genom. 2022, 15, 148. [Google Scholar] [CrossRef]
- Chu, G.; Xu, T.; Zhu, G.; Liu, S.; Niu, H.; Zhang, M. Identification of a Novel Protein-Based Signature to Improve Prognosis Prediction in Renal Clear Cell Carcinoma. Front. Mol. Biosci. 2021, 8, 623120. [Google Scholar] [CrossRef]
- Ma, W.; Chen, L.S.; Özbek, U.; Han, S.W.; Lin, C.; Paulovich, A.G.; Zhong, H.; Wang, P. Integrative Proteo-genomic Analysis to Construct CNA-protein Regulatory Map in Breast and Ovarian Tumors. Mol. Cell. Proteom. 2019, 18, S66–S81. [Google Scholar] [CrossRef]
- Tomczak, K.K.; Czerwińska, P.P.; Wiznerowicz, M.M. The Cancer Genome Atlas (TCGA): An immeasurable source of knowledge. Contemp. Oncol. 2015, 19, A68–A77. [Google Scholar] [CrossRef]
- Ouyang, W.; Winsnes, C.F.; Hjelmare, M.; Cesnik, A.J.; Åkesson, L.; Xu, H.; Sullivan, D.P.; Dai, S.; Lan, J.; Jinmo, P.; et al. Analysis of the Human Protein Atlas Image Classification competition. Nat. Methods 2019, 16, 1254–1261. [Google Scholar] [CrossRef]
- Shen, W.; Song, Z.; Zhong, X.; Huang, M.; Shen, D.; Gao, P.; Qian, X.; Wang, M.; He, X.; Wang, T.; et al. Sangerbox: A comprehensive, interaction-friendly clinical bioinformatics analysis platform. iMeta 2022, 1, e36. [Google Scholar] [CrossRef]
- Li, T.; Fan, J.; Wang, B.; Traugh, N.; Chen, Q.; Liu, J.S.; Li, B.; Liu, X.S. TIMER: AWeb Server for Comprehensive Analysis of Tumor-Infiltrating Immune Cells. Cancer Res. 2017, 77, e108–e110. [Google Scholar] [CrossRef] [PubMed]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The cBio Cancer Genomics Portal: An Open Platform for Exploring Multidimensional Cancer Genomics Data. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef] [PubMed]
- Sun, D.; Wang, J.; Han, Y.; Dong, X.; Ge, J.; Zheng, R.; Shi, X.; Wang, B.; Li, Z.; Ren, P.; et al. TISCH: A comprehensive web resource enabling interactive single-cell transcriptome visualization of tumor microenvironment. Nucleic Acids Res. 2021, 49, D1420–D1430. [Google Scholar] [CrossRef] [PubMed]
- Yuan, H.; Yan, M.; Zhang, G.; Liu, W.; Deng, C.; Liao, G.; Xu, L.; Luo, T.; Yan, H.; Long, Z.; et al. CancerSEA: A cancer single-cell state atlas. Nucleic Acids Res. 2019, 47, D900–D908. [Google Scholar] [CrossRef]
- Miheecheva, N.; Postovalova, E.; Lyu, Y.; Ramachandran, A.; Bagaev, A.; Svekolkin, V.; Galkin, I.; Zyrin, V.; Maximov, V.; Lozinsky, Y.; et al. Multiregional single-cell proteogenomic analysis of ccRCC reveals cytokine drivers of intratumor spatial heterogeneity. Cell Rep. 2022, 40, 111180. [Google Scholar] [CrossRef]
- Dong, L.; Lu, D.; Chen, R.; Lin, Y.; Zhu, H.; Zhang, Z.; Cai, S.; Cui, P.; Song, G.; Rao, D.; et al. Proteogenomic characterization identifies clinically relevant subgroups of intrahepatic cholangiocarcinoma. Cancer Cell 2022, 40, 70–87. [Google Scholar] [CrossRef]
- Pietzner, M.; Wheeler, E.; Carrasco-Zanini, J.; Cortes, A.; Koprulu, M.; Wörheide, M.A.; Oerton, E.; Cook, J.; Stewart, I.D.; Kerrison, N.D.; et al. Mapping the proteo-genomic convergence of human diseases. Science 2021, 374, eabj1541. [Google Scholar] [CrossRef]
- Dou, Y.; Kawaler, E.A.; Cui Zhou, D.; Gritsenko, M.A.; Huang, C.; Blumenberg, L.; Karpova, A.; Petyuk, V.A.; Savage, S.R.; Satpathy, S.; et al. Proteogenomic Characterization of Endometrial Carcinoma. Cell 2020, 180, 729–748. [Google Scholar] [CrossRef]
- Coscia, F.; Lengyel, E.; Duraiswamy, J.; Ashcroft, B.; Bassani-Sternberg, M.; Wierer, M.; Johnson, A.; Wroblewski, K.; Montag, A.; Yamada, S.D.; et al. Multi-level Proteomics Identifies CT45 as a Chemosensitivity Mediator and Immunotherapy Target in Ovarian Cancer. Cell 2018, 175, 159–170. [Google Scholar] [CrossRef]
- Steinhart, B.; Jordan, K.R.; Bapat, J.; Post, M.D.; Brubaker, L.W.; Bitler, B.G.; Wrobel, J. The Spatial Context of Tumor-Infiltrating Immune Cells Associates with Improved Ovarian Cancer Survival. Mol. Cancer Res. 2021, 19, 1973–1979. [Google Scholar] [CrossRef]
- Santos, J.M.; Heiniö, C.; Cervera-Carrascon, V.; Quixabeira, D.C.A.; Siurala, M.; Havunen, R.; Butzow, R.; Zafar, S.; de Gruijl, T.; Lassus, H.; et al. Oncolytic adenovirus shapes the ovarian tumor microenvironment for potent tumor-infiltrating lymphocyte tumor reactivity. J. Immunother. Cancer 2019, 8, e188. [Google Scholar] [CrossRef] [PubMed]
- Pierini, S.; Mishra, A.; Perales-Linares, R.; Uribe-Herranz, M.; Beghi, S.; Giglio, A.; Pustylnikov, S.; Costabile, F.; Rafail, S.; Amici, A.; et al. Combination of vasculature targeting, hypofractionated radiotherapy, and immune checkpoint inhibitor elicits potent antitumor immune response and blocks tumor progression. J. Immunother. Cancer 2021, 9, e1636. [Google Scholar] [CrossRef]
- Hao, D.; Liu, J.; Chen, M.; Li, J.; Wang, L.; Li, X.; Zhao, Q.; Di, L. Immunogenomic Analyses of Advanced Serous Ovarian Cancer Reveal Immune Score is a Strong Prognostic Factor and an Indicator of Chemosensitivity. Clin. Cancer Res. 2018, 24, 3560–3571. [Google Scholar] [CrossRef] [PubMed]
- Leary, A.; Tan, D.; Ledermann, J. Immune checkpoint inhibitors in ovarian cancer: Where do we stand? Ther. Adv. Med. Oncol. 2021, 13, 386371902. [Google Scholar] [CrossRef] [PubMed]
- Sharma, P.; Allison, J.P. The future of immune checkpoint therapy. Science 2015, 348, 56–61. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Zheng, M.; Wang, S.; Gao, L.; Gou, R.; Liu, O.; Dong, H.; Li, X.; Lin, B. Identification of a five-gene signature of the RGS gene family with prognostic value in ovarian cancer. Genomics 2021, 113, 2134–2144. [Google Scholar] [CrossRef]
- Pan, X.; Chen, Y.; Gao, S. Four genes relevant to pathological grade and prognosis in ovarian cancer. Cancer Biomark. 2020, 29, 169–178. [Google Scholar] [CrossRef]
- Teeuwssen, M.; Fodde, R. Wnt Signaling in Ovarian Cancer Stemness, EMT, and Therapy Resistance. J. Clin. Med. 2019, 8, 1658. [Google Scholar] [CrossRef]
- Skoda, A.M.; Simovic, D.; Karin, V.; Kardum, V.; Vranic, S.; Serman, L. The role of the Hedgehog signaling pathway in cancer: A comprehensive review. Bosn. J. Basic. Med. Sci. 2018, 18, 8–20. [Google Scholar] [CrossRef]
- O’Connell, C.E.; Vassilev, A. Combined Inhibition of p38MAPK and PIKfyve Synergistically Disrupts Autophagy to Selectively Target Cancer Cells. Cancer Res. 2021, 81, 2903–2917. [Google Scholar] [CrossRef]
- Gibieža, P.; Petrikaitė, V. The dual functions of Rab11 and Rab35 GTPases-regulation of cell division and promotion of tumorigenicity. Am. J. Cancer. Res. 2021, 11, 1861–1872. [Google Scholar] [PubMed]
- Ferro, E.; Bosia, C.; Campa, C.C. RAB11-Mediated Trafficking and Human Cancers: An Updated Review. Biology 2021, 10, 26. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Ao, X.; Ding, W.; Ponnusamy, M.; Wu, W.; Hao, X.; Yu, W.; Wang, Y.; Li, P.; Wang, J. Critical role of FOXO3a in carcinogenesis. Mol. Cancer 2018, 17, 104. [Google Scholar] [CrossRef] [PubMed]
- Tian, Y.; Qi, P.; Hu, X. Downregulated FOXO3a Associates with Poor Prognosis and Promotes Cell Invasion and Migration via WNT/β-catenin Signaling in Cervical Carcinoma. Front. Oncol. 2020, 10, 903. [Google Scholar] [CrossRef] [PubMed]
- Shi, L.; Lin, H.; Li, G.; Jin, R.; Xu, J.; Sun, Y.; Ma, W.; Yeh, S.; Cai, X.; Chang, C. Targeting Androgen Receptor (AR)→IL12A Signal Enhances Efficacy of Sorafenib plus NK Cells Immunotherapy to Better Suppress HCC Progression. Mol. Cancer Ther. 2016, 15, 731–742. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Li, S.; Zhang, Y.; Shi, S.; Qin, S.; Wang, C.; Du, J.; Ma, J.; Chen, H.; Cui, H. Androgen Plays a Carcinogenic Role in EOC via the PI3K/AKT Signaling Pathway in an AR-Dependent Manner. J. Cancer 2021, 12, 1815–1825. [Google Scholar] [CrossRef]
- Wen, Y.; Hou, Y.; Huang, Z.; Cai, J.; Wang, Z. SOX2 is required to maintain cancer stem cells in ovarian cancer. Cancer Sci. 2017, 108, 719–731. [Google Scholar] [CrossRef]
- Virant-Klun, I.; Kenda-Suster, N.; Smrkolj, S. Small putative NANOG, SOX2, and SSEA-4-positive stem cells resembling very small embryonic-like stem cells in sections of ovarian tissue in patients with ovarian cancer. J. Ovarian. Res. 2016, 9, 12. [Google Scholar] [CrossRef]
- Kortam, M.A.; Alawady, A.S.; Hamid Sadik, N.A.; Fathy, N. Fenofibrate mitigates testosterone induced benign prostatic hyperplasia via regulation of Akt/FOXO3a pathway and modulation of apoptosis and proliferation in rats. Arch. Biochem. Biophys. 2022, 723, 109237. [Google Scholar] [CrossRef]
- Walsh, C.S.; Ogawa, S.; Karahashi, H.; Scoles, D.R.; Pavelka, J.C.; Tran, H.; Miller, C.W.; Kawamata, N.; Ginther, C.; Dering, J.; et al. ERCC5 Is a Novel Biomarker of Ovarian Cancer Prognosis. J. Clin. Oncol. 2008, 26, 2952–2958. [Google Scholar] [CrossRef]
- Zuo, C.; Lv, X.; Liu, T.; Yang, L.; Yang, Z.; Yu, C.; Chen, H. Polymorphisms in ERCC4 and ERCC5 and risk of cancers: Systematic research synopsis, meta-analysis, and epidemiological evidence. Front. Oncol. 2022, 12, 951193. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jiang, J.; Chen, Z.; Wang, H.; Wang, Y.; Zheng, J.; Guo, Y.; Jiang, Y.; Mo, Z. Screening and Identification of a Prognostic Model of Ovarian Cancer by Combination of Transcriptomic and Proteomic Data. Biomolecules 2023, 13, 685. https://doi.org/10.3390/biom13040685
Jiang J, Chen Z, Wang H, Wang Y, Zheng J, Guo Y, Jiang Y, Mo Z. Screening and Identification of a Prognostic Model of Ovarian Cancer by Combination of Transcriptomic and Proteomic Data. Biomolecules. 2023; 13(4):685. https://doi.org/10.3390/biom13040685
Chicago/Turabian StyleJiang, Jinghang, Zhongyuan Chen, Honghong Wang, Yifu Wang, Jie Zheng, Yi Guo, Yonghua Jiang, and Zengnan Mo. 2023. "Screening and Identification of a Prognostic Model of Ovarian Cancer by Combination of Transcriptomic and Proteomic Data" Biomolecules 13, no. 4: 685. https://doi.org/10.3390/biom13040685
APA StyleJiang, J., Chen, Z., Wang, H., Wang, Y., Zheng, J., Guo, Y., Jiang, Y., & Mo, Z. (2023). Screening and Identification of a Prognostic Model of Ovarian Cancer by Combination of Transcriptomic and Proteomic Data. Biomolecules, 13(4), 685. https://doi.org/10.3390/biom13040685






