Regulation of Epithelial Sodium Transport by SARS-CoV-2 Is Closely Related with Fibrinolytic System-Associated Proteins
Abstract
1. Introduction
2. Relationship of SARS-CoV-2 Infection and Lung Impairment
2.1. ARDS Different from ‘Classic’ Occurs in COVID-19 Patients
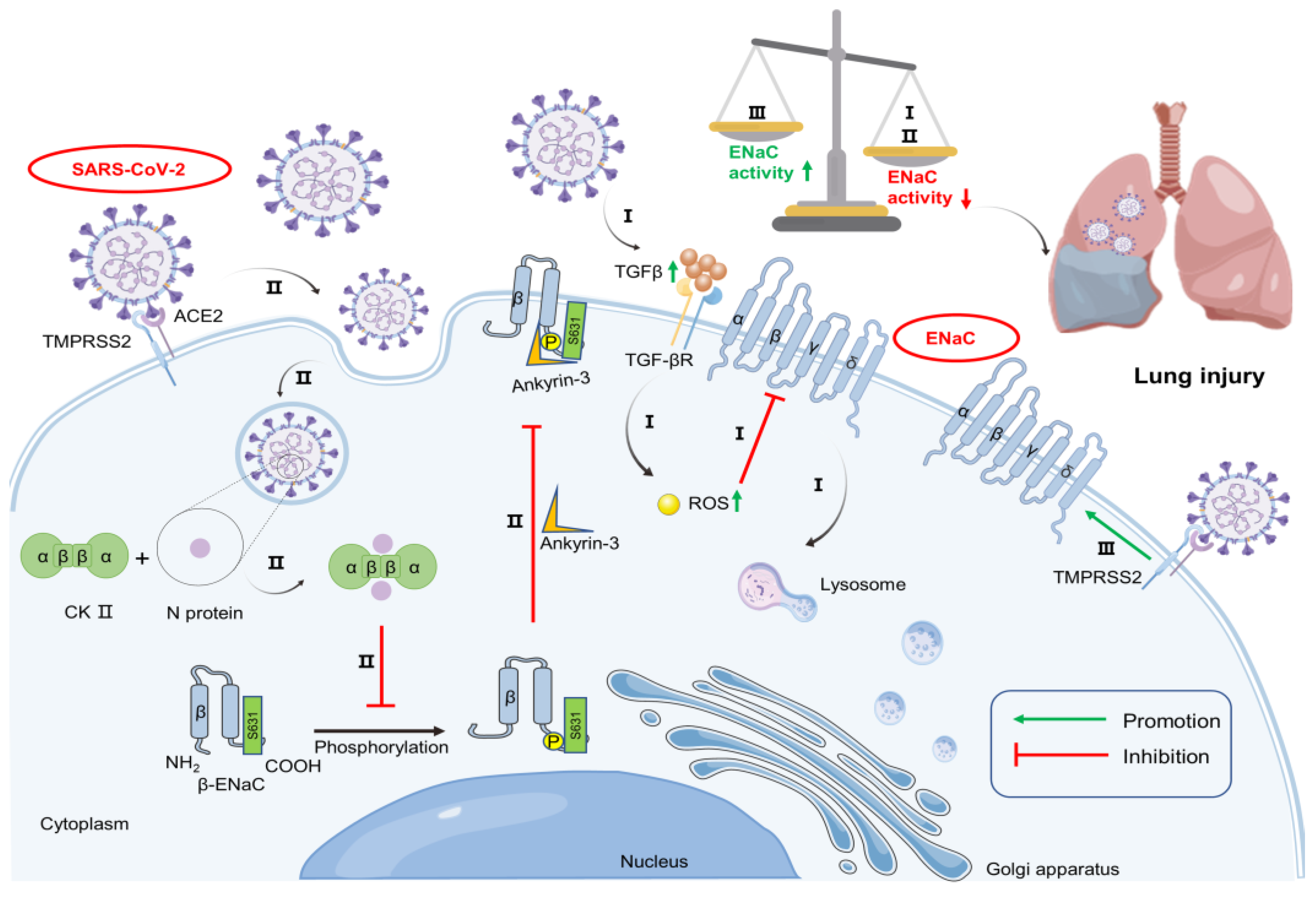
2.2. SARS-CoV-2 Promotes ENaC Degradation
2.3. SARS-CoV-2 May Change the Subcellular Localization of ENaC
2.4. SARS-CoV-2 Relieves the Inhibitory Effect of TMPRSS2 on ENaC Expression
2.5. Pro-Inflammatory Mediators Influence ENaC Expression under SARS-CoV-2 Infection
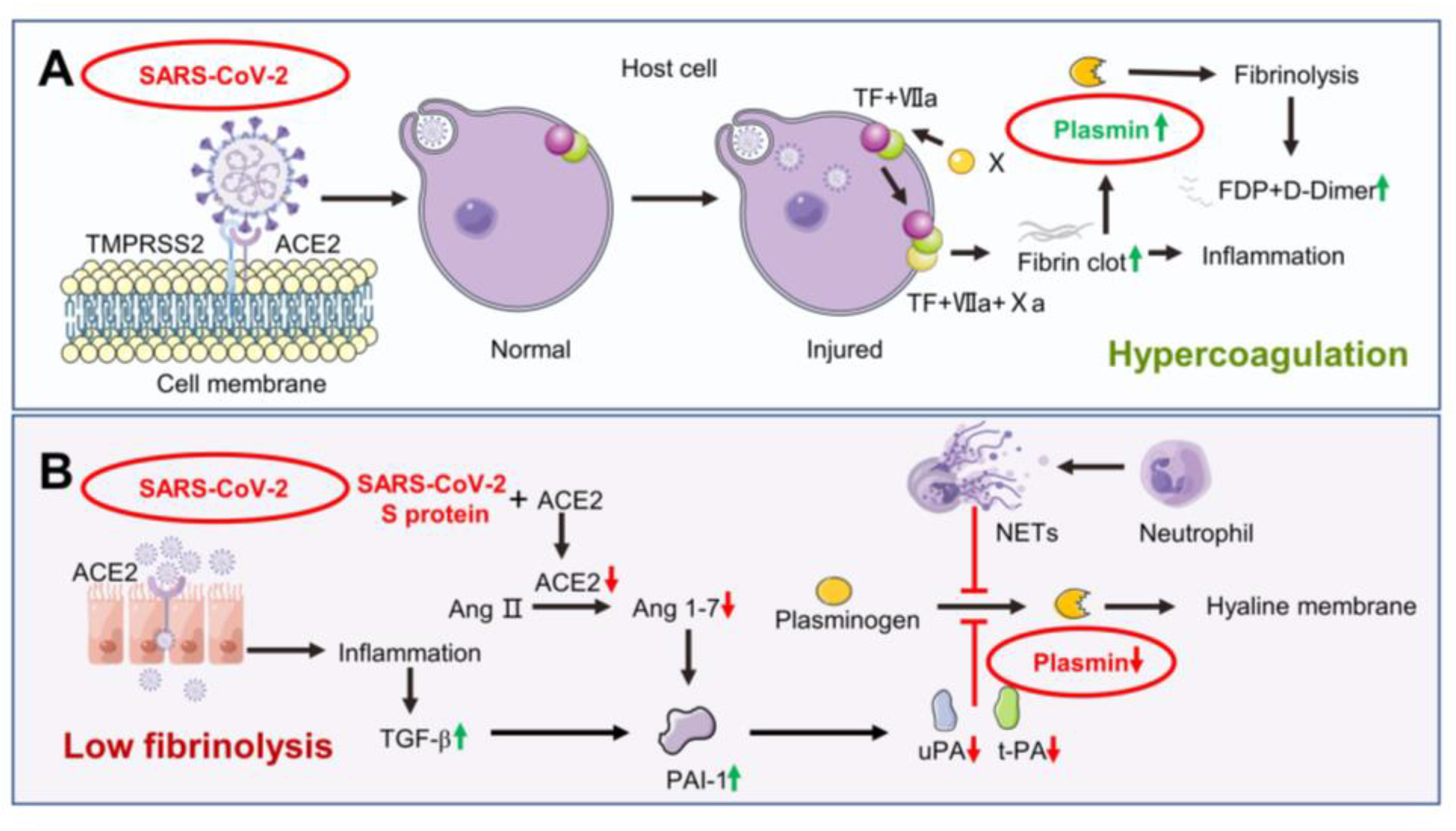
3. Dysfunction of Coagulation and Fibrinolysis System Caused by SARS-CoV-2 Infection
4. Regulation of Epithelial Sodium Transport by SARS-CoV-2 Related with Plasmin-Associated Proteins
4.1. Plasmin Participates in the Proteolytic Cleavage of ENaC
4.2. Plasmin Directly Regulates the Opening Probability of ENaC
4.3. Angiotensin II Reduces Plasmin Expression to Regulate ENaC Activity
4.4. Neutrophil Elastase May Inhibit Plasmin Expression to Regulate ENaC Activity
5. Emerging SARS-CoV-2 Therapeutics Associated with Fibrinolytic System Regulation of ENaC
5.1. Soluble ACE2
5.2. Protease Inhibitors
5.3. Immunoregulation Agents
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Chen, Y.; Liu, Q.; Guo, D. Emerging coronaviruses: Genome structure, replication, and pathogenesis. J. Med. Virol. 2020, 92, 2249. [Google Scholar] [CrossRef] [PubMed]
- Chan, J.F.; Kok, K.H.; Zhu, Z.; Chu, H.; To, K.K.; Yuan, S.; Yuen, K.Y. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg. Microbes Infect. 2020, 9, 221–236. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Krüger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.H.; Nitsche, A.; et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 2020, 181, 271–280.e278. [Google Scholar] [CrossRef] [PubMed]
- Pouya, F.; Imani Saber, Z.; Kerachian, M.A. Molecular aspects of co-morbidities in COVID-19 infection. Arch. Bone Jt. Surg. 2020, 8, 226–230. [Google Scholar] [CrossRef] [PubMed]
- Coutard, B.; Valle, C.; de Lamballerie, X.; Canard, B.; Seidah, N.G.; Decroly, E. The spike glycoprotein of the new coronavirus 2019-nCoV contains a furin-like cleavage site absent in CoV of the same clade. Antivir. Res. 2020, 176, 104742. [Google Scholar] [CrossRef]
- Walls, A.C.; Park, Y.J.; Tortorici, M.A.; Wall, A.; McGuire, A.T.; Veesler, D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell 2020, 181, 281–292. [Google Scholar] [CrossRef]
- Volz, E.; Hill, V.; McCrone, J.T.; Price, A.; Jorgensen, D.; O’Toole, Á.; Southgate, J.; Johnson, R.; Jackson, B.; Nascimento, F.F.; et al. Evaluating the Effects of SARS-CoV-2 Spike Mutation D614G on Transmissibility and Pathogenicity. Cell 2021, 184, 64–75.e11. [Google Scholar] [CrossRef]
- Liu, Y.; Liu, J.; Johnson, B.A.; Xia, H.; Ku, Z.; Schindewolf, C.; Widen, S.G.; An, Z.; Weaver, S.C.; Menachery, V.D.; et al. Delta spike P681R mutation enhances SARS-CoV-2 fitness over Alpha variant. Cell Rep. 2022, 39, 110829. [Google Scholar] [CrossRef]
- Yurkovetskiy, L.; Wang, X.; Pascal, K.E.; Tomkins-Tinch, C.; Nyalile, T.P.; Wang, Y.; Baum, A.; Diehl, W.E.; Dauphin, A.; Carbone, C.; et al. Structural and functional analysis of the D614G SARS-CoV-2 spike protein variant. Cell 2020, 183, 739–751.e738. [Google Scholar] [CrossRef]
- Thierry, A.R. Anti-protease treatments targeting plasmin(ogen) and neutrophil elastase may be beneficial in fighting COVID-19. Physiol. Rev. 2020, 100, 1597–1598. [Google Scholar] [CrossRef]
- Conway, E.M.; Mackman, N.; Warren, R.Q.; Wolberg, A.S.; Mosnier, L.O.; Campbell, R.A.; Gralinski, L.E.; Rondina, M.T.; van de Veerdonk, F.L.; Hoffmeister, K.M.; et al. Understanding COVID-19-associated coagulopathy. Nat. Rev. Immunol. 2022, 22, 639–649. [Google Scholar] [CrossRef] [PubMed]
- Ji, H.L.; Zhao, R.; Matalon, S.; Matthay, M.A. Elevated plasmin(ogen) as a common risk factor for COVID-19 susceptibility. Physiol. Rev. 2020, 100, 1065–1075. [Google Scholar] [CrossRef] [PubMed]
- Anand, P.; Puranik, A.; Aravamudan, M.; Venkatakrishnan, A.J.; Soundararajan, V. SARS-CoV-2 strategically mimics proteolytic activation of human ENaC. eLife 2020, 9, e58603. [Google Scholar] [CrossRef] [PubMed]
- Brown, E.F.; Mitaera, T.; Fronius, M. COVID-19 and liquid homeostasis in the lung-a perspective through the epithelial sodium channel (ENaC) lens. Cells 2022, 11, 1801. [Google Scholar] [CrossRef] [PubMed]
- Papa, G.; Mallery, D.L.; Albecka, A.; Welch, L.G.; Cattin-Ortolá, J.; Luptak, J.; Paul, D.; McMahon, H.T.; Goodfellow, I.G.; Carter, A.; et al. Furin cleavage of SARS-CoV-2 Spike promotes but is not essential for infection and cell-cell fusion. PLoS Pathog. 2021, 17, e1009246. [Google Scholar] [CrossRef]
- Ware, A.W.; Rasulov, S.R.; Cheung, T.T.; Lott, J.S.; McDonald, F.J. Membrane trafficking pathways regulating the epithelial Na+ channel. Am. J. Physiol. Ren. Physiol. 2020, 318, F1–F13. [Google Scholar] [CrossRef]
- Grant, S.N.; Lester, H.A. Regulation of epithelial sodium channel activity by SARS-CoV-1 and SARS-CoV-2 proteins. Biophys. J. 2021, 120, 2805–2813. [Google Scholar] [CrossRef]
- Matuschak, G.M.; Lechner, A.J. Acute lung injury and the acute respiratory distress syndrome: Pathophysiology and treatment. Mo. Med. 2010, 107, 252–258. [Google Scholar]
- Butt, Y.; Kurdowska, A.; Allen, T.C. Acute lung injury: A clinical and molecular review. Arch. Pathol. Lab. Med. 2016, 140, 345–350. [Google Scholar] [CrossRef]
- Gonzales, J.N.; Lucas, R.; Verin, A.D. The acute respiratory distress syndrome: Mechanisms and perspective therapeutic approaches. Austin J. Vasc. Med. 2015, 2, 1009. [Google Scholar]
- Meyer, N.J.; Gattinoni, L.; Calfee, C.S. Acute respiratory distress syndrome. Lancet 2021, 398, 622–637. [Google Scholar] [CrossRef] [PubMed]
- Cui, X.; Chen, W.; Zhou, H.; Gong, Y.; Zhu, B.; Lv, X.; Guo, H.; Duan, J.; Zhou, J.; Marcon, E.; et al. Pulmonary edema in COVID-19 patients: Mechanisms and treatment potential. Front. Pharmacol. 2021, 12, 664349. [Google Scholar] [CrossRef] [PubMed]
- Kamynina, E.; Staub, O. Concerted action of ENaC, Nedd4-2, and Sgk1 in transepithelial Na(+) transport. Am. J. Physiol. Ren. Physiol. 2002, 283, F377–F387. [Google Scholar] [CrossRef] [PubMed]
- Kashlan, O.B.; Kleyman, T.R. ENaC structure and function in the wake of a resolved structure of a family member. Am. J. Physiol. Ren. Physiol. 2011, 301, F684–F696. [Google Scholar] [CrossRef] [PubMed]
- Kellenberger, S.; Schild, L. Epithelial sodium channel/degenerin family of ion channels: A variety of functions for a shared structure. Physiol. Rev. 2002, 82, 735–767. [Google Scholar] [CrossRef]
- Musante, I.; Scudieri, P.; Venturini, A.; Guidone, D.; Caci, E.; Castellani, S.; Conese, M.; Galietta, L.J.V. Peripheral localization of the epithelial sodium channel in the apical membrane of bronchial epithelial cells. Exp. Physiol. 2019, 104, 866–875. [Google Scholar] [CrossRef]
- Jiang, Y.; Xia, M.; Xu, J.; Huang, Q.; Dai, Z.; Zhang, X. Dexmedetomidine alleviates pulmonary edema through the epithelial sodium channel (ENaC) via the PI3K/Akt/Nedd4-2 pathway in LPS-induced acute lung injury. Immunol. Res. 2021, 69, 162–175. [Google Scholar] [CrossRef]
- Caramaschi, S.; Kapp, M.E.; Miller, S.E.; Eisenberg, R.; Johnson, J.; Epperly, G.; Maiorana, A.; Silvestri, G.; Giannico, G.A. Histopathological findings and clinicopathologic correlation in COVID-19: A systematic review. Mod. Pathol. 2021, 34, 1614–1633. [Google Scholar] [CrossRef]
- Sakurai, A.; Sasaki, T.; Kato, S.; Hayashi, M.; Tsuzuki, S.I.; Ishihara, T.; Iwata, M.; Morise, Z.; Doi, Y. Natural history of asymptomatic SARS-CoV-2 infection. N. Engl. J. Med. 2020, 383, 885–886. [Google Scholar] [CrossRef]
- Arons, M.M.; Hatfield, K.M.; Reddy, S.C.; Kimball, A.; James, A.; Jacobs, J.R.; Taylor, J.; Spicer, K.; Bardossy, A.C.; Oakley, L.P.; et al. Presymptomatic SARS-CoV-2 infections and transmission in a skilled nursing facility. N. Engl. J. Med. 2020, 382, 2081–2090. [Google Scholar] [CrossRef]
- Ranieri, V.M.; Rubenfeld, G.D.; Thompson, B.T.; Ferguson, N.D.; Caldwell, E.; Fan, E.; Camporota, L.; Slutsky, A.S. Acute respiratory distress syndrome: The Berlin definition. JAMA 2012, 307, 2526–2533. [Google Scholar] [CrossRef] [PubMed]
- Kushimoto, S.; Endo, T.; Yamanouchi, S.; Sakamoto, T.; Ishikura, H.; Kitazawa, Y.; Taira, Y.; Okuchi, K.; Tagami, T.; Watanabe, A.; et al. Relationship between extravascular lung water and severity categories of acute respiratory distress syndrome by the Berlin definition. Crit. Care 2013, 17, R132. [Google Scholar] [CrossRef]
- Shi, R.; Lai, C.; Teboul, J.L.; Dres, M.; Moretto, F.; De Vita, N.; Pham, T.; Bonny, V.; Mayaux, J.; Vaschetto, R.; et al. COVID-19 ARDS is characterized by higher extravascular lung water than non-COVID-19 ARDS: The PiCCOVID study. Crit. Care 2021, 25, 186. [Google Scholar] [CrossRef] [PubMed]
- Rasch, S.; Schmidle, P.; Sancak, S.; Herner, A.; Huberle, C.; Schulz, D.; Mayr, U.; Schneider, J.; Spinner, C.D.; Geisler, F.; et al. Increased extravascular lung water index (EVLWI) reflects rapid non-cardiogenic oedema and mortality in COVID-19 associated ARDS. Sci. Rep. 2021, 11, 11524. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Vicente, A.; Hong, N.; Garvin, J.L. Effects of reactive oxygen species on renal tubular transport. Am. J. Physiol. Ren. Physiol. 2019, 317, F444–F455. [Google Scholar] [CrossRef] [PubMed]
- Goodson, P.; Kumar, A.; Jain, L.; Kundu, K.; Murthy, N.; Koval, M.; Helms, M.N. Nadph oxidase regulates alveolar epithelial sodium channel activity and lung fluid balance in vivo via O2− signaling. Am. J. Physiol. Lung Cell. Mol. Physiol. 2012, 302, L410–L419. [Google Scholar] [CrossRef] [PubMed]
- Eaton, A.F.; Yue, Q.; Eaton, D.C.; Bao, H.F. ENaC activity and expression is decreased in the lungs of protein kinase C-α knockout mice. Am. J. Physiol. Lung Cell. Mol. Physiol. 2014, 307, L374–L385. [Google Scholar] [CrossRef]
- Peters, D.M.; Vadász, I.; Wujak, L.; Wygrecka, M.; Olschewski, A.; Becker, C.; Herold, S.; Papp, R.; Mayer, K.; Rummel, S.; et al. TGF-β directs trafficking of the epithelial sodium channel ENaC which has implications for ion and fluid transport in acute lung injury. Proc. Natl. Acad. Sci. USA 2014, 111, E374–E383. [Google Scholar] [CrossRef]
- Chen, D.; Zheng, Q.; Sun, L.; Ji, M.; Li, Y.; Deng, H.; Zhang, H. ORF3a of SARS-CoV-2 promotes lysosomal exocytosis-mediated viral egress. Dev. Cell 2021, 56, 3250–3263.e3255. [Google Scholar] [CrossRef]
- Vaz de Paula, C.B.; Nagashima, S.; Liberalesso, V.; Collete, M.; da Silva, F.P.G.; Oricil, A.G.G.; Barbosa, G.S.; da Silva, G.V.C.; Wiedmer, D.B.; da Silva Dezidério, F.; et al. COVID-19: Immunohistochemical analysis of TGF-β signaling pathways in pulmonary fibrosis. Int. J. Mol. Sci. 2021, 23, 168. [Google Scholar] [CrossRef]
- Huntington, K.E.; Carlsen, L.; So, E.Y.; Piesche, M.; Liang, O.; El-Deiry, W.S. Integrin/TGF-β1 inhibitor GLPG-0187 blocks SARS-CoV-2 delta and omicron pseudovirus infection of airway epithelial cells in vitro, which could attenuate disease severity. Pharmaceuticals 2022, 15, 618. [Google Scholar] [CrossRef] [PubMed]
- Abd El-Aziz, T.M.; Soares, A.G.; Mironova, E.; Boiko, N.; Kaur, A.; Archer, C.R.; Stockand, J.D.; Berman, J.M. Mechanisms and consequences of casein kinase II and ankyrin-3 regulation of the epithelial Na(+) channel. Sci. Rep. 2021, 11, 14600. [Google Scholar] [CrossRef] [PubMed]
- Bouhaddou, M.; Memon, D.; Meyer, B.; White, K.M.; Rezelj, V.V.; Correa Marrero, M.; Polacco, B.J.; Melnyk, J.E.; Ulferts, S.; Kaake, R.M.; et al. The global phosphorylation landscape of SARS-CoV-2 infection. Cell 2020, 182, 685–712.e619. [Google Scholar] [CrossRef] [PubMed]
- Berman, J.M.; Mironova, E.; Stockand, J.D. Physiological regulation of the epithelial Na(+) channel by casein kinase II. Am. J. Physiol. Ren. Physiol. 2018, 314, F367–F372. [Google Scholar] [CrossRef]
- Borgo, C.; D’Amore, C.; Sarno, S.; Salvi, M.; Ruzzene, M. Protein kinase CK2: A potential therapeutic target for diverse human diseases. Signal Transduct. Target. Ther. 2021, 6, 183. [Google Scholar] [CrossRef] [PubMed]
- Donaldson, S.H.; Hirsh, A.; Li, D.C.; Holloway, G.; Chao, J.; Boucher, R.C.; Gabriel, S.E. Regulation of the epithelial sodium channel by serine proteases in human airways. J. Biol. Chem. 2002, 277, 8338–8345. [Google Scholar] [CrossRef]
- Shen, L.W.; Qian, M.Q.; Yu, K.; Narva, S.; Yu, F.; Wu, Y.L.; Zhang, W. Inhibition of Influenza A virus propagation by benzoselenoxanthenes stabilizing TMPRSS2 Gene G-quadruplex and hence down-regulating TMPRSS2 expression. Sci. Rep. 2020, 10, 7635. [Google Scholar] [CrossRef]
- Muhanna, D.; Arnipalli, S.R.; Kumar, S.B.; Ziouzenkova, O. Osmotic Adaptation by Na(+)-Dependent Transporters and ACE2: Correlation with Hemostatic Crisis in COVID-19. Biomedicines 2020, 8, 460. [Google Scholar] [CrossRef]
- Noori, M.; Nejadghaderi, S.A.; Sullman, M.J.M.; Carson-Chahhoud, K.; Ardalan, M.; Kolahi, A.A.; Safiri, S. How SARS-CoV-2 might affect potassium balance via impairing epithelial sodium channels? Mol. Biol. Rep. 2021, 48, 6655–6661. [Google Scholar] [CrossRef]
- Hamet, P.; Pausova, Z.; Attaoua, R.; Hishmih, C.; Haloui, M.; Shin, J.; Paus, T.; Abrahamowicz, M.; Gaudet, D.; Santucci, L.; et al. SARS-CoV-2 receptor ACE2 gene is associated with hypertension and severity of COVID 19: Interaction with sex, obesity, and smoking. Am. J. Hypertens. 2021, 34, 367–376. [Google Scholar] [CrossRef]
- Yang, L.; Wang, J.; Hui, P.; Yarovinsky, T.O.; Badeti, S.; Pham, K.; Liu, C. Potential role of IFN-α in COVID-19 patients and its underlying treatment options. Appl. Microbiol. Biotechnol. 2021, 105, 4005–4015. [Google Scholar] [CrossRef] [PubMed]
- Nemeth, Z.; Hildebrandt, E.; Parsa, N.; Fleming, A.B.; Wasson, R.; Pittman, K.; Bell, X.; Granger, J.P.; Ryan, M.J.; Drummond, H.A. Epithelial sodium channels in macrophage migration and polarization: Role of proinflammatory cytokines TNFα and IFNγ. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2022, 323, R763–R775. [Google Scholar] [CrossRef] [PubMed]
- Mustafa, S.B.; Hernandez, T.F.; Johnson-Pais, T.L.; Kumar, P.A.; Petershack, J.A.; Henson, B.M.; Seidner, S.R. IL-1 promotes α-epithelial Sodium Channel (α-ENaC) expression in murine lung epithelial cells: Involvement of NF-κB. J. Cell Commun. Signal. 2020, 14, 303–314. [Google Scholar] [CrossRef] [PubMed]
- Hadjadj, J.; Yatim, N.; Barnabei, L.; Corneau, A.; Boussier, J.; Smith, N.; Péré, H.; Charbit, B.; Bondet, V.; Chenevier-Gobeaux, C.; et al. Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients. Science 2020, 369, 718–724. [Google Scholar] [CrossRef]
- Yamagata, T.; Yamagata, Y.; Nishimoto, T.; Hirano, T.; Nakanishi, M.; Minakata, Y.; Ichinose, M.; Dagenais, A.; Berthiaume, Y. The regulation of amiloride-sensitive epithelial sodium channels by tumor necrosis factor-alpha in injured lungs and alveolar type II cells. Respir. Physiol. Neurobiol. 2009, 166, 16–23. [Google Scholar] [CrossRef]
- Ware, L.B.; Bastarache, J.A.; Wang, L. Coagulation and fibrinolysis in human acute lung injury--new therapeutic targets? Keio J. Med. 2005, 54, 142–149. [Google Scholar] [CrossRef]
- Barton, L.M.; Duval, E.J.; Stroberg, E.; Ghosh, S.; Mukhopadhyay, S. COVID-19 autopsies, Oklahoma, USA. Am. J. Clin. Pathol. 2020, 153, 725–733. [Google Scholar] [CrossRef]
- Menter, T.; Haslbauer, J.D.; Nienhold, R.; Savic, S.; Hopfer, H.; Deigendesch, N.; Frank, S.; Turek, D.; Willi, N.; Pargger, H.; et al. Postmortem examination of COVID-19 patients reveals diffuse alveolar damage with severe capillary congestion and variegated findings in lungs and other organs suggesting vascular dysfunction. Histopathology 2020, 77, 198–209. [Google Scholar] [CrossRef]
- Colling, M.E.; Kanthi, Y. COVID-19-associated coagulopathy: An exploration of mechanisms. Vasc. Med. 2020, 25, 471–478. [Google Scholar] [CrossRef]
- Edler, C.; Schröder, A.S.; Aepfelbacher, M.; Fitzek, A.; Heinemann, A.; Heinrich, F.; Klein, A.; Langenwalder, F.; Lütgehetmann, M.; Meißner, K.; et al. Dying with SARS-CoV-2 infection-an autopsy study of the first consecutive 80 cases in Hamburg, Germany. Int. J. Leg. Med. 2020, 134, 1275–1284. [Google Scholar] [CrossRef]
- Lippi, G.; Sanchis-Gomar, F.; Henry, B.M. COVID-19: Unravelling the clinical progression of nature’s virtually perfect biological weapon. Ann. Transl. Med. 2020, 8, 693. [Google Scholar] [CrossRef] [PubMed]
- Cui, S.; Chen, S.; Li, X.; Liu, S.; Wang, F. Prevalence of venous thromboembolism in patients with severe novel coronavirus pneumonia. J. Thromb. Haemost. JTH 2020, 18, 1421–1424. [Google Scholar] [CrossRef] [PubMed]
- Ozolina, A.; Sarkele, M.; Sabelnikovs, O.; Skesters, A.; Jaunalksne, I.; Serova, J.; Ievins, T.; Bjertnaes, L.J.; Vanags, I. Activation of Coagulation and Fibrinolysis in Acute Respiratory Distress Syndrome: A Prospective Pilot Study. Front. Med. 2016, 3, 64. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Wang, Y.; Liu, B.; Cheng, Y.; Qian, H.; Yang, H.; Li, X.; Yang, G.; Zheng, X.; Shen, F. SN50 attenuates alveolar hypercoagulation and fibrinolysis inhibition in acute respiratory distress syndrome mice through inhibiting NF-κB p65 translocation. Respir. Res. 2020, 21, 130. [Google Scholar] [CrossRef]
- Sebag, S.C.; Bastarache, J.A.; Ware, L.B. Therapeutic modulation of coagulation and fibrinolysis in acute lung injury and the acute respiratory distress syndrome. Curr. Pharm. Biotechnol. 2011, 12, 1481–1496. [Google Scholar] [CrossRef]
- Bhandary, Y.P.; Shetty, S.K.; Marudamuthu, A.S.; Ji, H.L.; Neuenschwander, P.F.; Boggaram, V.; Morris, G.F.; Fu, J.; Idell, S.; Shetty, S. Regulation of lung injury and fibrosis by p53-mediated changes in urokinase and plasminogen activator inhibitor-1. Am. J. Pathol. 2013, 183, 131–143. [Google Scholar] [CrossRef]
- Della-Morte, D.; Pacifici, F.; Ricordi, C.; Massoud, R.; Rovella, V.; Proietti, S.; Iozzo, M.; Lauro, D.; Bernardini, S.; Bonassi, S.; et al. Low level of plasminogen increases risk for mortality in COVID-19 patients. Cell Death Dis. 2021, 12, 773. [Google Scholar] [CrossRef]
- Juneja, G.K.; Castelo, M.; Yeh, C.H.; Cerroni, S.E.; Hansen, B.E.; Chessum, J.E.; Abraham, J.; Cani, E.; Dwivedi, D.J.; Fraser, D.D.; et al. Biomarkers of coagulation, endothelial function, and fibrinolysis in critically ill patients with COVID-19: A single-center prospective longitudinal study. J. Thromb. Haemost. JTH 2021, 19, 1546–1557. [Google Scholar] [CrossRef]
- Friedrich, C.; Neugebauer, L.; Zamora, M.; Robles, J.P.; Martinez de la Escalera, G.; Clapp, C.; Bertsch, T.; Triebel, J. Plasmin generates vasoinhibin-like peptides by cleaving prolactin and placental lactogen. Mol. Cell. Endocrinol. 2021, 538, 111471. [Google Scholar] [CrossRef]
- Gomez-Builes, J.C.; Acuna, S.A.; Nascimento, B.; Madotto, F.; Rizoli, S.B. Harmful or physiologic: Diagnosing fibrinolysis shutdown in a trauma cohort with rotational thromboelastometry. Anesth. Analg. 2018, 127, 840–849. [Google Scholar] [CrossRef]
- Creel-Bulos, C.; Auld, S.C.; Caridi-Scheible, M.; Barker, N.A.; Friend, S.; Gaddh, M.; Kempton, C.L.; Maier, C.L.; Nahab, F.; Sniecinski, R. Fibrinolysis shutdown and thrombosis in a COVID-19 ICU. Shock 2021, 55, 316–320. [Google Scholar] [CrossRef] [PubMed]
- Han, H.; Yang, L.; Liu, R.; Liu, F.; Wu, K.L.; Li, J.; Liu, X.H.; Zhu, C.L. Prominent changes in blood coagulation of patients with SARS-CoV-2 infection. Clin. Chem. Lab. Med. 2020, 58, 1116–1120. [Google Scholar] [CrossRef]
- Kleyman, T.R.; Kashlan, O.B.; Hughey, R.P. Epithelial Na(+) channel regulation by extracellular and intracellular factors. Annu. Rev. Physiol. 2018, 80, 263–281. [Google Scholar] [CrossRef] [PubMed]
- Sheng, S.; Carattino, M.D.; Bruns, J.B.; Hughey, R.P.; Kleyman, T.R. Furin cleavage activates the epithelial Na+ channel by relieving Na+ self-inhibition. Am. J. Physiol. Ren. Physiol. 2006, 290, F1488–F1496. [Google Scholar] [CrossRef] [PubMed]
- Hughey, R.P.; Bruns, J.B.; Kinlough, C.L.; Harkleroad, K.L.; Tong, Q.; Carattino, M.D.; Johnson, J.P.; Stockand, J.D.; Kleyman, T.R. Epithelial sodium channels are activated by furin-dependent proteolysis. J. Biol. Chem. 2004, 279, 18111–18114. [Google Scholar] [CrossRef]
- Adebamiro, A.; Cheng, Y.; Rao, U.S.; Danahay, H.; Bridges, R.J. A segment of gamma ENaC mediates elastase activation of Na+ transport. J. Gen. Physiol. 2007, 130, 611–629. [Google Scholar] [CrossRef]
- Alli, A.A.; Song, J.Z.; Al-Khalili, O.; Bao, H.F.; Ma, H.P.; Alli, A.A.; Eaton, D.C. Cathepsin B is secreted apically from Xenopus 2F3 cells and cleaves the epithelial sodium channel (ENaC) to increase its activity. J. Biol. Chem. 2012, 287, 30073–30083. [Google Scholar] [CrossRef]
- Mansley, M.K.; Niklas, C.; Nacken, R.; Mandery, K.; Glaeser, H.; Fromm, M.F.; Korbmacher, C.; Bertog, M. Prostaglandin E2 stimulates the epithelial sodium channel (ENaC) in cultured mouse cortical collecting duct cells in an autocrine manner. J. Gen. Physiol. 2020, 152, e201912525. [Google Scholar] [CrossRef]
- Zhao, R.; Ali, G.; Nie, H.G.; Chang, Y.; Bhattarai, D.; Su, X.; Zhao, X.; Matthay, M.A.; Ji, H.L. Plasmin improves blood-gas barrier function in oedematous lungs by cleaving epithelial sodium channels. Br. J. Pharmacol. 2020, 177, 3091–3106. [Google Scholar] [CrossRef]
- Abdel Hameid, R.; Cormet-Boyaka, E.; Kuebler, W.M.; Uddin, M.; Berdiev, B.K. SARS-CoV-2 may hijack GPCR signaling pathways to dysregulate lung ion and fluid transport. Am. J. Physiol. Lung Cell. Mol. Physiol. 2021, 320, L430–L435. [Google Scholar] [CrossRef]
- Haerteis, S.; Krappitz, M.; Diakov, A.; Krappitz, A.; Rauh, R.; Korbmacher, C. Plasmin and chymotrypsin have distinct preferences for channel activating cleavage sites in the γ subunit of the human epithelial sodium channel. J. Gen. Physiol. 2012, 140, 375–389. [Google Scholar] [CrossRef] [PubMed]
- Jackson, C.B.; Farzan, M.; Chen, B.; Choe, H. Mechanisms of SARS-CoV-2 entry into cells. Nat. Rev. Mol. Cell Biol. 2022, 23, 3–20. [Google Scholar] [CrossRef] [PubMed]
- Ridker, P.M.; Gaboury, C.L.; Conlin, P.R.; Seely, E.W.; Williams, G.H.; Vaughan, D.E. Stimulation of plasminogen activator inhibitor in vivo by infusion of angiotensin II. Evidence of a potential interaction between the renin-angiotensin system and fibrinolytic function. Circulation 1993, 87, 1969–1973. [Google Scholar] [CrossRef] [PubMed]
- Sarzani, R.; Giulietti, F.; Di Pentima, C.; Giordano, P.; Spannella, F. Disequilibrium between the classic renin-angiotensin system and its opposing arm in SARS-CoV-2-related lung injury. Am. J. Physiol. Lung Cell. Mol. Physiol. 2020, 319, L325–L336. [Google Scholar] [CrossRef]
- Yoshida, M.; Naito, Y.; Urano, T.; Takada, A.; Takada, Y. L-158,809 and (D-Ala(7))-angiotensin I/II (1-7) decrease PAI-1 release from human umbilical vein endothelial cells. Thromb. Res. 2002, 105, 531–536. [Google Scholar] [CrossRef]
- Zhang, X.; Li, S.; Niu, S. ACE2 and COVID-19 and the resulting ARDS. Postgrad. Med. J. 2020, 96, 403–407. [Google Scholar] [CrossRef]
- Vaughan, D.E.; Lazos, S.A.; Tong, K. Angiotensin II regulates the expression of plasminogen activator inhibitor-1 in cultured endothelial cells. A potential link between the renin-angiotensin system and thrombosis. J. Clin. Investig. 1995, 95, 995–1001. [Google Scholar] [CrossRef]
- Nakamura, S.; Nakamura, I.; Ma, L.; Vaughan, D.E.; Fogo, A.B. Plasminogen activator inhibitor-1 expression is regulated by the angiotensin type 1 receptor in vivo. Kidney Int. 2000, 58, 251–259. [Google Scholar] [CrossRef]
- Abassi, Z.A.; Skorecki, K.; Heyman, S.N.; Kinaneh, S.; Armaly, Z. COVID-19 infection and mortality: A physiologist’s perspective enlightening clinical features and plausible interventional strategies. Am. J. Physiol. Lung Cell. Mol. Physiol. 2020, 318, L1020–L1022. [Google Scholar] [CrossRef]
- Chen, D.; Li, X.; Song, Q.; Hu, C.; Su, F.; Dai, J.; Ye, Y.; Huang, J.; Zhang, X. Assessment of hypokalemia and clinical characteristics in patients with coronavirus disease 2019 in Wenzhou, China. JAMA Netw. Open 2020, 3, e2011122. [Google Scholar] [CrossRef]
- Veras, F.P.; Pontelli, M.C.; Silva, C.M.; Toller-Kawahisa, J.E.; de Lima, M.; Nascimento, D.C.; Schneider, A.H.; Caetité, D.; Tavares, L.A.; Paiva, I.M.; et al. SARS-CoV-2-triggered neutrophil extracellular traps mediate COVID-19 pathology. J. Exp. Med. 2020, 217, e20201129. [Google Scholar] [CrossRef]
- Radermecker, C.; Detrembleur, N.; Guiot, J.; Cavalier, E.; Henket, M.; d’Emal, C.; Vanwinge, C.; Cataldo, D.; Oury, C.; Delvenne, P.; et al. Neutrophil extracellular traps infiltrate the lung airway, interstitial, and vascular compartments in severe COVID-19. J. Exp. Med. 2020, 217, e20201012. [Google Scholar] [CrossRef] [PubMed]
- Barnes, B.J.; Adrover, J.M.; Baxter-Stoltzfus, A.; Borczuk, A.; Cools-Lartigue, J.; Crawford, J.M.; Daßler-Plenker, J.; Guerci, P.; Huynh, C.; Knight, J.S.; et al. Targeting potential drivers of COVID-19: Neutrophil extracellular traps. J. Exp. Med. 2020, 217, e20200652. [Google Scholar] [CrossRef] [PubMed]
- Barbosa da Cruz, D.; Helms, J.; Aquino, L.R.; Stiel, L.; Cougourdan, L.; Broussard, C.; Chafey, P.; Riès-Kautt, M.; Meziani, F.; Toti, F.; et al. DNA-bound elastase of neutrophil extracellular traps degrades plasminogen, reduces plasmin formation, and decreases fibrinolysis: Proof of concept in septic shock plasma. FASEB J. 2019, 33, 14270–14280. [Google Scholar] [CrossRef] [PubMed]
- Zoufaly, A.; Poglitsch, M.; Aberle, J.H.; Hoepler, W.; Seitz, T.; Traugott, M.; Grieb, A.; Pawelka, E.; Laferl, H.; Wenisch, C.; et al. Human recombinant soluble ACE2 in severe COVID-19. Lancet Respir. Med. 2020, 8, 1154–1158. [Google Scholar] [CrossRef]
- Alhenc-Gelas, F.; Drueke, T.B. Blockade of SARS-CoV-2 infection by recombinant soluble ACE2. Kidney Int. 2020, 97, 1091–1093. [Google Scholar] [CrossRef]
- Iwanami, J.; Mogi, M.; Tsukuda, K.; Wang, X.L.; Nakaoka, H.; Ohshima, K.; Chisaka, T.; Bai, H.Y.; Kanno, H.; Min, L.J.; et al. Role of angiotensin-converting enzyme 2/angiotensin-(1-7)/Mas axis in the hypotensive effect of azilsartan. Hypertens. Res. 2014, 37, 616–620. [Google Scholar] [CrossRef]
- Monteil, V.; Kwon, H.; Prado, P.; Hagelkrüys, A.; Wimmer, R.A.; Stahl, M.; Leopoldi, A.; Garreta, E.; Hurtado Del Pozo, C.; Prosper, F.; et al. Inhibition of SARS-CoV-2 infections in engineered human tissues using clinical-grade soluble human ACE2. Cell 2020, 181, 905–913.e907. [Google Scholar] [CrossRef]
- Khodavirdipour, A.; Piri, M.; Jabbari, S.; Khalaj-Kondori, M. Potential of CRISPR/Cas13 system in treatment and diagnosis of COVID-19. Glob. Med. Genet. 2021, 8, 7–10. [Google Scholar] [CrossRef]
- Wu, C.; Zheng, M.; Yang, Y.; Gu, X.; Yang, K.; Li, M.; Liu, Y.; Zhang, Q.; Zhang, P.; Wang, Y.; et al. Furin: A potential therapeutic target for COVID-19. iScience 2020, 23, 101642. [Google Scholar] [CrossRef]
- Hoffmann, M.; Hofmann-Winkler, H.; Smith, J.C.; Krüger, N.; Arora, P.; Sørensen, L.K.; Søgaard, O.S.; Hasselstrøm, J.B.; Winkler, M.; Hempel, T.; et al. Camostat mesylate inhibits SARS-CoV-2 activation by TMPRSS2-related proteases and its metabolite GBPA exerts antiviral activity. EBioMedicine 2021, 65, 103255. [Google Scholar] [CrossRef] [PubMed]
- Hempel, T.; Raich, L.; Olsson, S.; Azouz, N.P.; Klingler, A.M.; Hoffmann, M.; Pöhlmann, S.; Rothenberg, M.E.; Noé, F. Molecular mechanism of inhibiting the SARS-CoV-2 cell entry facilitator TMPRSS2 with camostat and nafamostat. Chem. Sci. 2021, 12, 983–992. [Google Scholar] [CrossRef] [PubMed]
- Kodama, K.; Imai, T.; Asai, Y.; Kozu, Y.; Hayashi, K.; Shimizu, T.; Gon, Y.; Ootsuka, S. Incidence and risk factors for hyperkalaemia in patients treated for COVID-19 with nafamostat mesylate. J. Clin. Pharm. Ther. 2022, 47, 1070–1078. [Google Scholar] [CrossRef] [PubMed]
- Ansarin, K.; Tolouian, R.; Ardalan, M.; Taghizadieh, A.; Varshochi, M.; Teimouri, S.; Vaezi, T.; Valizadeh, H.; Saleh, P.; Safiri, S.; et al. Effect of bromhexine on clinical outcomes and mortality in COVID-19 patients: A randomized clinical trial. BioImpacts 2020, 10, 209–215. [Google Scholar] [CrossRef]
- Shapira, T.; Monreal, I.A.; Dion, S.P.; Buchholz, D.W.; Imbiakha, B.; Olmstead, A.D.; Jager, M.; Désilets, A.; Gao, G.; Martins, M.; et al. A TMPRSS2 inhibitor acts as a pan-SARS-CoV-2 prophylactic and therapeutic. Nature 2022, 605, 340–348. [Google Scholar] [CrossRef]
- Satarker, S.; Tom, A.A.; Shaji, R.A.; Alosious, A.; Luvis, M.; Nampoothiri, M. JAK-STAT pathway inhibition and their implications in COVID-19 therapy. Postgrad. Med. 2021, 133, 489–507. [Google Scholar] [CrossRef]
- Choy, E.H.S.; Miceli-Richard, C.; González-Gay, M.A.; Sinigaglia, L.; Schlichting, D.E.; Meszaros, G.; de la Torre, I.; Schulze-Koops, H. The effect of JAK1/JAK2 inhibition in rheumatoid arthritis: Efficacy and safety of baricitinib. Clin. Exp. Rheumatol. 2019, 37, 694–704. [Google Scholar]
- Tahsini Tekantapeh, S.; Ghojazadeh, M.; Ghamari, A.A.; Mohammadi, A.; Soleimanpour, H. Therapeutic and anti-inflammatory effects of baricitinib on mortality, ICU transfer, clinical improvement, and CRS-related laboratory parameters of hospitalized patients with moderate to severe COVID-19 pneumonia: A systematic review and meta-analysis. Expert Rev. Respir. Med. 2022, 16, 1109–1132. [Google Scholar] [CrossRef]
- Marconi, V.C.; Ramanan, A.V.; de Bono, S.; Kartman, C.E.; Krishnan, V.; Liao, R.; Piruzeli, M.L.B.; Goldman, J.D.; Alatorre-Alexander, J.; de Cassia Pellegrini, R.; et al. Efficacy and safety of baricitinib for the treatment of hospitalised adults with COVID-19 (COV-BARRIER): A randomised, double-blind, parallel-group, placebo-controlled phase 3 trial. Lancet. Respir. Med. 2021, 9, 1407–1418. [Google Scholar] [CrossRef]
- Levy, G.; Guglielmelli, P.; Langmuir, P.; Constantinescu, S.N. JAK inhibitors and COVID-19. J. Immunother. Cancer 2022, 10, e002838. [Google Scholar] [CrossRef]
- Gudowska-Sawczuk, M.; Mroczko, B. The role of nuclear factor kappa B (NF-κB) in development and treatment of COVID-19: Review. Int. J. Mol. Sci. 2022, 23, 5283. [Google Scholar] [CrossRef] [PubMed]
- Sharma, V.K.; Prateeksha; Singh, S.P.; Singh, B.N.; Rao, C.V.; Barik, S.K. Nanocurcumin potently inhibits SARS-CoV-2 spike protein-induced cytokine storm by deactivation of MAPK/NF-κB signaling in epithelial cells. ACS Appl. Bio Mater. 2022, 5, 483–491. [Google Scholar] [CrossRef] [PubMed]
- Ran, Y.; Su, W.; Gao, F.; Ding, Z.; Yang, S.; Ye, L.; Chen, X.; Tian, G.; Xi, J.; Liu, Z. Curcumin ameliorates white matter injury after ischemic stroke by inhibiting microglia/macrophage pyroptosis through NF-κB suppression and NLRP3 inflammasome inhibition. Oxidative Med. Cell. Longev. 2021, 2021, 1552127. [Google Scholar] [CrossRef] [PubMed]
- Fathimath Muneesa, M.; Barki, R.R.; Shaikh, S.B.; Bhandary, Y.P. Curcumin intervention during progressive fibrosis controls inflammatory cytokines and the fibrinolytic system in pulmonary fibrosis. Toxicol. Appl. Pharmacol. 2022, 449, 116116. [Google Scholar] [CrossRef] [PubMed]
- Zahedipour, F.; Hosseini, S.A.; Sathyapalan, T.; Majeed, M.; Jamialahmadi, T.; Al-Rasadi, K.; Banach, M.; Sahebkar, A. Potential effects of curcumin in the treatment of COVID-19 infection. Phytother. Res. 2020, 34, 2911–2920. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, T.; Zhai, Y.; Xue, H.; Zhou, W.; Ding, Y.; Nie, H. Regulation of Epithelial Sodium Transport by SARS-CoV-2 Is Closely Related with Fibrinolytic System-Associated Proteins. Biomolecules 2023, 13, 578. https://doi.org/10.3390/biom13040578
Wang T, Zhai Y, Xue H, Zhou W, Ding Y, Nie H. Regulation of Epithelial Sodium Transport by SARS-CoV-2 Is Closely Related with Fibrinolytic System-Associated Proteins. Biomolecules. 2023; 13(4):578. https://doi.org/10.3390/biom13040578
Chicago/Turabian StyleWang, Tingyu, Yiman Zhai, Hao Xue, Wei Zhou, Yan Ding, and Hongguang Nie. 2023. "Regulation of Epithelial Sodium Transport by SARS-CoV-2 Is Closely Related with Fibrinolytic System-Associated Proteins" Biomolecules 13, no. 4: 578. https://doi.org/10.3390/biom13040578
APA StyleWang, T., Zhai, Y., Xue, H., Zhou, W., Ding, Y., & Nie, H. (2023). Regulation of Epithelial Sodium Transport by SARS-CoV-2 Is Closely Related with Fibrinolytic System-Associated Proteins. Biomolecules, 13(4), 578. https://doi.org/10.3390/biom13040578





