Factors and Methods for the Detection of Gene Expression Regulation
Abstract
:1. Introduction
2. TFs
2.1. Genome-Wide Methods for Identifying TFBS
2.1.1. Chromatin Immunoprecipitation
2.1.2. Cleavage under Targets and Release Using Nuclease and Cleavage under Targets and Tagmentation
2.1.3. DNA Affinity Purification Sequencing
2.2. Detection Methods for TFs
3. Chromatin Accessibility
3.1. Detection Methods for Chromatin Accessibility
3.1.1. DNase I Hypersensitive Site Sequencing Subsubsection
3.1.2. Micrococcal Nuclease Sequencing
3.1.3. Sequencing-Based Assays for the Detection of Transposase-Accessible Chromatin
3.1.4. Nucleosome Occupancy and Methylome Sequencing
4. Histone Modifications
4.1. Acetylation
4.2. Methylation
4.3. Phosphorylation
4.4. Methods for Identifying Specific Histone Modification Binding Sites
4.5. Methods for Identifying Histone Modifications
5. DNA Methylation
5.1. Detection Methods for DNA Methylation
5.1.1. Bisulfite Sequencing and Its Derivatives
5.1.2. TET-Assisted Pyridine Borane Sequencing and Its Derivatives
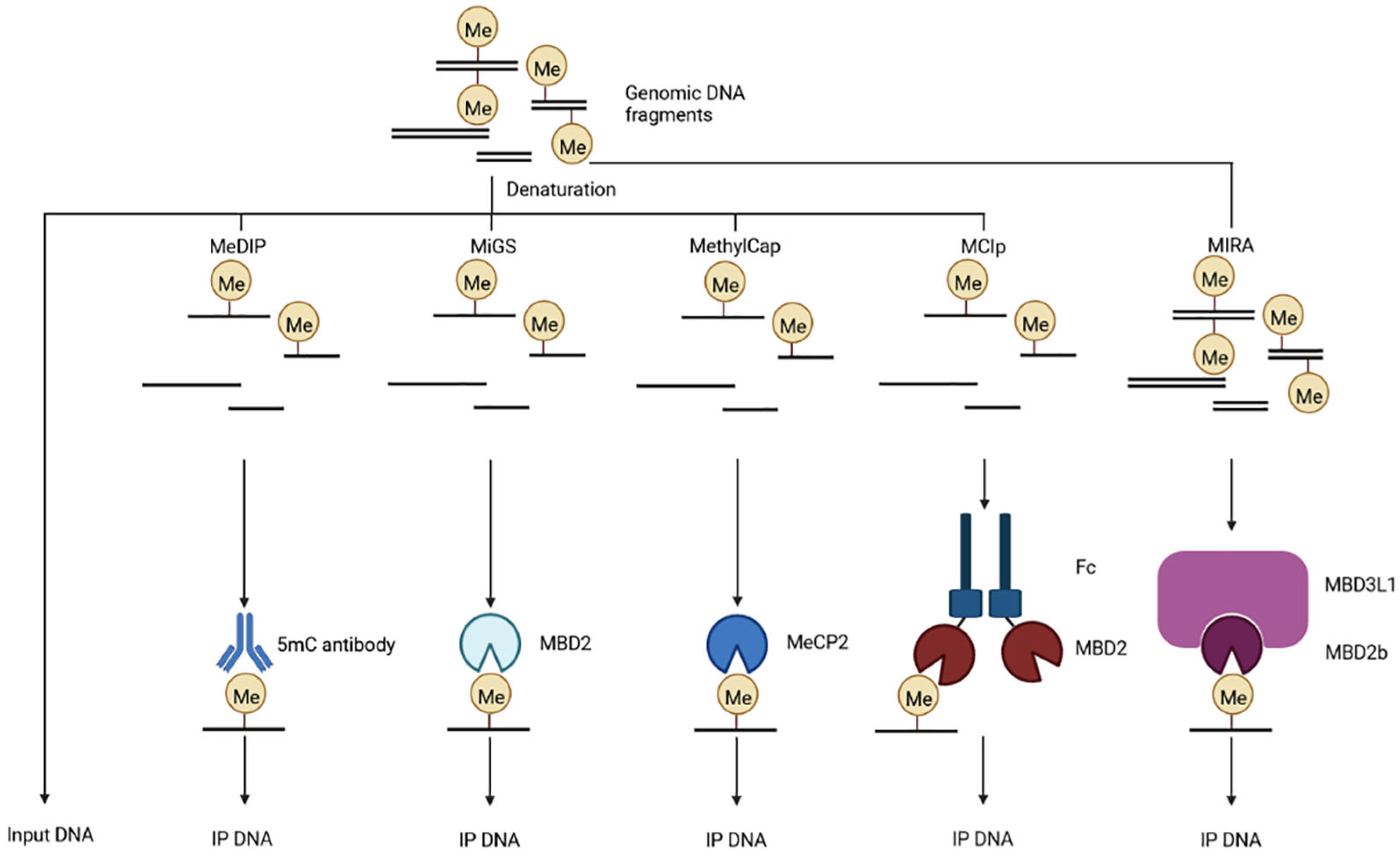
5.1.3. Methylated DNA Immunoprecipitation
5.1.4. Nanopore Sequencing
5.1.5. Single-Molecule, Real-Time (SMRT) Sequencing
6. RNA Modifications
6.1. Quantitative Methods of Detecting m6A Modifications
6.2. Localization-Detection Methods for m6A Modifications
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Goldberg, A.D.; Allis, C.D.; Bernstein, E. Epigenetics: A landscape takes shape. Cell 2007, 128, 635–638. [Google Scholar] [CrossRef] [PubMed]
- Brodsky, S.; Jana, T.; Mittelman, K.; Chapal, M.; Kumar, D.K.; Carmi, M.; Barkai, N. Intrinsically disordered regions direct transcription factor in vivo binding specificity. Mol. Cell 2020, 79, 459–471.e4. [Google Scholar] [CrossRef] [PubMed]
- Solomon, M.J.; Larsen, P.L.; Varshavsky, A. Mapping protein-DNA interactions in vivo with formaldehyde: Evidence that histone H4 is retained on a highly transcribed gene. Cell 1988, 53, 937–947. [Google Scholar] [CrossRef] [PubMed]
- Ti, L.; Se, J.; Ra, Y. Chromatin immunoprecipitation and microarray-based analysis of protein location. Nat. Protoc. 2006, 1, 729–748. [Google Scholar] [CrossRef]
- Yamakawa, T.; Itakura, K. Chromatin immunoprecipitation assay using micrococcal nucleases in mammalian cells. J. Vis. Exp. JoVE 2019, 147, e59375. [Google Scholar] [CrossRef]
- Park, P.J. ChIP-Seq: Advantages and challenges of a maturing technology. Nat. Rev. Genet. 2009, 10, 669–680. [Google Scholar] [CrossRef]
- Johnson, D.S.; Mortazavi, A.; Myers, R.M.; Wold, B. Genome-wide mapping of in vivo protein-DNA interactions. Science 2007, 316, 1497–1502. [Google Scholar] [CrossRef]
- Dirks, R.A.M.; Thomas, P.C.; Wu, H.; Jones, R.C.; Stunnenberg, H.G.; Marks, H. A Plug and play microfluidic platform for standardized sensitive low-input chromatin immunoprecipitation. Genome Res. 2021, 31, 919–933. [Google Scholar] [CrossRef]
- Wang, Q.; Xiong, H.; Ai, S.; Yu, X.; Liu, Y.; Zhang, J.; He, A. CoBATCH for high-throughput single-cell epigenomic profiling. Mol. Cell 2019, 76, 206–216.e7. [Google Scholar] [CrossRef]
- Ai, S.; Xiong, H.; Li, C.C.; Luo, Y.; Shi, Q.; Liu, Y.; Yu, X.; Li, C.; He, A. Profiling chromatin states using single-cell ItChIP-Seq. Nat. Cell Biol. 2019, 21, 1164–1172. [Google Scholar] [CrossRef]
- Schmid, M.; Durussel, T.; Laemmli, U.K. ChIC and ChEC: Genomic mapping of chromatin proteins. Mol. Cell 2004, 16, 147–157. [Google Scholar] [CrossRef] [PubMed]
- Skene, P.J.; Henikoff, S. An efficient targeted nuclease strategy for high-resolution mapping of DNA Binding Sites. eLife 2017, 6, e21856. [Google Scholar] [CrossRef] [PubMed]
- Skene, P.J.; Henikoff, J.G.; Henikoff, S. Targeted in situ genome-wide profiling with high efficiency for low cell numbers. Nat. Protoc. 2018, 13, 1006–1019. [Google Scholar] [CrossRef] [PubMed]
- Jain, D.; Baldi, S.; Zabel, A.; Straub, T.; Becker, P.B. Active promoters give rise to false positive “Phantom Peaks” in ChIP-Seq experiments. Nucleic Acids Res. 2015, 43, 6959–6968. [Google Scholar] [CrossRef]
- Baranello, L.; Kouzine, F.; Sanford, S.; Levens, D. ChIP bias as a function of cross-linking time. Chromosome Res. Int. J. Mol. Supramol. Evol. Asp. Chromosome Biol. 2016, 24, 175–181. [Google Scholar] [CrossRef]
- Hainer, S.J.; Bošković, A.; McCannell, K.N.; Rando, O.J.; Fazzio, T.G. Profiling of pluripotency factors in single cells and early embryos. Cell 2019, 177, 1319–1329.e11. [Google Scholar] [CrossRef]
- Kaya-Okur, H.S.; Wu, S.J.; Codomo, C.A.; Pledger, E.S.; Bryson, T.D.; Henikoff, J.G.; Ahmad, K.; Henikoff, S. CUT&Tag for efficient epigenomic profiling of small samples and single cells. Nat. Commun. 2019, 10, 1930. [Google Scholar] [CrossRef]
- Picelli, S.; Björklund, A.K.; Reinius, B.; Sagasser, S.; Winberg, G.; Sandberg, R. Tn5 transposase and tagmentation procedures for massively scaled sequencing projects. Genome Res. 2014, 24, 2033–2040. [Google Scholar] [CrossRef]
- Kaya-Okur, H.S.; Janssens, D.H.; Henikoff, J.G.; Ahmad, K.; Henikoff, S. Efficient low-cost chromatin profiling with CUT&Tag. Nat. Protoc. 2020, 15, 3264–3283. [Google Scholar] [CrossRef]
- Wu, S.J.; Furlan, S.N.; Mihalas, A.B.; Kaya-Okur, H.S.; Feroze, A.H.; Emerson, S.N.; Zheng, Y.; Carson, K.; Cimino, P.J.; Keene, C.D.; et al. Single-cell CUT&Tag analysis of chromatin modifications in differentiation and tumor progression. Nat. Biotechnol. 2021, 39, 819–824. [Google Scholar] [CrossRef]
- Bartosovic, M.; Kabbe, M.; Castelo-Branco, G. Single-Cell CUT&Tag profiles histone modifications and transcription factors in complex tissues. Nat. Biotechnol. 2021, 39, 825–835. [Google Scholar] [CrossRef] [PubMed]
- O’Malley, R.C.; Huang, S.-S.C.; Song, L.; Lewsey, M.G.; Bartlett, A.; Nery, J.R.; Galli, M.; Gallavotti, A.; Ecker, J.R. Cistrome and epicistrome features shape the regulatory DNA landscape. Cell 2016, 165, 1280–1292. [Google Scholar] [CrossRef] [PubMed]
- Huberman, L.B.; Wu, V.W.; Kowbel, D.J.; Lee, J.; Daum, C.; Grigoriev, I.V.; O’Malley, R.C.; Glass, N.L. DNA affinity purification sequencing and transcriptional profiling reveal new aspects of nitrogen regulation in a filamentous fungus. Proc. Natl. Acad. Sci. USA 2021, 118, e2009501118. [Google Scholar] [CrossRef] [PubMed]
- Lai, X.; Stigliani, A.; Lucas, J.; Hugouvieux, V.; Parcy, F.; Zubieta, C. Genome-wide binding of SEPALLATA3 and AGAMOUS complexes determined by sequential DNA-affinity purification sequencing. Nucleic Acids Res. 2020, 48, 9637–9648. [Google Scholar] [CrossRef]
- Bartlett, A.; O’Malley, R.C.; Huang, S.-S.C.; Galli, M.; Nery, J.R.; Gallavotti, A.; Ecker, J.R. Mapping genome-wide transcription-factor binding sites using DAP-Seq. Nat. Protoc. 2017, 12, 1659–1672. [Google Scholar] [CrossRef]
- Sui, H.; Imamichi, T. A DNA pull-down assay with diversity forms of competitor for detecting or evaluating protein-DNA interactions. Method. Mol. Biol. 2023, 2599, 1–10. [Google Scholar] [CrossRef]
- Nassar, L.R.; Barber, G.P.; Benet-Pagès, A.; Casper, J.; Clawson, H.; Diekhans, M.; Fischer, C.; Gonzalez, J.N.; Hinrichs, A.S.; Lee, B.T.; et al. The UCSC genome browser database: 2023 update. Nucleic Acids Res. 2022, 51, gkac1072. [Google Scholar] [CrossRef]
- Castro-Mondragon, J.A.; Riudavets-Puig, R.; Rauluseviciute, I.; Lemma, R.B.; Turchi, L.; Blanc-Mathieu, R.; Lucas, J.; Boddie, P.; Khan, A.; Pérez, N.M.; et al. JASPAR 2022: The 9th release of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 2022, 50, D165–D173. [Google Scholar] [CrossRef]
- Ying, Y.; Ma, X.; Fang, J.; Chen, S.; Wang, W.; Li, J.; Xie, H.; Wu, J.; Xie, B.; Liu, B.; et al. EGR2-mediated regulation of M6A reader IGF2BP proteins drive RCC tumorigenesis and metastasis via enhancing S1PR3 MRNA stabilization. Cell Death Dis. 2021, 12, 750. [Google Scholar] [CrossRef]
- Solberg, N.; Krauss, S. Luciferase assay to study the activity of a cloned promoter DNA fragment. Method. Mol. Biol. Clifton NJ 2013, 977, 65–78. [Google Scholar] [CrossRef]
- Luger, K.; Mäder, A.W.; Richmond, R.K.; Sargent, D.F.; Richmond, T.J. Crystal structure of the nucleosome core particle at 2.8 a resolution. Nature 1997, 389, 251–260. [Google Scholar] [CrossRef] [PubMed]
- Fyodorov, D.V.; Zhou, B.-R.; Skoultchi, A.I.; Bai, Y. Emerging roles of linker histones in regulating chromatin structure and function. Nat. Rev. Mol. Cell Biol. 2018, 19, 192–206. [Google Scholar] [CrossRef] [PubMed]
- Venkatesh, S.; Workman, J.L. Histone exchange, chromatin structure and the regulation of transcription. Nat. Rev. Mol. Cell Biol. 2015, 16, 178–189. [Google Scholar] [CrossRef] [PubMed]
- Bell, O.; Tiwari, V.K.; Thomä, N.H.; Schübeler, D. Determinants and dynamics of genome accessibility. Nat. Rev. Genet. 2011, 12, 554–564. [Google Scholar] [CrossRef] [PubMed]
- Klemm, S.L.; Shipony, Z.; Greenleaf, W.J. Chromatin accessibility and the regulatory epigenome. Nat. Rev. Genet. 2019, 20, 207–220. [Google Scholar] [CrossRef]
- Stalder, J.; Larsen, A.; Engel, J.D.; Dolan, M.; Groudine, M.; Weintraub, H. Tissue-Specific DNA cleavages in the globin chromatin domain introduced by DNAase I. Cell 1980, 20, 451–460. [Google Scholar] [CrossRef]
- Thurman, R.E.; Rynes, E.; Humbert, R.; Vierstra, J.; Maurano, M.T.; Haugen, E.; Sheffield, N.C.; Stergachis, A.B.; Wang, H.; Vernot, B.; et al. The accessible chromatin landscape of the human genome. Nature 2012, 489, 75–82. [Google Scholar] [CrossRef]
- Meuleman, W.; Muratov, A.; Rynes, E.; Halow, J.; Lee, K.; Bates, D.; Diegel, M.; Dunn, D.; Neri, F.; Teodosiadis, A.; et al. Index and biological spectrum of human DNase I hypersensitive sites. Nature 2020, 584, 244–251. [Google Scholar] [CrossRef]
- Keene, M.A.; Corces, V.; Lowenhaupt, K.; Elgin, S.C. DNase I hypersensitive sites in drosophila chromatin occur at the 5′ ends of regions of transcription. Proc. Natl. Acad. Sci. USA 1981, 78, 143–146. [Google Scholar] [CrossRef]
- Boyle, A.P.; Davis, S.; Shulha, H.P.; Meltzer, P.; Margulies, E.H.; Weng, Z.; Furey, T.S.; Crawford, G.E. High-resolution mapping and characterization of open chromatin across the genome. Cell 2008, 132, 311–322. [Google Scholar] [CrossRef]
- Tsompana, M.; Buck, M.J. Chromatin accessibility: A window into the genome. Epigenetics Chromatin 2014, 7, 33. [Google Scholar] [CrossRef] [PubMed]
- Jin, W.; Tang, Q.; Wan, M.; Cui, K.; Zhang, Y.; Ren, G.; Ni, B.; Sklar, J.; Przytycka, T.M.; Childs, R.; et al. Genome-wide detection of DNase I hypersensitive sites in single cells and FFPE tissue samples. Nature 2015, 528, 142–146. [Google Scholar] [CrossRef] [PubMed]
- Gao, W.; Ku, W.L.; Pan, L.; Perrie, J.; Zhao, T.; Hu, G.; Wu, Y.; Zhu, J.; Ni, B.; Zhao, K. Multiplex indexing approach for the detection of DNase I hypersensitive sites in single cells. Nucleic Acids Res. 2021, 49, e56. [Google Scholar] [CrossRef] [PubMed]
- Rizzo, J.M.; Sinha, S. Analyzing the global chromatin structure of keratinocytes by MNase-Seq. Method. Mol. Biol. Clifton NJ 2014, 1195, 49–59. [Google Scholar] [CrossRef]
- Mieczkowski, J.; Cook, A.; Bowman, S.K.; Mueller, B.; Alver, B.H.; Kundu, S.; Deaton, A.M.; Urban, J.A.; Larschan, E.; Park, P.J.; et al. MNase titration reveals differences between nucleosome occupancy and chromatin accessibility. Nat. Commun. 2016, 7, 11485. [Google Scholar] [CrossRef]
- Jiang, C.; Pugh, B.F. Nucleosome positioning and gene regulation: Advances through genomics. Nat. Rev. Genet. 2009, 10, 161–172. [Google Scholar] [CrossRef]
- Dingwall, C.; Lomonossoff, G.P.; Laskey, R.A. High sequence specificity of micrococcal nuclease. Nucleic Acids Res. 1981, 9, 2659–2673. [Google Scholar] [CrossRef]
- Hörz, W.; Altenburger, W. Sequence specific cleavage of DNA by micrococcal nuclease. Nucleic Acids Res. 1981, 9, 2643–2658. [Google Scholar] [CrossRef]
- Lai, B.; Gao, W.; Cui, K.; Xie, W.; Tang, Q.; Jin, W.; Hu, G.; Ni, B.; Zhao, K. Principles of nucleosome organization revealed by single-cell micrococcal nuclease sequencing. Nature 2018, 562, 281–285. [Google Scholar] [CrossRef]
- Buenrostro, J.D.; Giresi, P.G.; Zaba, L.C.; Chang, H.Y.; Greenleaf, W.J. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Method. 2013, 10, 1213–1218. [Google Scholar] [CrossRef]
- Luo, L.; Gribskov, M.; Wang, S. Bibliometric review of ATAC-Seq and its application in gene expression. Brief. Bioinform. 2022, 23, bbac061. [Google Scholar] [CrossRef] [PubMed]
- Buenrostro, J.D.; Wu, B.; Chang, H.Y.; Greenleaf, W.J. ATAC-Seq: A method for assaying chromatin accessibility genome-wide. Curr. Protoc. Mol. Biol. 2015, 109, 21.29.1–21.29.9. [Google Scholar] [CrossRef] [PubMed]
- Cusanovich, D.A.; Daza, R.; Adey, A.; Pliner, H.A.; Christiansen, L.; Gunderson, K.L.; Steemers, F.J.; Trapnell, C.; Shendure, J. Multiplex single cell profiling of chromatin accessibility by combinatorial cellular indexing. Science 2015, 348, 910–914. [Google Scholar] [CrossRef]
- Buenrostro, J.D.; Wu, B.; Litzenburger, U.M.; Ruff, D.; Gonzales, M.L.; Snyder, M.P.; Chang, H.Y.; Greenleaf, W.J. Single-cell chromatin accessibility reveals principles of regulatory variation. Nature 2015, 523, 486–490. [Google Scholar] [CrossRef] [PubMed]
- Buenrostro, J.D.; Corces, M.R.; Lareau, C.A.; Wu, B.; Schep, A.N.; Aryee, M.J.; Majeti, R.; Chang, H.Y.; Greenleaf, W.J. Integrated single-cell analysis maps the continuous regulatory landscape of human hematopoietic differentiation. Cell 2018, 173, 1535–1548.e16. [Google Scholar] [CrossRef] [PubMed]
- Cusanovich, D.A.; Hill, A.J.; Aghamirzaie, D.; Daza, R.M.; Pliner, H.A.; Berletch, J.B.; Filippova, G.N.; Huang, X.; Christiansen, L.; DeWitt, W.S.; et al. A single-cell atlas of in vivo mammalian chromatin accessibility. Cell 2018, 174, 1309–1324.e18. [Google Scholar] [CrossRef] [PubMed]
- Mezger, A.; Klemm, S.; Mann, I.; Brower, K.; Mir, A.; Bostick, M.; Farmer, A.; Fordyce, P.; Linnarsson, S.; Greenleaf, W. High-throughput chromatin accessibility profiling at single-cell resolution. Nat. Commun. 2018, 9, 3647. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Miragaia, R.J.; Natarajan, K.N.; Teichmann, S.A. A rapid and robust method for single cell chromatin accessibility profiling. Nat. Commun. 2018, 9, 5345. [Google Scholar] [CrossRef]
- Lareau, C.A.; Duarte, F.M.; Chew, J.G.; Kartha, V.K.; Burkett, Z.D.; Kohlway, A.S.; Pokholok, D.; Aryee, M.J.; Steemers, F.J.; Lebofsky, R.; et al. Droplet-based combinatorial indexing for massive-scale single-cell chromatin accessibility. Nat. Biotechnol. 2019, 37, 916–924. [Google Scholar] [CrossRef]
- Kelly, T.K.; Liu, Y.; Lay, F.D.; Liang, G.; Berman, B.P.; Jones, P.A. Genome-wide mapping of nucleosome positioning and DNA methylation within individual DNA molecules. Genome Res. 2012, 22, 2497–2506. [Google Scholar] [CrossRef]
- Rhie, S.K.; Schreiner, S.; Farnham, P.J. Defining regulatory elements in the human genome using nucleosome occupancy and methylome sequencing (NOMe-Seq). Methods Mol. Biol. Clifton NJ 2018, 1766, 209–229. [Google Scholar] [CrossRef]
- Lay, F.D.; Kelly, T.K.; Jones, P.A. Nucleosome occupancy and methylome sequencing (NOMe-Seq). Methods Mol. Biol. Clifton NJ 2018, 1708, 267–284. [Google Scholar] [CrossRef]
- Kouzarides, T. Chromatin modifications and their function. Cell 2007, 128, 693–705. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Sabari, B.R.; Garcia, B.A.; Allis, C.D.; Zhao, Y. SnapShot: Histone modifications. Cell 2014, 159, 458–458.e1. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Tang, Z.; Huang, H.; Zhou, G.; Cui, C.; Weng, Y.; Liu, W.; Kim, S.; Lee, S.; Perez-Neut, M.; et al. Metabolic regulation of gene expression by histone lactylation. Nature 2019, 574, 575–580. [Google Scholar] [CrossRef]
- Allfrey, V.G.; Faulkner, R.; Mirsky, A.E. Acetylation and methylation of histones and their possible role in the regulation of rna synthesis. Proc. Natl. Acad. Sci. USA 1964, 51, 786–794. [Google Scholar] [CrossRef]
- Stillman, B. Histone modifications: Insights into their influence on gene expression. Cell 2018, 175, 6–9. [Google Scholar] [CrossRef]
- Lawrence, M.; Daujat, S.; Schneider, R. Lateral thinking: How histone modifications regulate gene expression. Trends Genet. TIG 2016, 32, 42–56. [Google Scholar] [CrossRef]
- Tessarz, P.; Kouzarides, T. Histone core modifications regulating nucleosome structure and dynamics. Nat. Rev. Mol. Cell Biol. 2014, 15, 703–708. [Google Scholar] [CrossRef]
- Sterner, D.E.; Berger, S.L. Acetylation of histones and transcription-related factors. Microbiol. Mol. Biol. Rev. MMBR 2000, 64, 435–459. [Google Scholar] [CrossRef]
- D’Mello, S.R. Histone deacetylases 1, 2 and 3 in nervous system development. Curr. Opin. Pharmacol. 2020, 50, 74–81. [Google Scholar] [CrossRef]
- Morgan, M.A.J.; Shilatifard, A. Reevaluating the roles of histone-modifying enzymes and their associated chromatin modifications in transcriptional regulation. Nat. Genet. 2020, 52, 1271–1281. [Google Scholar] [CrossRef]
- Zhang, Y.; Reinberg, D. Transcription regulation by histone methylation: Interplay between different covalent modifications of the core histone tails. Genes Dev. 2001, 15, 2343–2360. [Google Scholar] [CrossRef]
- Maurer-Stroh, S.; Dickens, N.J.; Hughes-Davies, L.; Kouzarides, T.; Eisenhaber, F.; Ponting, C.P. The tudor domain “Royal Family”: Tudor, plant agenet, chromo, PWWP and MBT domains. Trends Biochem. Sci. 2003, 28, 69–74. [Google Scholar] [CrossRef]
- Wysocka, J.; Swigut, T.; Xiao, H.; Milne, T.A.; Kwon, S.Y.; Landry, J.; Kauer, M.; Tackett, A.J.; Chait, B.T.; Badenhorst, P.; et al. A PHD Finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature 2006, 442, 86–90. [Google Scholar] [CrossRef]
- Fischle, W.; Tseng, B.S.; Dormann, H.L.; Ueberheide, B.M.; Garcia, B.A.; Shabanowitz, J.; Hunt, D.F.; Funabiki, H.; Allis, C.D. Regulation of HP1-chromatin binding by histone H3 methylation and phosphorylation. Nature 2005, 438, 1116–1122. [Google Scholar] [CrossRef]
- Nakato, R.; Sakata, T. Methods for ChIP-Seq Analysis: A practical workflow and advanced applications. Method. San Diego Calif 2021, 187, 44–53. [Google Scholar] [CrossRef] [PubMed]
- Ford, B.R.; Vignali, P.D.A.; Rittenhouse, N.L.; Scharping, N.E.; Peralta, R.; Lontos, K.; Frisch, A.T.; Delgoffe, G.M.; Poholek, A.C. Tumor microenvironmental signals reshape chromatin landscapes to limit the functional potential of exhausted T cells. Sci. Immunol. 2022, 7, eabj9123. [Google Scholar] [CrossRef] [PubMed]
- Yashar, W.M.; Kong, G.; VanCampen, J.; Curtiss, B.M.; Coleman, D.J.; Carbone, L.; Yardimci, G.G.; Maxson, J.E.; Braun, T.P. GoPeaks: Histone modification peak calling for CUT&Tag. Genome Biol. 2022, 23, 144. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Wang, C.; Liu, W.; Li, J.; Li, C.; Kou, X.; Chen, J.; Zhao, Y.; Gao, H.; Wang, H.; et al. Distinct features of H3K4me3 and H3K27me3 chromatin domains in pre-implantation embryos. Nature 2016, 537, 558–562. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Z.-F.; Arnaudo, A.M.; Garcia, B.A. Mass spectrometric analysis of histone proteoforms. Annu. Rev. Anal. Chem. Palo Alto Calif 2014, 7, 113–128. [Google Scholar] [CrossRef]
- Britton, L.-M.P.; Gonzales-Cope, M.; Zee, B.M.; Garcia, B.A. Breaking the histone code with quantitative mass spectrometry. Expert Rev. Proteomics 2011, 8, 631–643. [Google Scholar] [CrossRef]
- Boyne, M.T.; Pesavento, J.J.; Mizzen, C.A.; Kelleher, N.L. Precise Characterization of Human Histones in the H2A gene family by top down mass spectrometry. J. Proteome Res. 2006, 5, 248–253. [Google Scholar] [CrossRef]
- Taverna, S.D.; Ueberheide, B.M.; Liu, Y.; Tackett, A.J.; Diaz, R.L.; Shabanowitz, J.; Chait, B.T.; Hunt, D.F.; Allis, C.D. Long-distance combinatorial linkage between methylation and acetylation on histone H3 N termini. Proc. Natl. Acad. Sci. USA 2007, 104, 2086–2091. [Google Scholar] [CrossRef]
- Raiber, E.-A.; Hardisty, R.; van Delft, P.; Balasubramanian, S. Mapping and elucidating the function of modified bases in DNA. Nat. Rev. Chem. 2017, 1, 0069. [Google Scholar] [CrossRef]
- Lyko, F. The DNA methyltransferase family: A versatile toolkit for epigenetic regulation. Nat. Rev. Genet. 2018, 19, 81–92. [Google Scholar] [CrossRef]
- Wu, X.; Zhang, Y. TET-mediated active DNA demethylation: Mechanism, function and beyond. Nat. Rev. Genet. 2017, 18, 517–534. [Google Scholar] [CrossRef] [PubMed]
- Schübeler, D. Function and information content of DNA methylation. Nature 2015, 517, 321–326. [Google Scholar] [CrossRef] [PubMed]
- Bird, A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002, 16, 6–21. [Google Scholar] [CrossRef] [PubMed]
- Wade, P.A. Methyl CpG-binding proteins and transcriptional repression. BioEssays News Rev. Mol. Cell. Dev. Biol. 2001, 23, 1131–1137. [Google Scholar] [CrossRef]
- Jones, P.A. Functions of DNA methylation: Islands, start sites, gene bodies and beyond. Nat. Rev. Genet. 2012, 13, 484–492. [Google Scholar] [CrossRef] [PubMed]
- Seale, K.; Horvath, S.; Teschendorff, A.; Eynon, N.; Voisin, S. Making sense of the ageing methylome. Nat. Rev. Genet. 2022, 23, 585–605. [Google Scholar] [CrossRef] [PubMed]
- Domingo-Relloso, A.; Makhani, K.; Riffo-Campos, A.L.; Tellez-Plaza, M.; Klein, K.O.; Subedi, P.; Zhao, J.; Moon, K.A.; Bozack, A.K.; Haack, K.; et al. Arsenic exposure, blood DNA methylation, and cardiovascular disease. Circ. Res. 2022, 131, e51–e69. [Google Scholar] [CrossRef]
- Ling, C.; Rönn, T. Epigenetics in human obesity and Type 2 Diabetes. Cell Metab. 2019, 29, 1028–1044. [Google Scholar] [CrossRef] [PubMed]
- Jeffries, M.A.; Donica, M.; Baker, L.W.; Stevenson, M.E.; Annan, A.C.; Humphrey, M.B.; James, J.A.; Sawalha, A.H. Genome-wide DNA methylation study identifies significant epigenomic changes in osteoarthritic cartilage. Arthritis Rheumatol. Hoboken NJ 2014, 66, 2804–2815. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Silva, T.C.; Young, J.I.; Gomez, L.; Schmidt, M.A.; Hamilton-Nelson, K.L.; Kunkle, B.W.; Chen, X.; Martin, E.R.; Wang, L. Epigenome-wide meta-analysis of DNA methylation differences in prefrontal cortex implicates the immune processes in Alzheimer’s Disease. Nat. Commun. 2020, 11, 6114. [Google Scholar] [CrossRef]
- Periyasamy, P.; Shinohara, T. Age-related cataracts: Role of unfolded protein response, Ca2+ mobilization, epigenetic DNA modifications, and loss of Nrf2/Keap1 dependent cytoprotection. Prog. Retin. Eye Res. 2017, 60, 1–19. [Google Scholar] [CrossRef]
- Laird, P.W. The power and the promise of DNA methylation markers. Nat. Rev. Cancer 2003, 3, 253–266. [Google Scholar] [CrossRef]
- Kim, H.; Wang, X.; Jin, P. Developing DNA methylation-based diagnostic biomarkers. J. Genet. Genomics Yi Chuan Xue Bao 2018, 45, 87–97. [Google Scholar] [CrossRef]
- Koch, A.; Joosten, S.C.; Feng, Z.; de Ruijter, T.C.; Draht, M.X.; Melotte, V.; Smits, K.M.; Veeck, J.; Herman, J.G.; Van Neste, L.; et al. Analysis of DNA methylation in cancer: Location revisited. Nat. Rev. Clin. Oncol. 2018, 15, 459–466. [Google Scholar] [CrossRef]
- Cokus, S.J.; Feng, S.; Zhang, X.; Chen, Z.; Merriman, B.; Haudenschild, C.D.; Pradhan, S.; Nelson, S.F.; Pellegrini, M.; Jacobsen, S.E. Shotgun bisulfite sequencing of the arabidopsis genome reveals DNA methylation patterning. Nature 2008, 452, 215–219. [Google Scholar] [CrossRef] [PubMed]
- Frommer, M.; McDonald, L.E.; Millar, D.S.; Collis, C.M.; Watt, F.; Grigg, G.W.; Molloy, P.L.; Paul, C.L. A Genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc. Natl. Acad. Sci. USA 1992, 89, 1827–1831. [Google Scholar] [CrossRef] [PubMed]
- Booth, M.J.; Branco, M.R.; Ficz, G.; Oxley, D.; Krueger, F.; Reik, W.; Balasubramanian, S. Quantitative sequencing of 5-Methylcytosine and 5-Hydroxymethylcytosine at single-base resolution. Science 2012, 336, 934–937. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Wu, X.; Zhang, Y. Base-resolution profiling of active DNA demethylation using MAB-Seq and CaMAB-Seq. Nat. Protoc. 2016, 11, 1081–1100. [Google Scholar] [CrossRef] [PubMed]
- Meissner, A.; Mikkelsen, T.S.; Gu, H.; Wernig, M.; Hanna, J.; Sivachenko, A.; Zhang, X.; Bernstein, B.E.; Nusbaum, C.; Jaffe, D.B.; et al. Genome-Scale DNA methylation maps of pluripotent and differentiated cells. Nature 2008, 454, 766–770. [Google Scholar] [CrossRef]
- Meissner, A.; Gnirke, A.; Bell, G.W.; Ramsahoye, B.; Lander, E.S.; Jaenisch, R. Reduced representation bisulfite sequencing for comparative high-resolution DNA methylation analysis. Nucleic Acids Res. 2005, 33, 5868–5877. [Google Scholar] [CrossRef]
- Shareef, S.J.; Bevill, S.M.; Raman, A.T.; Aryee, M.J.; van Galen, P.; Hovestadt, V.; Bernstein, B.E. Extended-representation bisulfite sequencing of gene regulatory elements in multiplexed samples and single cells. Nat. Biotechnol. 2021, 39, 1086–1094. [Google Scholar] [CrossRef]
- Olova, N.; Krueger, F.; Andrews, S.; Oxley, D.; Berrens, R.V.; Branco, M.R.; Reik, W. Comparison of whole-genome bisulfite sequencing library preparation strategies identifies sources of biases affecting DNA methylation data. Genome Biol. 2018, 19, 33. [Google Scholar] [CrossRef]
- Liu, Y.; Siejka-Zielińska, P.; Velikova, G.; Bi, Y.; Yuan, F.; Tomkova, M.; Bai, C.; Chen, L.; Schuster-Böckler, B.; Song, C.-X. Bisulfite-free direct detection of 5-Methylcytosine and 5-Hydroxymethylcytosine at base resolution. Nat. Biotechnol. 2019, 37, 424–429. [Google Scholar] [CrossRef]
- Liu, Y.; Hu, Z.; Cheng, J.; Siejka-Zielińska, P.; Chen, J.; Inoue, M.; Ahmed, A.A.; Song, C.-X. Subtraction-free and bisulfite-free specific sequencing of 5-Methylcytosine and its oxidized derivatives at base resolution. Nat. Commun. 2021, 12, 618. [Google Scholar] [CrossRef]
- Siejka-Zielińska, P.; Cheng, J.; Jackson, F.; Liu, Y.; Soonawalla, Z.; Reddy, S.; Silva, M.; Puta, L.; McCain, M.V.; Culver, E.L.; et al. Cell-free DNA TAPS provides multimodal information for early cancer detection. Sci. Adv. 2021, 7, eabh0534. [Google Scholar] [CrossRef] [PubMed]
- Lo, Y.M.D.; Han, D.S.C.; Jiang, P.; Chiu, R.W.K. Epigenetics, fragmentomics, and topology of cell-free DNA in liquid biopsies. Science 2021, 372, eaaw3616. [Google Scholar] [CrossRef] [PubMed]
- Wan, J.C.M.; Massie, C.; Garcia-Corbacho, J.; Mouliere, F.; Brenton, J.D.; Caldas, C.; Pacey, S.; Baird, R.; Rosenfeld, N. Liquid Biopsies come of age: Towards implementation of circulating tumour DNA. Nat. Rev. Cancer 2017, 17, 223–238. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.; Siejka-Zielińska, P.; Liu, Y.; Chandran, A.; Kriaucionis, S.; Song, C.-X. Endonuclease enrichment TAPS for cost-effective genome-wide base-resolution DNA methylation detection. Nucleic Acids Res. 2021, 49, e76. [Google Scholar] [CrossRef]
- Weber, M.; Davies, J.J.; Wittig, D.; Oakeley, E.J.; Haase, M.; Lam, W.L.; Schübeler, D. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human Cells. Nat. Genet. 2005, 37, 853–862. [Google Scholar] [CrossRef] [PubMed]
- Shen, S.Y.; Burgener, J.M.; Bratman, S.V.; De Carvalho, D.D. Preparation of CfMeDIP-Seq libraries for methylome profiling of plasma cell-free DNA. Nat. Protoc. 2019, 14, 2749–2780. [Google Scholar] [CrossRef]
- Shen, S.Y.; Singhania, R.; Fehringer, G.; Chakravarthy, A.; Roehrl, M.H.A.; Chadwick, D.; Zuzarte, P.C.; Borgida, A.; Wang, T.T.; Li, T.; et al. Sensitive tumour detection and classification using plasma cell-free DNA methylomes. Nature 2018, 563, 579–583. [Google Scholar] [CrossRef]
- Taiwo, O.; Wilson, G.A.; Morris, T.; Seisenberger, S.; Reik, W.; Pearce, D.; Beck, S.; Butcher, L.M. Methylome analysis using MeDIP-Seq with low DNA concentrations. Nat. Protoc. 2012, 7, 617–636. [Google Scholar] [CrossRef]
- Harris, R.A.; Wang, T.; Coarfa, C.; Nagarajan, R.P.; Hong, C.; Downey, S.L.; Johnson, B.E.; Fouse, S.D.; Delaney, A.; Zhao, Y.; et al. Comparison of sequencing-based methods to profile DNA methylation and identification of monoallelic epigenetic modifications. Nat. Biotechnol. 2010, 28, 1097–1105. [Google Scholar] [CrossRef]
- Jeltsch, A.; Broche, J.; Lungu, C.; Bashtrykov, P. Biotechnological applications of MBD domain proteins for DNA methylation analysis. J. Mol. Biol. 2019, 432, 1816–1823. [Google Scholar] [CrossRef]
- Serre, D.; Lee, B.H.; Ting, A.H. MBD-isolated genome sequencing provides a high-throughput and comprehensive survey of DNA methylation in the human genome. Nucleic Acids Res. 2010, 38, 391–399. [Google Scholar] [CrossRef] [PubMed]
- Brinkman, A.B.; Simmer, F.; Ma, K.; Kaan, A.; Zhu, J.; Stunnenberg, H.G. Whole-genome DNA methylation profiling using MethylCap-Seq. Method. San Diego Calif 2010, 52, 232–236. [Google Scholar] [CrossRef]
- Gebhard, C.; Schwarzfischer, L.; Pham, T.-H.; Schilling, E.; Klug, M.; Andreesen, R.; Rehli, M. Genome-wide profiling of CpG methylation identifies novel targets of aberrant hypermethylation in myeloid leukemia. Cancer Res. 2006, 66, 6118–6128. [Google Scholar] [CrossRef] [PubMed]
- Rauch, T.; Li, H.; Wu, X.; Pfeifer, G.P. MIRA-assisted microarray analysis, a new technology for the determination of DNA methylation patterns, identifies frequent methylation of homeodomain-containing genes in lung cancer cells. Cancer Res. 2006, 66, 7939–7947. [Google Scholar] [CrossRef] [PubMed]
- Jung, M.; Kadam, S.; Xiong, W.; Rauch, T.A.; Jin, S.-G.; Pfeifer, G.P. MIRA-Seq for DNA methylation analysis of CpG islands. Epigenomics 2015, 7, 695–706. [Google Scholar] [CrossRef]
- Simpson, J.T.; Workman, R.E.; Zuzarte, P.C.; David, M.; Dursi, L.J.; Timp, W. Detecting DNA cytosine methylation using nanopore sequencing. Nat. Method. 2017, 14, 407–410. [Google Scholar] [CrossRef]
- Laszlo, A.H.; Derrington, I.M.; Brinkerhoff, H.; Langford, K.W.; Nova, I.C.; Samson, J.M.; Bartlett, J.J.; Pavlenok, M.; Gundlach, J.H. Detection and mapping of 5-Methylcytosine and 5-Hydroxymethylcytosine with nanopore MspA. Proc. Natl. Acad. Sci. USA 2013, 110, 18904–18909. [Google Scholar] [CrossRef]
- Schreiber, J.; Wescoe, Z.L.; Abu-Shumays, R.; Vivian, J.T.; Baatar, B.; Karplus, K.; Akeson, M. Error rates for nanopore discrimination among cytosine, methylcytosine, and hydroxymethylcytosine along individual DNA strands. Proc. Natl. Acad. Sci. USA 2013, 110, 18910–18915. [Google Scholar] [CrossRef]
- Rand, A.C.; Jain, M.; Eizenga, J.M.; Musselman-Brown, A.; Olsen, H.E.; Akeson, M.; Paten, B. Mapping DNA methylation with high-throughput nanopore sequencing. Nat. Method. 2017, 14, 411–413. [Google Scholar] [CrossRef]
- Wescoe, Z.L.; Schreiber, J.; Akeson, M. Nanopores discriminate among Five C5-Cytosine variants in DNA. J. Am. Chem. Soc. 2014, 136, 16582–16587. [Google Scholar] [CrossRef]
- Branton, D.; Deamer, D.W.; Marziali, A.; Bayley, H.; Benner, S.A.; Butler, T.; Di Ventra, M.; Garaj, S.; Hibbs, A.; Huang, X.; et al. The potential and challenges of nanopore sequencing. Nat. Biotechnol. 2008, 26, 1146–1153. [Google Scholar] [CrossRef] [PubMed]
- Eid, J.; Fehr, A.; Gray, J.; Luong, K.; Lyle, J.; Otto, G.; Peluso, P.; Rank, D.; Baybayan, P.; Bettman, B.; et al. Real-time DNA sequencing from single polymerase molecules. Science 2009, 323, 133–138. [Google Scholar] [CrossRef] [PubMed]
- Tse, O.Y.O.; Jiang, P.; Cheng, S.H.; Peng, W.; Shang, H.; Wong, J.; Chan, S.L.; Poon, L.C.Y.; Leung, T.Y.; Chan, K.C.A.; et al. Genome-wide detection of cytosine methylation by single molecule real-time sequencing. Proc. Natl. Acad. Sci. USA 2021, 118, e2019768118. [Google Scholar] [CrossRef] [PubMed]
- Song, C.-X.; Clark, T.A.; Lu, X.-Y.; Kislyuk, A.; Dai, Q.; Turner, S.W.; He, C.; Korlach, J. Sensitive and specific single-molecule sequencing of 5-Hydroxymethylcytosine. Nat. Method. 2011, 9, 75–77. [Google Scholar] [CrossRef]
- Ye, P.; Luan, Y.; Chen, K.; Liu, Y.; Xiao, C.; Xie, Z. MethSMRT: An integrative database for DNA N6-Methyladenine and N4-Methylcytosine generated by single-molecular real-time sequencing. Nucleic Acids Res. 2017, 45, D85–D89. [Google Scholar] [CrossRef] [PubMed]
- Wenger, A.M.; Peluso, P.; Rowell, W.J.; Chang, P.-C.; Hall, R.J.; Concepcion, G.T.; Ebler, J.; Fungtammasan, A.; Kolesnikov, A.; Olson, N.D.; et al. Accurate circular consensus long-read sequencing improves variant detection and assembly of a human genome. Nat. Biotechnol. 2019, 37, 1155–1162. [Google Scholar] [CrossRef]
- Choy, L.Y.L.; Peng, W.; Jiang, P.; Cheng, S.H.; Yu, S.C.Y.; Shang, H.; Olivia Tse, O.Y.; Wong, J.; Wong, V.W.S.; Wong, G.L.H.; et al. Single-molecule sequencing enables long cell-free DNA detection and direct methylation analysis for cancer patients. Clin. Chem. 2022, 68, 1151–1163. [Google Scholar] [CrossRef]
- Boccaletto, P.; Machnicka, M.A.; Purta, E.; Piatkowski, P.; Baginski, B.; Wirecki, T.K.; de Crécy-Lagard, V.; Ross, R.; Limbach, P.A.; Kotter, A.; et al. MODOMICS: A database of RNA modification pathways. 2017 Update. Nucleic Acids Res. 2018, 46, D303–D307. [Google Scholar] [CrossRef]
- Liu, J.; Yue, Y.; Han, D.; Wang, X.; Fu, Y.; Zhang, L.; Jia, G.; Yu, M.; Lu, Z.; Deng, X.; et al. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat. Chem. Biol. 2014, 10, 93–95. [Google Scholar] [CrossRef]
- Bawankar, P.; Lence, T.; Paolantoni, C.; Haussmann, I.U.; Kazlauskiene, M.; Jacob, D.; Heidelberger, J.B.; Richter, F.M.; Nallasivan, M.P.; Morin, V.; et al. Hakai is required for stabilization of core components of the M6A MRNA methylation machinery. Nat. Commun. 2021, 12, 3778. [Google Scholar] [CrossRef]
- Yue, Y.; Liu, J.; Cui, X.; Cao, J.; Luo, G.; Zhang, Z.; Cheng, T.; Gao, M.; Shu, X.; Ma, H.; et al. VIRMA mediates preferential M6A MRNA methylation in 3′UTR and near stop codon and associates with alternative polyadenylation. Cell Discov. 2018, 4, 10. [Google Scholar] [CrossRef] [PubMed]
- Wen, J.; Lv, R.; Ma, H.; Shen, H.; He, C.; Wang, J.; Jiao, F.; Liu, H.; Yang, P.; Tan, L.; et al. Zc3h13 regulates nuclear RNA M6A methylation and mouse embryonic stem cell self-renewal. Mol. Cell 2018, 69, 1028–1038.e6. [Google Scholar] [CrossRef] [PubMed]
- Pendleton, K.E.; Chen, B.; Liu, K.; Hunter, O.V.; Xie, Y.; Tu, B.P.; Conrad, N.K. The U6 SnRNA M6A methyltransferase METTL16 regulates SAM synthetase intron retention. Cell 2017, 169, 824–835.e14. [Google Scholar] [CrossRef] [PubMed]
- Jia, G.; Fu, Y.; Zhao, X.; Dai, Q.; Zheng, G.; Yang, Y.; Yi, C.; Lindahl, T.; Pan, T.; Yang, Y.-G.; et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 2011, 7, 885–887. [Google Scholar] [CrossRef]
- Zheng, G.; Dahl, J.A.; Niu, Y.; Fedorcsak, P.; Huang, C.-M.; Li, C.J.; Vågbø, C.B.; Shi, Y.; Wang, W.-L.; Song, S.-H.; et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol. Cell 2013, 49, 18–29. [Google Scholar] [CrossRef]
- Ueda, Y.; Ooshio, I.; Fusamae, Y.; Kitae, K.; Kawaguchi, M.; Jingushi, K.; Hase, H.; Harada, K.; Hirata, K.; Tsujikawa, K. AlkB Homolog 3-Mediated TRNA demethylation promotes protein synthesis in cancer cells. Sci. Rep. 2017, 7, 42271. [Google Scholar] [CrossRef]
- Roignant, J.-Y.; Soller, M. M6A in MRNA: An ancient mechanism for fine-tuning gene expression. Trends Genet. TIG 2017, 33, 380–390. [Google Scholar] [CrossRef]
- Haussmann, I.U.; Bodi, Z.; Sanchez-Moran, E.; Mongan, N.P.; Archer, N.; Fray, R.G.; Soller, M. M6A potentiates Sxl Alternative pre-MRNA splicing for robust drosophila sex determination. Nature 2016, 540, 301–304. [Google Scholar] [CrossRef]
- Li, Q.; Li, X.; Tang, H.; Jiang, B.; Dou, Y.; Gorospe, M.; Wang, W. NSUN2-mediated M5C methylation and METTL3/METTL14-mediated M6A methylation cooperatively enhance P21 translation. J. Cell. Biochem. 2017, 118, 2587–2598. [Google Scholar] [CrossRef] [PubMed]
- Qi, S.-T.; Ma, J.-Y.; Wang, Z.-B.; Guo, L.; Hou, Y.; Sun, Q.-Y. N6-methyladenosine sequencing highlights the involvement of MRNA methylation in oocyte meiotic maturation and embryo development by regulating translation in Xenopus laevis. J. Biol. Chem. 2016, 291, 23020–23026. [Google Scholar] [CrossRef]
- Batista, P.J.; Molinie, B.; Wang, J.; Qu, K.; Zhang, J.; Li, L.; Bouley, D.M.; Lujan, E.; Haddad, B.; Daneshvar, K.; et al. M(6)A RNA modification controls cell fate transition in mammalian embryonic stem cells. Cell Stem Cell 2014, 15, 707–719. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.; Dai, Q.; Zheng, G.; He, C.; Parisien, M.; Pan, T. N(6)-Methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature 2015, 518, 560–564. [Google Scholar] [CrossRef] [PubMed]
- Geula, S.; Moshitch-Moshkovitz, S.; Dominissini, D.; Mansour, A.A.; Kol, N.; Salmon-Divon, M.; Hershkovitz, V.; Peer, E.; Mor, N.; Manor, Y.S.; et al. Stem cells. M6A MRNA methylation facilitates resolution of naïve pluripotency toward differentiation. Science 2015, 347, 1002–1006. [Google Scholar] [CrossRef] [PubMed]
- Zhong, S.; Li, H.; Bodi, Z.; Button, J.; Vespa, L.; Herzog, M.; Fray, R.G. MTA is an arabidopsis messenger RNA adenosine methylase and interacts with a homolog of a sex-specific splicing factor. Plant Cell 2008, 20, 1278–1288. [Google Scholar] [CrossRef] [PubMed]
- Hongay, C.F.; Orr-Weaver, T.L. Drosophila inducer of MEiosis 4 (IME4) is required for notch signaling during oogenesis. Proc. Natl. Acad. Sci. USA 2011, 108, 14855–14860. [Google Scholar] [CrossRef]
- Frye, M.; Harada, B.T.; Behm, M.; He, C. RNA modifications modulate gene expression during development. Science 2018, 361, 1346–1349. [Google Scholar] [CrossRef]
- Wang, T.; Kong, S.; Tao, M.; Ju, S. The potential role of RNA N6-methyladenosine in cancer progression. Mol. Cancer 2020, 19, 88. [Google Scholar] [CrossRef]
- Barbieri, I.; Kouzarides, T. Role of RNA modifications in cancer. Nat. Rev. Cancer 2020, 20, 303–322. [Google Scholar] [CrossRef]
- Li, T.; Hu, P.-S.; Zuo, Z.; Lin, J.-F.; Li, X.; Wu, Q.-N.; Chen, Z.-H.; Zeng, Z.-L.; Wang, F.; Zheng, J.; et al. METTL3 facilitates tumor progression via an M6A-IGF2BP2-dependent mechanism in colorectal carcinoma. Mol. Cancer 2019, 18, 112. [Google Scholar] [CrossRef]
- Shen, C.; Sheng, Y.; Zhu, A.C.; Robinson, S.; Jiang, X.; Dong, L.; Chen, H.; Su, R.; Yin, Z.; Li, W.; et al. RNA demethylase ALKBH5 selectively promotes tumorigenesis and cancer stem cell self-renewal in acute myeloid leukemia. Cell Stem Cell 2020, 27, 64–80.e9. [Google Scholar] [CrossRef]
- Bodi, Z.; Fray, R.G. Detection and quantification of N 6-methyladenosine in messenger RNA by TLC. Method. Mol. Biol. Clifton NJ 2017, 1562, 79–87. [Google Scholar] [CrossRef]
- Huang, T.; Guo, J.; Lv, Y.; Zheng, Y.; Feng, T.; Gao, Q.; Zeng, W. Meclofenamic acid represses spermatogonial proliferation through modulating M6A RNA modification. J. Anim. Sci. Biotechnol. 2019, 10, 63. [Google Scholar] [CrossRef] [PubMed]
- Wetzel, C.; Limbach, P.A. Mass spectrometry of modified RNAs: Recent developments. The Analyst 2016, 141, 16–23. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.-Y.; Song, J.; Liu, Y.; Song, C.-X.; Yi, C. Mapping the epigenetic modifications of DNA and RNA. Protein Cell 2020, 11, 792–808. [Google Scholar] [CrossRef] [PubMed]
- Meyer, K.D.; Saletore, Y.; Zumbo, P.; Elemento, O.; Mason, C.E.; Jaffrey, S.R. Comprehensive Analysis of MRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell 2012, 149, 1635–1646. [Google Scholar] [CrossRef]
- Chen, K.; Lu, Z.; Wang, X.; Fu, Y.; Luo, G.-Z.; Liu, N.; Han, D.; Dominissini, D.; Dai, Q.; Pan, T.; et al. High-resolution N(6)-Methyladenosine (m(6) A) map using photo-crosslinking-assisted m(6) a sequencing. Angew. Chem. Int. Ed. Engl. 2015, 54, 1587–1590. [Google Scholar] [CrossRef]
- Grozhik, A.V.; Linder, B.; Olarerin-George, A.O.; Jaffrey, S.R. Mapping M6A at individual-nucleotide resolution using crosslinking and immunoprecipitation (MiCLIP). Method. Mol. Biol. Clifton NJ 2017, 1562, 55–78. [Google Scholar] [CrossRef]
- Linder, B.; Grozhik, A.V.; Olarerin-George, A.O.; Meydan, C.; Mason, C.E.; Jaffrey, S.R. Single-nucleotide-resolution mapping of M6A and M6Am throughout the transcriptome. Nat. Method. 2015, 12, 767–772. [Google Scholar] [CrossRef]
- Hu, L.; Liu, S.; Peng, Y.; Ge, R.; Su, R.; Senevirathne, C.; Harada, B.T.; Dai, Q.; Wei, J.; Zhang, L.; et al. M6A RNA modifications are measured at single-base resolution across the mammalian transcriptome. Nat. Biotechnol. 2022, 40, 1210–1219. [Google Scholar] [CrossRef]





Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, M.; Li, Q.; Liu, L. Factors and Methods for the Detection of Gene Expression Regulation. Biomolecules 2023, 13, 304. https://doi.org/10.3390/biom13020304
Wang M, Li Q, Liu L. Factors and Methods for the Detection of Gene Expression Regulation. Biomolecules. 2023; 13(2):304. https://doi.org/10.3390/biom13020304
Chicago/Turabian StyleWang, Mengyuan, Qian Li, and Lingbo Liu. 2023. "Factors and Methods for the Detection of Gene Expression Regulation" Biomolecules 13, no. 2: 304. https://doi.org/10.3390/biom13020304
APA StyleWang, M., Li, Q., & Liu, L. (2023). Factors and Methods for the Detection of Gene Expression Regulation. Biomolecules, 13(2), 304. https://doi.org/10.3390/biom13020304




