Copper Binding and Redox Activity of α-Synuclein in Membrane-Like Environment
Abstract
1. Introduction
2. Materials and Methods
2.1. Protein and Reagents
2.2. Unilamellar Vesicle Preparation
2.3. NMR, EPR, XAS and CD Spectroscopic Measurements
2.4. XAS Data Analysis
2.5. Catalytic Oxidation of 4-Methyl Catechol and Dopamine by Cu2+ and Cu+ Ions Bound to the Full-Length αSyn
2.6. HPLC-ESI/MS Analysis of Protein Oxidative Modification
3. Results and Discussion
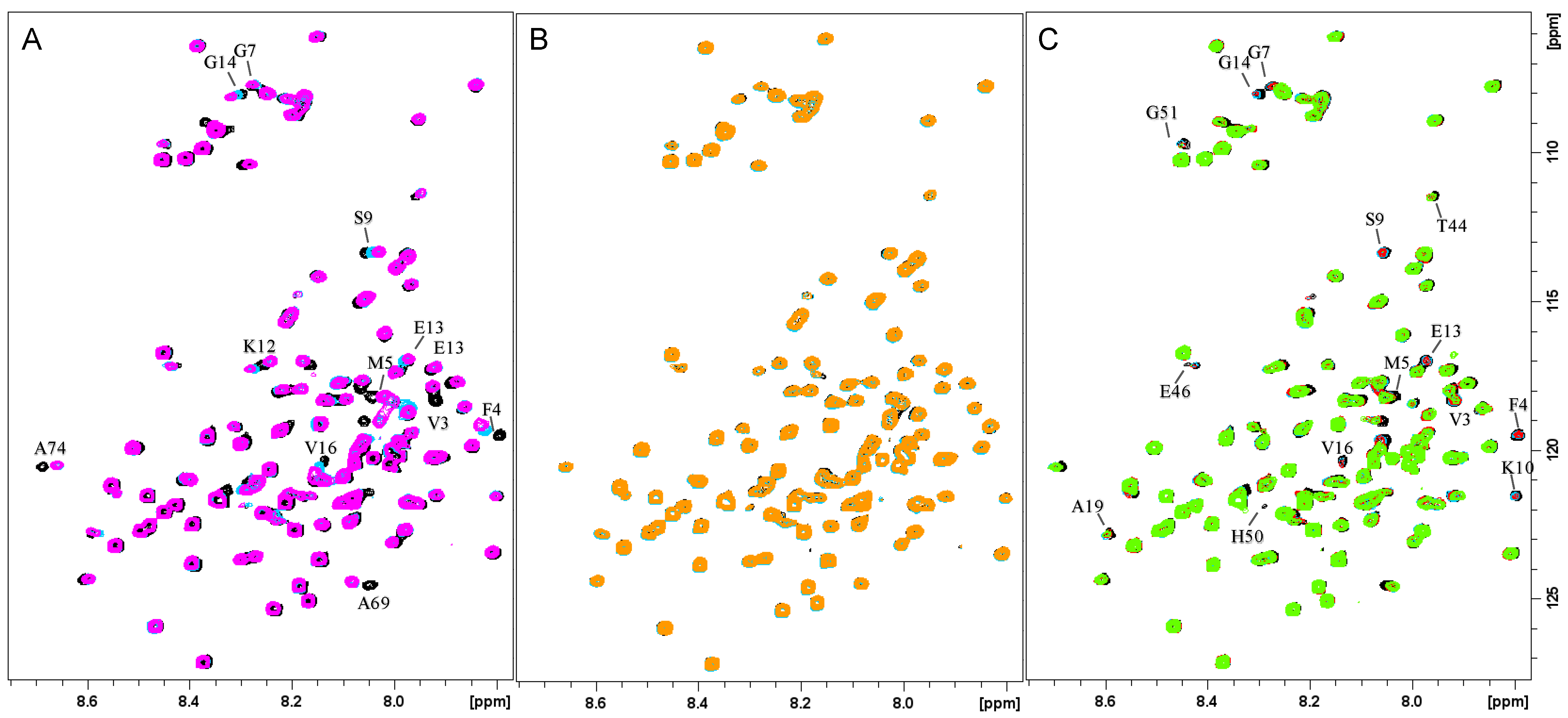
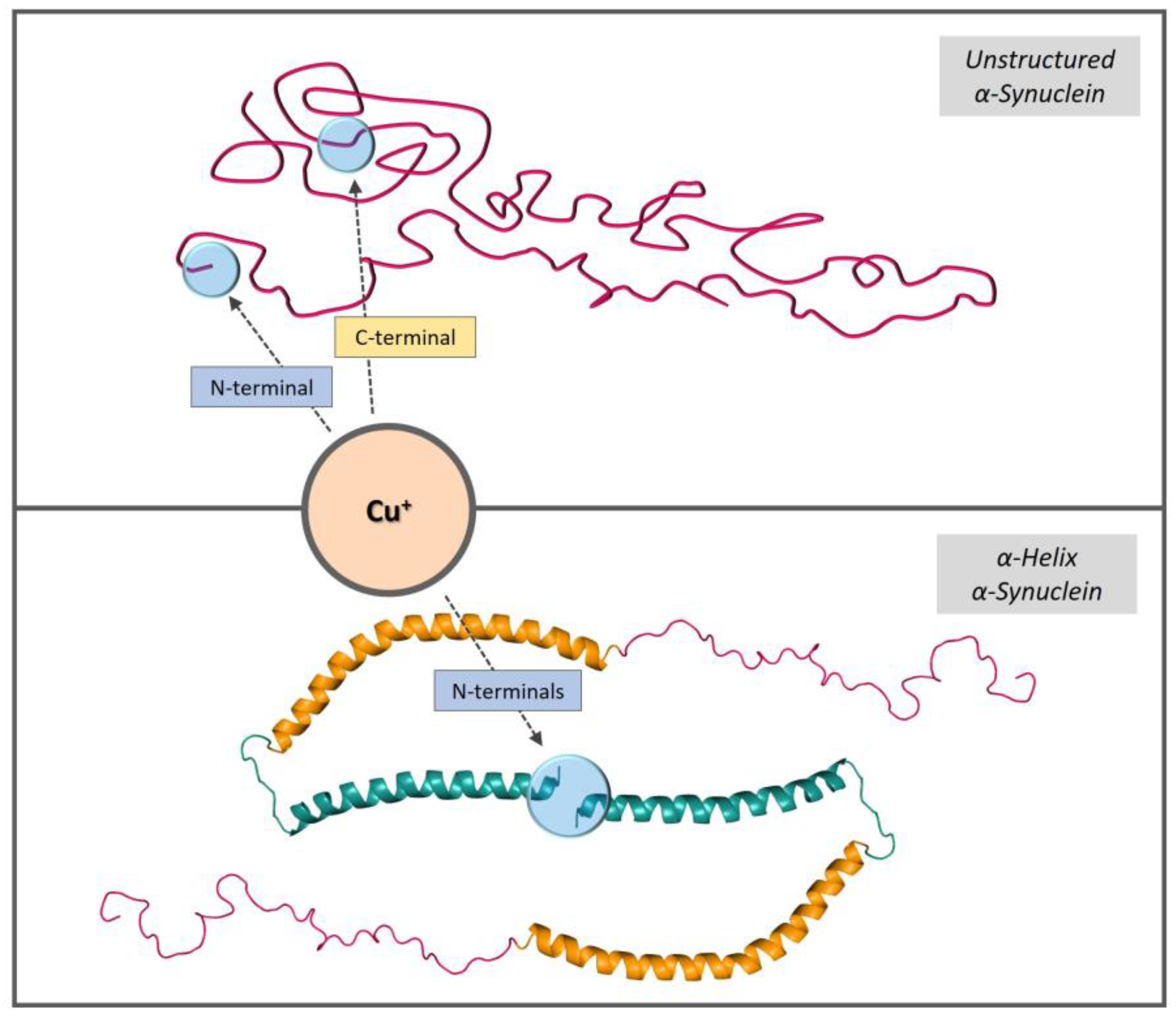
3.1. Characterization of Cu+/Ag+ Binding to Membrane-Bound αSyn
3.2. Characterization of Cu2+ Binding to Membrane-Bound αSyn
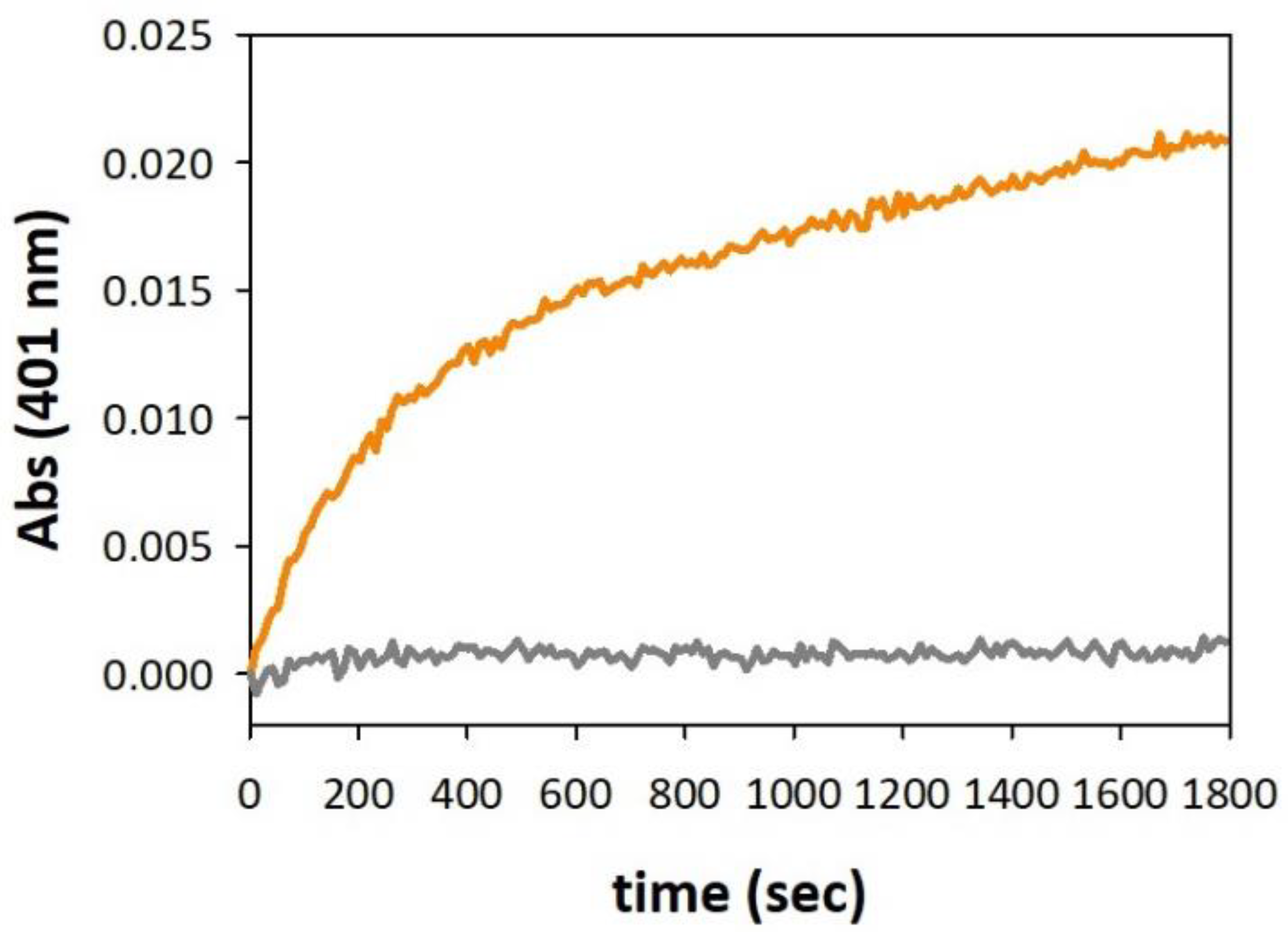
3.3. Oxidative Reactivity of Copper–Full-Length αSyn Complex in Membrane Environment
3.4. HPLC-ESI/MS Analysis of Protein Oxidative Modification
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wakabayashi, K.; Tanji, K.; Mori, F.; Takahashi, H. The Lewy body in Parkinson’s disease: Molecules implicated in the formation and degradation of alpha-synuclein aggregates. Neuropathology 2007, 27, 494–506. [Google Scholar] [CrossRef]
- Runwal, G.; Edwards, R.H. The Membrane Interactions of Synuclein: Physiology and Pathology. Annu. Rev. Pathol. 2021, 16, 465–485. [Google Scholar] [CrossRef]
- Burré, J.; Sharma, M.; Tsetsenis, T.; Buchman, V.; Etherton, M.R.; Südhof, T.C. Alpha-synuclein promotes SNARE-complex assembly in vivo and in vitro. Science 2010, 329, 1663–1667. [Google Scholar] [CrossRef]
- Bartels, T.; Ahlstrom, L.S.; Leftin, A.; Kamp, F.; Haass, C.; Brown, M.F.; Beyer, K. The N-terminus of the intrinsically disordered protein α-synuclein triggers membrane binding and helix folding. Biophys. J. 2010, 99, 2116–2124. [Google Scholar] [CrossRef]
- Hashimoto, M.; Takenouchi, T.; Mallory, M.; Masliah, E.; Takeda, A. The role of NAC in amyloidogenesis in Alzheimer’s disease. Am. J. Pathol. 2000, 156, 734–736. [Google Scholar] [CrossRef]
- Davidson, W.S.; Jonas, A.; Clayton, D.F.; George, J.M. Stabilization of alpha-synuclein secondary structure upon binding to synthetic membranes. J. Biol. Chem. 1998, 273, 9443–9449. [Google Scholar] [CrossRef]
- Rasia, R.M.; Bertoncini, C.W.; Marsh, D.; Hoyer, W.; Cherny, D.; Zweckstetter, M.; Griesinger, C.; Jovin, T.M.; Fernández, C.O. Structural characterization of copper(II) binding to alpha-synuclein: Insights into the bioinorganic chemistry of Parkinson’s disease. Proc. Natl. Acad. Sci. USA 2005, 102, 4294–4299. [Google Scholar] [CrossRef]
- Myhre, O.; Utkilen, H.; Duale, N.; Brunborg, G.; Hofer, T. Metal dyshomeostasis and inflammation in Alzheimer’s and Parkinson’s diseases: Possible impact of environmental exposures. Oxid. Med. Cell. Longev. 2013, 2013, 726954. [Google Scholar] [CrossRef] [PubMed]
- Bisaglia, M.; Bubacco, L. Copper Ions and Parkinson’s Disease: Why Is Homeostasis So Relevant? Biomolecules 2020, 10, 195. [Google Scholar] [CrossRef]
- Valensin, D.; Dell’Acqua, S.; Kozlowski, H.; Casella, L. Coordination and redox properties of copper interaction with α-synuclein. J. Inorg. Biochem. 2016, 163, 292–300. [Google Scholar] [CrossRef]
- Abeyawardhane, D.L.; Heitger, D.R.; Fernández, R.D.; Forney, A.K.; Lucas, H.R. C-Terminal Cu(II) Coordination to α-Synuclein Enhances Aggregation. ACS Chem. Neurosci. 2019, 10, 1402–1410. [Google Scholar] [CrossRef] [PubMed]
- Anderson, J.P.; Walker, D.E.; Goldstein, J.M.; de Laat, R.; Banducci, K.; Caccavello, R.J.; Barbour, R.; Huang, J.; Kling, K.; Lee, M.; et al. Phosphorylation of Ser-129 is the dominant pathological modification of alpha-synuclein in familial and sporadic Lewy body disease. J. Biol. Chem. 2006, 281, 29739–29752. [Google Scholar] [CrossRef] [PubMed]
- Moriarty, G.M.; Minetti, C.A.S.A.; Remeta, D.P.; Baum, J. A Revised Picture of the Cu(II)−α-Synuclein Complex: The Role of N-Terminal Acetylation. Biochemistry 2014, 53, 2815–2817. [Google Scholar] [CrossRef]
- Dudzik, C.G.; Walter, E.D.; Millhauser, G.L. Coordination Features and Affinity of the Cu2+ Site in the α-Synuclein Protein of Parkinson’s Disease. Biochemistry 2011, 50, 1771–1777. [Google Scholar] [CrossRef] [PubMed]
- Camponeschi, F.; Valensin, D.; Tessari, I.; Bubacco, L.; Dell’Acqua, S.; Casella, L.; Monzani, E.; Gaggelli, E.; Valensin, G. Copper(I)-α-synuclein interaction: Structural description of two independent and competing metal binding sites. Inorg. Chem. 2013, 52, 1358–1367. [Google Scholar] [CrossRef]
- Miotto, M.C.; Valiente-Gabioud, A.A.; Rossetti, G.; Zweckstetter, M.; Carloni, P.; Selenko, P.; Griesinger, C.; Binolfi, A.; Fernandez, C.O. Copper binding to the N-terminally acetylated, naturally occurring form of alpha-synuclein induces local helical folding. J. Am. Chem. Soc. 2015, 137, 6444–6447. [Google Scholar] [CrossRef]
- Binolfi, A.; Valiente-Gabioud, A.A.; Duran, R.; Zweckstetter, M.; Griesinger, C.; Fernandez, C.O. Exploring the structural details of Cu(I) binding to α-synuclein by NMR spectroscopy. J. Am. Chem. Soc. 2011, 133, 194–196. [Google Scholar] [CrossRef]
- Gentile, I.; Garro, H.A.; Delgado Ocaña, S.; Gonzalez, N.; Strohäker, T.; Schibich, D.; Quintanar, L.; Sambrotta, L.; Zweckstetter, M.; Griesinger, C.; et al. Interaction of Cu(i) with the Met-X(3)-Met motif of alpha-synuclein: Binding ligands, affinity and structural features. Metallomics 2018, 10, 1383–1389. [Google Scholar] [CrossRef]
- De Ricco, R.; Valensin, D.; Dell’Acqua, S.; Casella, L.; Gaggelli, E.; Valensin, G.; Bubacco, L.; Mangani, S. Differences in the binding of copper(I) to α- and β-synuclein. Inorg. Chem. 2015, 54, 265–272. [Google Scholar] [CrossRef]
- Monzani, E.; Nicolis, S.; Dell’Acqua, S.; Capucciati, A.; Bacchella, C.; Zucca, F.A.; Mosharov, E.V.; Sulzer, D.; Zecca, L.; Casella, L. Dopamine, Oxidative Stress and Protein-Quinone Modifications in Parkinson’s and Other Neurodegenerative Diseases. Angew. Chem. Int. Ed. Engl. 2019, 58, 6512–6527. [Google Scholar] [CrossRef]
- Pirota, V.; Dell’Acqua, S.; Monzani, E.; Nicolis, S.; Casella, L. Copper-Aβ Peptides and Oxidation of Catecholic Substrates: Reactivity and Endogenous Peptide Damage. Chem.–A Eur. J. 2016, 22, 16964–16973. [Google Scholar] [CrossRef] [PubMed]
- Bacchella, C.; Gentili, S.; Bellotti, D.; Quartieri, E.; Draghi, S.; Baratto, M.C.; Remelli, M.; Valensin, D.; Monzani, E.; Nicolis, S.; et al. Binding and Reactivity of Copper to R1 and R3 Fragments of tau Protein. Inorg. Chem. 2020, 59, 274–286. [Google Scholar] [CrossRef] [PubMed]
- Dell’Acqua, S.; Bacchella, C.; Monzani, E.; Nicolis, S.; Di Natale, G.; Rizzarelli, E.; Casella, L. Prion Peptides Are Extremely Sensitive to Copper Induced Oxidative Stress. Inorg. Chem. 2017, 56, 11317–11325. [Google Scholar] [CrossRef]
- Bacchella, C.; Dell’Acqua, S.; Nicolis, S.; Monzani, E.; Casella, L. The reactivity of copper complexes with neuronal peptides promoted by catecholamines and its impact on neurodegeneration. Coord. Chem. Rev. 2022, 471, 214756. [Google Scholar] [CrossRef]
- Dell’Acqua, S.; Pirota, V.; Anzani, C.; Rocco, M.M.; Nicolis, S.; Valensin, D.; Monzani, E.; Casella, L. Reactivity of copper-[small alpha]-synuclein peptide complexes relevant to Parkinson’s disease. Metallomics 2015, 7, 1091–1102. [Google Scholar] [CrossRef]
- Dell’Acqua, S.; Pirota, V.; Monzani, E.; Camponeschi, F.; De Ricco, R.; Valensin, D.; Casella, L. Copper(I) Forms a Redox-Stable 1:2 Complex with α-Synuclein N-Terminal Peptide in a Membrane-Like Environment. Inorg. Chem. 2016, 55, 6100–6106. [Google Scholar] [CrossRef]
- Wang, H.; Mörman, C.; Sternke-Hoffmann, R.; Huang, C.Y.; Prota, A.; Ma, P.; Luo, J. Cu(2+) ions modulate the interaction between α-synuclein and lipid membranes. J. Inorg. Biochem. 2022, 236, 111945. [Google Scholar] [CrossRef]
- Huang, C.; Ren, G.; Zhou, H.; Wang, C.C. A new method for purification of recombinant human alpha-synuclein in Escherichia coli. Protein Expr. Purif. 2005, 42, 173–177. [Google Scholar] [CrossRef]
- Dudzik, C.G.; Walter, E.D.; Abrams, B.S.; Jurica, M.S.; Millhauser, G.L. Coordination of copper to the membrane-bound form of α-synuclein. Biochemistry 2013, 52, 53–60. [Google Scholar] [CrossRef]
- Weinreb, P.H.; Zhen, W.; Poon, A.W.; Conway, K.A.; Lansbury, P.T., Jr. NACP, a protein implicated in Alzheimer’s disease and learning, is natively unfolded. Biochemistry 1996, 35, 13709–13715. [Google Scholar] [CrossRef]
- Savitzky, A.; Golay, M.J.E. Smoothing and Differentiation of Data by Simplified Least Squares Procedures. Anal. Chem. 1964, 36, 1627–1639. [Google Scholar] [CrossRef]
- Keller, R.; Wüthrich, K. A New Software for the Analysis of Protein NMR Spectra. 2002. Available online: http://cara.nmr-software.org/downloads/PosterDavos2002.pdf (accessed on 28 January 2023).
- Hwang, T.L.; Shaka, A.J. Water Suppression That Works. Excitation Sculpting Using Arbitrary Wave-Forms and Pulsed-Field Gradients. J. Magn. Reson.-Ser. A 1995, 112, 275–279. [Google Scholar] [CrossRef]
- Rakhit, G.; Antholine, W.E.; Froncisz, W.; Hyde, J.S.; Pilbrow, J.R.; Sinclair, G.R.; Sarkar, B. Direct Evidence of Nitrogen Coupling in the Copper(II) Complex of Bovine Serum Albumin by S-Band Electron Spin Resonance Technique. J. Inorg. Biochem. 1985, 25, 217–224. [Google Scholar] [CrossRef] [PubMed]
- Ravel, B.; Newville, M. Athena, Artemis, Hephaestus: Data analysis for X-ray absorption spectroscopy using IFEFFIT. J. Synchrotron Radiat. 2005, 12, 537–541. [Google Scholar] [CrossRef] [PubMed]
- Puchkova, L.V.; Broggini, M.; Polishchuk, E.V.; Ilyechova, E.Y.; Polishchuk, R.S. Silver Ions as a Tool for Understanding Different Aspects of Copper Metabolism. Nutrients 2019, 11, 1364. [Google Scholar] [CrossRef]
- Veronesi, G.; Gallon, T.; Deniaud, A.; Boff, B.; Gateau, C.; Lebrun, C.; Vidaud, C.; Rollin-Genetet, F.; Carrière, M.; Kieffer, I.; et al. XAS Investigation of Silver(I) Coordination in Copper(I) Biological Binding Sites. Inorg. Chem. 2015, 54, 11688–11696. [Google Scholar] [CrossRef]
- Blackburn, N.J.; Strange, R.W.; Reedijk, J.; Volbeda, A.; Farooq, A.; Zubieta, J.; Karlin, K.D. X-ray Absorption Edge Spectroscopy of Copper(I) Complexes. Coordination Geometry of Copper(I) in the Reduced Forms of Copper Proteins and Their Derivatives with Carbon Monoxide. Inorg. Chem. 1989, 28, 1349–1357. [Google Scholar] [CrossRef]
- Zhang, R.; McEwen, J.S. Local Environment Sensitivity of the Cu K-Edge XANES Features in Cu-SSZ-13: Analysis from First-Principles. J. Phys. Chem. Lett. 2018, 9, 3035–3042. [Google Scholar] [CrossRef]
- Barrett, M.L.; Harvey, I.; Sundararajan, M.; Surendran, R.; Hall, J.F.; Ellis, M.J.; Hough, M.A.; Strange, R.W.; Hillier, I.H.; Hasnain, S.S. Atomic resolution crystal structures, EXAFS, and quantum chemical studies of rusticyanin and its two mutants provide insight into its unusual properties. Biochemistry 2006, 45, 2927–2939. [Google Scholar] [CrossRef]
- Singh, S.K.; Roberts, S.A.; McDevitt, S.F.; Weichsel, A.; Wildner, G.F.; Grass, G.B.; Rensing, C.; Montfort, W.R. Crystal structures of multicopper oxidase CueO bound to copper(I) and silver(I): Functional role of a methionine-rich sequence. J. Biol. Chem. 2011, 286, 37849–37857. [Google Scholar] [CrossRef]
- Pickering, I.J.; George, G.N.; Dameron, C.T.; Kurz, B.; Winge, D.R.; Dance, I.G. X-ray Absorption Spectroscopy of Cuprous-Thiolate Clusters in Proteins and Model Systems. J. Am. Chem. Soc. 1993, 115, 9498–9505. [Google Scholar] [CrossRef]
- Banci, L.; Bertini, I.; Del Conte, R.; Mangani, S.; Meyer-Klaucke, W. X-ray absorption and NMR spectroscopic studies of CopZ, a copper chaperone in Bacillus subtilis: The coordination properties of the copper ion. Biochemistry 2003, 42, 2467–2474. [Google Scholar] [CrossRef]
- Sarret, G.; Favier, A.; Covès, J.; Hazemann, J.L.; Mergeay, M.; Bersch, B. CopK from Cupriavidus metallidurans CH34 binds Cu(I) in a tetrathioether site: Characterization by X-ray absorption and NMR spectroscopy. J. Am. Chem. Soc. 2010, 132, 3770–3777. [Google Scholar] [CrossRef]
- Perlmutter, J.D.; Braun, A.R.; Sachs, J.N. Curvature dynamics of alpha-synuclein familial Parkinson disease mutants: Molecular simulations of the micelle- and bilayer-bound forms. J. Biol. Chem. 2009, 284, 7177–7189. [Google Scholar] [CrossRef]
- Bisaglia, M.; Tessari, I.; Pinato, L.; Bellanda, M.; Giraudo, S.; Fasano, M.; Bergantino, E.; Bubacco, L.; Mammi, S. A topological model of the interaction between alpha-synuclein and sodium dodecyl sulfate micelles. Biochemistry 2005, 44, 329–339. [Google Scholar] [CrossRef]
- Rocha, S.; Kumar, R.; Nordén, B.; Wittung-Stafshede, P. Orientation of α-Synuclein at Negatively Charged Lipid Vesicles: Linear Dichroism Reveals Time-Dependent Changes in Helix Binding Mode. J. Am. Chem. Soc. 2021, 143, 18899–18906. [Google Scholar] [CrossRef]
- Lokappa, S.B.; Suk, J.E.; Balasubramanian, A.; Samanta, S.; Situ, A.J.; Ulmer, T.S. Sequence and membrane determinants of the random coil-helix transition of α-synuclein. J. Mol. Biol. 2014, 426, 2130–2144. [Google Scholar] [CrossRef]
- Jain, N.; Bhasne, K.; Hemaswasthi, M.; Mukhopadhyay, S. Structural and dynamical insights into the membrane-bound α-synuclein. PLoS ONE 2013, 8, e83752. [Google Scholar] [CrossRef]
- Ulmer, T.S.; Bax, A.; Cole, N.B.; Nussbaum, R.L. Structure and dynamics of micelle-bound human alpha-synuclein. J. Biol. Chem. 2005, 280, 9595–9603. [Google Scholar] [CrossRef]
- Chandra, S.; Chen, X.; Rizo, J.; Jahn, R.; Südhof, T.C. A broken alpha -helix in folded alpha -Synuclein. J. Biol. Chem. 2003, 278, 15313–15318. [Google Scholar] [CrossRef]
- Eliezer, D.; Kutluay, E.; Bussell, R., Jr.; Browne, G. Conformational properties of alpha-synuclein in its free and lipid-associated states. J. Mol. Biol. 2001, 307, 1061–1073. [Google Scholar] [CrossRef] [PubMed]
- Lucas, H.R.; Lee, J.C. Copper(ii) enhances membrane-bound [small alpha]-synuclein helix formation. Metallomics 2011, 3, 280–283. [Google Scholar] [CrossRef] [PubMed]
- Binolfi, A.; Rodriguez, E.E.; Valensin, D.; D’Amelio, N.; Ippoliti, E.; Obal, G.; Duran, R.; Magistrato, A.; Pritsch, O.; Zweckstetter, M.; et al. Bioinorganic chemistry of Parkinson’s disease: Structural determinants for the copper-mediated amyloid formation of alpha-synuclein. Inorg. Chem. 2010, 49, 10668–10679. [Google Scholar] [CrossRef] [PubMed]
- Valensin, D.; Camponeschi, F.; Luczkowski, M.; Baratto, M.C.; Remelli, M.; Valensin, G.; Kozlowski, H. The role of His-50 of α-synuclein in binding Cu(II): pH dependence, speciation, thermodynamics and structure. Metallomics 2011, 3, 292–302. [Google Scholar] [CrossRef]
- Calvo, J.S.; Mulpuri, N.V.; Dao, A.; Qazi, N.K.; Meloni, G. Membrane insertion exacerbates the α-Synuclein-Cu(II) dopamine oxidase activity: Metallothionein-3 targets and silences all α-synuclein-Cu(II) complexes. Free Radic. Biol. Med. 2020, 158, 149–161. [Google Scholar] [CrossRef]
- Bacchella, C.; Nicolis, S.; Dell’Acqua, S.; Rizzarelli, E.; Monzani, E.; Casella, L. Membrane Binding Strongly Affecting the Dopamine Reactivity Induced by Copper Prion and Copper/Amyloid-beta (Abeta) Peptides. A Ternary Copper/Abeta/Prion Peptide Complex Stabilized and Solubilized in Sodium Dodecyl Sulfate Micelles. Inorg. Chem. 2020, 59, 900–912. [Google Scholar] [CrossRef]




| Atom Type | N * | Distance (Å) | 2σ2 (Å2) | R-Factor | χν2 | ΔE0 (eV) | |
|---|---|---|---|---|---|---|---|
| αSyn1–15 SDS-Cu+ | S | 2 | 2.16 (1) | 0.003 (1) | 0.011 | 6 | −4 (2) |
| S | 2 | 2.68 (1) | 0.005 (1) | ||||
| C1 | 4 | 2.35 (1) | 0.013 (1) | ||||
| C2 | 4 | 2.44 (1) | 0.013 (1) | ||||
| αSyn SDS-Cu+ | S | 2 | 2.12 (1) | 0.007 (1) | 0.042 | 11 | −2 (2) |
| S | 2 | 3.00 (1) | 0.015 (1) | ||||
| αSyn UVs-Cu+ | S | 4 | 2.28 (1) | 0.011 (1) | 0.022 | 29 | 6 (2) |
| Time/min | Modified Residues (%) | ||||||
|---|---|---|---|---|---|---|---|
| Met1 | Met5 | His50 | Met116 | Tyr125 | Met127 | Tyr133 | |
| (a) | |||||||
| 30 | 38 | 85 | 1 | 23 | 19 | - | - |
| 240 | 53 | 72 | 21 * | 72 | 27 | 22 | 29 |
| (b) | |||||||
| 30 | - | - | - | <1 | - | - | - |
| (c) | |||||||
| 30 | - | 9 | 8 | <1 | - | <1 | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bacchella, C.; Camponeschi, F.; Kolkowska, P.; Kola, A.; Tessari, I.; Baratto, M.C.; Bisaglia, M.; Monzani, E.; Bubacco, L.; Mangani, S.; et al. Copper Binding and Redox Activity of α-Synuclein in Membrane-Like Environment. Biomolecules 2023, 13, 287. https://doi.org/10.3390/biom13020287
Bacchella C, Camponeschi F, Kolkowska P, Kola A, Tessari I, Baratto MC, Bisaglia M, Monzani E, Bubacco L, Mangani S, et al. Copper Binding and Redox Activity of α-Synuclein in Membrane-Like Environment. Biomolecules. 2023; 13(2):287. https://doi.org/10.3390/biom13020287
Chicago/Turabian StyleBacchella, Chiara, Francesca Camponeschi, Paulina Kolkowska, Arian Kola, Isabella Tessari, Maria Camilla Baratto, Marco Bisaglia, Enrico Monzani, Luigi Bubacco, Stefano Mangani, and et al. 2023. "Copper Binding and Redox Activity of α-Synuclein in Membrane-Like Environment" Biomolecules 13, no. 2: 287. https://doi.org/10.3390/biom13020287
APA StyleBacchella, C., Camponeschi, F., Kolkowska, P., Kola, A., Tessari, I., Baratto, M. C., Bisaglia, M., Monzani, E., Bubacco, L., Mangani, S., Casella, L., Dell’Acqua, S., & Valensin, D. (2023). Copper Binding and Redox Activity of α-Synuclein in Membrane-Like Environment. Biomolecules, 13(2), 287. https://doi.org/10.3390/biom13020287











