The Epigenetic Regulation of RNA N6-Methyladenosine Methylation in Glycolipid Metabolism
Abstract
1. Introduction
2. m6A Methylation
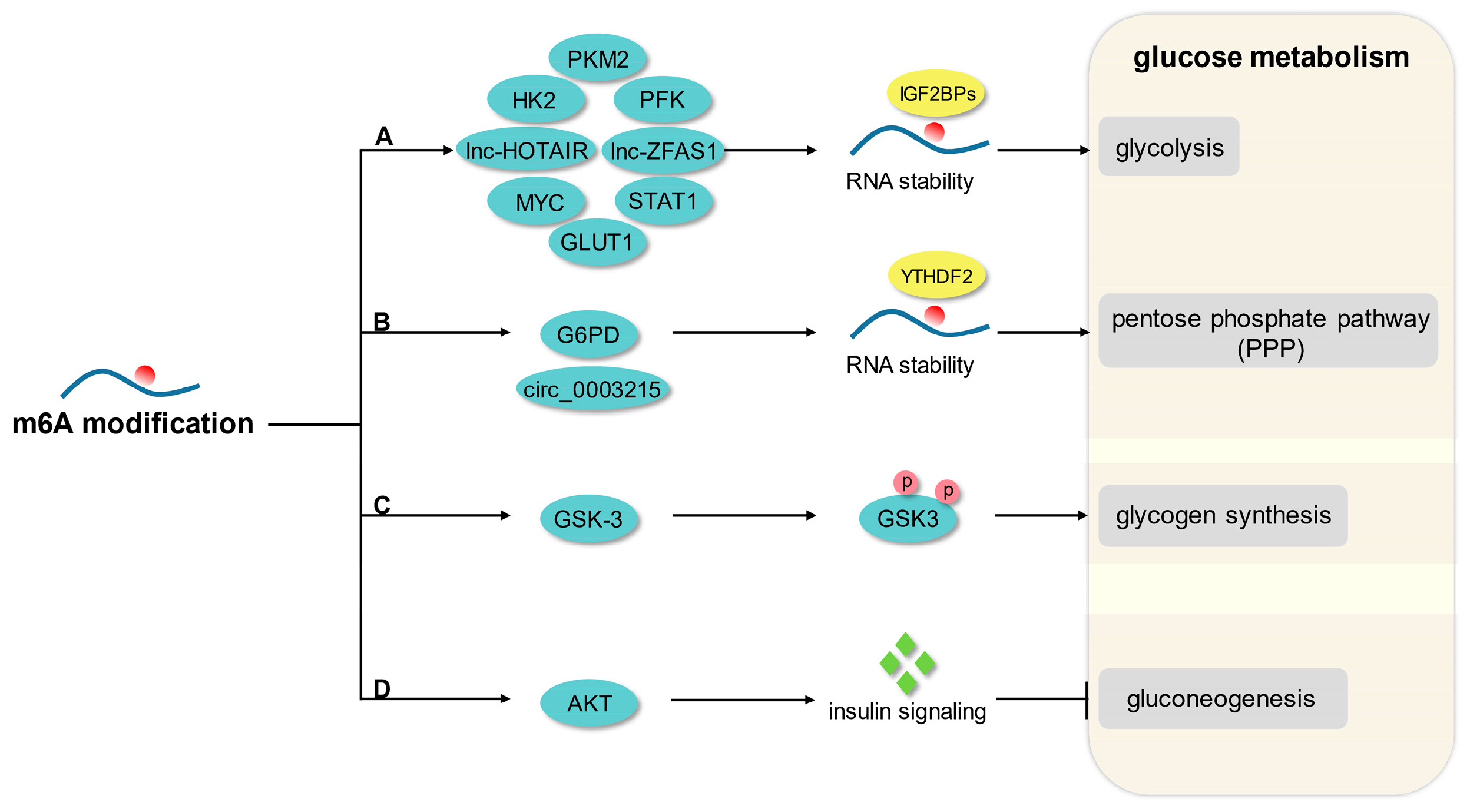
3. m6A Modification and Glucose Metabolism
3.1. Glycolysis
3.2. Pentose Phosphate Pathway
3.3. Glycogen Synthesis and Gluconeogenesis
4. m6A Modification and Lipid Metabolism
4.1. Triglyceride Metabolism
4.2. Cholesterol Metabolism
4.3. Plasma Lipoprotein Metabolism
5. m6A Modification in Glucolipid Metabolic Disease (GLMD)
5.1. Obesity
5.2. Diabetes Mellitus (DM)
5.3. Hyperlipemia
5.4. Nonalcoholic Fatty Liver Disorders (NAFLD)
5.5. Hypertension
5.6. Atherosclerosis (AS)
6. Conclusions and Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Jia, G.; Fu, Y.; Zhao, X.; Dai, Q.; Zheng, G.; Yang, Y.; Yi, C.; Lindahl, T.; Pan, T.; Yang, Y.G.; et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 2011, 7, 885–887. [Google Scholar] [CrossRef] [PubMed]
- Zheng, G.; Dahl, J.A.; Niu, Y.; Fedorcsak, P.; Huang, C.M.; Li, C.J.; Vagbo, C.B.; Shi, Y.; Wang, W.L.; Song, S.H.; et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol. Cell 2013, 49, 18–29. [Google Scholar] [CrossRef] [PubMed]
- Wei, C.; Gershowitz, A.; Moss, B. Methylated nucleotides block 5′ terminus of HeLa cell messenger RNA. Cell 1975, 4, 379–386. [Google Scholar] [CrossRef] [PubMed]
- Bokar, J.; Shambaugh, M.; Polayes, D.; Matera, A.; Rottman, F. Purification and cDNA cloning of the AdoMet-binding subunit of the human mRNA (N6-adenosine)-methyltransferase. RNA 1997, 3, 1233–1247. [Google Scholar]
- He, L.; Li, H.; Wu, A.; Peng, Y.; Shu, G.; Yin, G. Functions of N6-methyladenosine and its role in cancer. Mol. Cancer 2019, 18, 176. [Google Scholar] [CrossRef]
- Huang, H.; Weng, H.; Chen, J. m6A Modification in Coding and Non-coding RNAs: Roles and Therapeutic Implications in Cancer. Cancer Cell 2020, 37, 270–288. [Google Scholar] [CrossRef]
- Li, A.; Chen, Y.-S.; Ping, X.-L.; Yang, X.; Xiao, W.; Yang, Y.; Sun, H.-Y.; Zhu, Q.; Baidya, P.; Wang, X.; et al. Cytoplasmic m6A reader YTHDF3 promotes mRNA translation. Cell Res. 2017, 27, 444–447. [Google Scholar] [CrossRef]
- Śledź, P.; Jinek, M. Structural insights into the molecular mechanism of the m6A writer complex. eLife 2016, 5, e18434. [Google Scholar] [CrossRef]
- Wang, X.; Lu, Z.; Gomez, A.; Hon, G.C.; Yue, Y.; Han, D.; Fu, Y.; Parisien, M.; Dai, Q.; Jia, G.; et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 2013, 505, 117–120. [Google Scholar] [CrossRef]
- Xiao, W.; Adhikari, S.; Dahal, U.; Chen, Y.-S.; Hao, Y.-J.; Sun, B.-F.; Sun, H.-Y.; Li, A.; Ping, X.-L.; Lai, W.-Y.; et al. Nuclear m6A Reader YTHDC1 Regulates mRNA Splicing. Mol. Cell 2016, 61, 507–519. [Google Scholar] [CrossRef]
- Cai, Z.; Deng, X.; Zhao, L.; Wang, X.; Yang, L.; Yuan, G. The relationship between Schistosoma and glycolipid metabolism. Microb. Pathog. 2021, 159, 105120. [Google Scholar] [CrossRef]
- Wu, J.; Frazier, K.; Zhang, J.; Gan, Z.; Wang, T.; Zhong, X. Emerging role of m6A RNA methylation in nutritional physiology and metabolism. Obes. Rev. 2019, 21, e12942. [Google Scholar] [CrossRef]
- Dominissini, D.; Moshitch-Moshkovitz, S.; Schwartz, S.; Salmon-Divon, M.; Ungar, L.; Osenberg, S.; Cesarkas, K.; Jacob-Hirsch, J.; Amariglio, N.; Kupiec, M.; et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 2012, 485, 201–206. [Google Scholar] [CrossRef]
- Meyer, K.D.; Saletore, Y.; Zumbo, P.; Elemento, O.; Mason, C.E.; Jaffrey, S.R. Comprehensive Analysis of mRNA Methylation Reveals Enrichment in 3′ UTRs and near Stop Codons. Cell 2012, 149, 1635–1646. [Google Scholar] [CrossRef]
- Yang, Y.; Fan, X.; Mao, M.; Song, X.; Wu, P.; Zhang, Y.; Jin, Y.; Yang, Y.; Chen, L.-L.; Wang, Y.; et al. Extensive translation of circular RNAs driven by N6-methyladenosine. Cell Res. 2017, 27, 626–641. [Google Scholar] [CrossRef]
- Wang, P.; Doxtader, K.A.; Nam, Y. Structural Basis for Cooperative Function of Mettl3 and Mettl14 Methyltransferases. Mol. Cell 2016, 63, 306–317. [Google Scholar] [CrossRef]
- Zhou, K.I.; Pan, T. Structures of the m6A Methyltransferase Complex: Two Subunits with Distinct but Coordinated Roles. Mol. Cell 2016, 63, 183–185. [Google Scholar] [CrossRef]
- Yue, Y.; Liu, J.; Cui, X.; Cao, J.; Luo, G.; Zhang, Z.; Cheng, T.; Gao, M.; Shu, X.; Ma, H.; et al. VIRMA mediates preferential m6A mRNA methylation in 3′UTR and near stop codon and associates with alternative polyadenylation. Cell Discov. 2018, 4, 10. [Google Scholar] [CrossRef]
- Patil, D.P.; Chen, C.-K.; Pickering, B.F.; Chow, A.; Jackson, C.; Guttman, M.; Jaffrey, S.R. m6A RNA methylation promotes XIST-mediated transcriptional repression. Nature 2016, 537, 369–373. [Google Scholar] [CrossRef]
- Wen, J.; Lv, R.; Ma, H.; Shen, H.; He, C.; Wang, J.; Jiao, F.; Liu, H.; Yang, P.; Tan, L.; et al. Zc3h13 Regulates Nuclear RNA m6A Methylation and Mouse Embryonic Stem Cell Self-Renewal. Mol. Cell 2018, 69, 1028–1038.e6. [Google Scholar] [CrossRef]
- Schwartz, S.; Agarwala, S.D.; Mumbach, M.; Jovanovic, M.; Mertins, P.; Shishkin, A.; Tabach, Y.; Mikkelsen, T.S.; Satija, R.; Ruvkun, G.; et al. High-Resolution Mapping Reveals a Conserved, Widespread, Dynamic mRNA Methylation Program in Yeast Meiosis. Cell 2013, 155, 1409–1421. [Google Scholar] [CrossRef]
- Wei, J.; Liu, F.; Lu, Z.; Fei, Q.; Ai, Y.; He, P.C.; Shi, H.; Cui, X.; Su, R.; Klungland, A.; et al. Differential m6A, m6Am, and m1A Demethylation Mediated by FTO in the Cell Nucleus and Cytoplasm. Mol. Cell 2018, 71, 973–985.e5. [Google Scholar] [CrossRef]
- Tang, C.; Klukovich, R.; Peng, H.; Wang, Z.; Yu, T.; Zhang, Y.; Zheng, H.; Klungland, A.; Yan, W. ALKBH5-dependent m6A demethylation controls splicing and stability of long 3′-UTR mRNAs in male germ cells. Proc. Natl. Acad. Sci. USA 2017, 115, E325–E333. [Google Scholar] [CrossRef]
- Wang, X.; Zhao, B.S.; Roundtree, I.A.; Lu, Z.; Han, D.; Ma, H.; Weng, X.; Chen, K.; Shi, H.; He, C. N6-methyladenosine Modulates Messenger RNA Translation Efficiency. Cell 2015, 161, 1388–1399. [Google Scholar] [CrossRef]
- Roundtree, I.A.; Luo, G.-Z.; Zhang, Z.; Wang, X.; Zhou, T.; Cui, Y.; Sha, J.; Huang, X.; Guerrero, L.; Xie, P.; et al. YTHDC1 mediates nuclear export of N6-methyladenosine methylated mRNAs. eLife 2017, 6, e31311. [Google Scholar] [CrossRef]
- Hsu, P.J.; Zhu, Y.; Ma, H.; Guo, Y.; Shi, X.; Liu, Y.; Qi, M.; Lu, Z.; Shi, H.; Wang, J.; et al. Ythdc2 is an N6-methyladenosine binding protein that regulates mammalian spermatogenesis. Cell Res. 2017, 27, 1115–1127. [Google Scholar] [CrossRef]
- Meyer, K.D.; Patil, D.P.; Zhou, J.; Zinoviev, A.; Skabkin, M.A.; Elemento, O.; Pestova, T.V.; Qian, S.-B.; Jaffrey, S.R. 5′ UTR m6A Promotes Cap-Independent Translation. Cell 2015, 163, 999–1010. [Google Scholar] [CrossRef]
- Huang, H.; Weng, H.; Sun, W.; Qin, X.; Shi, H.; Wu, H.; Zhao, B.S.; Mesquita, A.; Liu, C.; Yuan, C.L.; et al. Recognition of RNA N6-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat. Cell Biol. 2018, 20, 285–295, Correction in Nat. Cell Biol. 2018, 20, 1098; Correction in Nat. Cell Biol. 2020, 22, 1288. [Google Scholar] [CrossRef]
- Wu, B.; Su, S.; Patil, D.P.; Liu, H.; Gan, J.; Jaffrey, S.R.; Ma, J. Molecular basis for the specific and multivariant recognitions of RNA substrates by human hnRNP A2/B1. Nat. Commun. 2018, 9, 420. [Google Scholar] [CrossRef]
- Boucheé, C.; Serdy, S.; Kahn, C.R.; Goldfine, A. The Cellular Fate of Glucose and Its Relevance in Type 2 Diabetes. Endocr. Rev. 2004, 25, 807–830. [Google Scholar] [CrossRef]
- Butterfield, D.A.; Halliwell, B. Oxidative stress, dysfunctional glucose metabolism and Alzheimer disease. Nat. Rev. Neurosci. 2019, 20, 148–160. [Google Scholar] [CrossRef] [PubMed]
- Shen, C.; Xuan, B.; Yan, T.; Ma, Y.; Xu, P.; Tian, X.; Zhang, X.; Cao, Y.; Ma, D.; Zhu, X.; et al. m6A-dependent glycolysis enhances colorectal cancer progression. Mol. Cancer 2020, 19, 72. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Wang, Y.; Liu, Y.; Xie, L.; Ge, J.; Yu, G.; Zhao, G. N6-Methyladenosine Modification of PTTG3P Contributes to Colorectal Cancer Proliferation via YAP1. Front. Oncol. 2021, 11, 669731. [Google Scholar] [CrossRef]
- Du, L.; Li, Y.; Kang, M.; Feng, M.; Ren, Y.; Dai, H.; Wang, Y.; Wang, Y.; Tang, B. USP48 Is Upregulated by Mettl14 to Attenuate Hepatocellular Carcinoma via Regulating SIRT6 Stabilization. Cancer Res. 2021, 81, 3822–3834. [Google Scholar] [CrossRef]
- Lin, J.-X.; Lian, N.-Z.; Gao, Y.-X.; Zheng, Q.-L.; Yang, Y.-H.; Ma, Y.-B.; Xiu, Z.-S.; Qiu, Q.-Z.; Wang, H.-G.; Zheng, C.-H.; et al. m6A methylation mediates LHPP acetylation as a tumour aerobic glycolysis suppressor to improve the prognosis of gastric cancer. Cell Death Dis. 2022, 13, 463. [Google Scholar] [CrossRef]
- Lyu, Y.; Zhang, Y.; Wang, Y.; Luo, Y.; Ding, H.; Li, P.; Ni, G. HIF-1α Regulated WTAP Overexpression Promoting the Warburg Effect of Ovarian Cancer by m6A-Dependent Manner. J. Immunol. Res. 2022, 2022, 6130806. [Google Scholar] [CrossRef]
- Ou, B.; Liu, Y.; Yang, X.; Xu, X.; Yan, Y.; Zhang, J. C5aR1-positive neutrophils promote breast cancer glycolysis through WTAP-dependent m6A methylation of ENO1. Cell Death Dis. 2021, 12, 737. [Google Scholar] [CrossRef]
- Yu, H.; Zhao, K.; Zeng, H.; Li, Z.; Chen, K.; Zhang, Z.; Li, E.; Wu, Z. N6-methyladenosine (m6A) methyltransferase WTAP accelerates the Warburg effect of gastric cancer through regulating HK2 stability. Biomed. Pharmacother. 2020, 133, 111075. [Google Scholar] [CrossRef]
- Li, Y.; He, L.; Wang, Y.; Tan, Y.; Zhang, F. N6-methyladenosine methyltransferase KIAA1429 elevates colorectal cancer aerobic glycolysis via HK2-dependent manner. Bioengineered 2022, 13, 11923–11932. [Google Scholar] [CrossRef]
- Yang, D.; Chang, S.; Li, F.; Ma, M.; Yang, J.; Lv, X.; Huangfu, L.; Jia, C. m6A transferase KIAA1429-stabilized LINC00958 accelerates gastric cancer aerobic glycolysis through targeting GLUT1. IUBMB Life 2021, 73, 1325–1333. [Google Scholar] [CrossRef]
- Wang, Q.; Xie, H.; Peng, H.; Yan, J.; Han, L.; Ye, G. ZC3H13 Inhibits the Progression of Hepatocellular Carcinoma through m6A-PKM2-Mediated Glycolysis and Enhances Chemosensitivity. J. Oncol. 2021, 2021, 1328444. [Google Scholar] [CrossRef]
- Sun, X.; Li, Q.; Yang, L. Sevoflurane Inhibits lncRNA HOTAIR-Modulated Stability of HK2 mRNA in a m6A-Dependent Manner to Dampen Aerobic Glycolysis and Proliferation in Lung Cancer. BioMed. Res. Int. 2022, 2022, 4668774. [Google Scholar] [CrossRef]
- Wang, H.; Liang, Z.; Gou, Y.; Li, Z.; Cao, Y.; Jiao, N.; Tan, J.; Yu, Y.; Zhang, Z. FTO-dependent N(6)-Methyladenosine regulates the progression of endometriosis via the ATG5/PKM2 Axis. Cell. Signal. 2022, 98, 110406. [Google Scholar] [CrossRef]
- Huang, J.; Sun, W.; Wang, Z.; Lv, C.; Zhang, T.; Zhang, D.; Dong, W.; Shao, L.; He, L.; Ji, X.; et al. FTO suppresses glycolysis and growth of papillary thyroid cancer via decreasing stability of APOE mRNA in an N6-methyladenosine-dependent manner. J. Exp. Clin. Cancer Res. 2022, 41, 42. [Google Scholar] [CrossRef]
- Yang, X.; Shao, F.; Guo, D.; Wang, W.; Wang, J.; Zhu, R.; Gao, Y.; He, J.; Lu, Z. WNT/β-catenin-suppressed FTO expression increases m6A of c-Myc mRNA to promote tumor cell glycolysis and tumorigenesis. Cell Death Dis. 2021, 12, 462. [Google Scholar] [CrossRef]
- Qing, Y.; Dong, L.; Gao, L.; Li, C.; Li, Y.; Han, L.; Prince, E.; Tan, B.; Deng, X.; Wetzel, C.; et al. R-2-hydroxyglutarate attenuates aerobic glycolysis in leukemia by targeting the FTO/m6A/PFKP/LDHB axis. Mol. Cell 2021, 81, 922–939.e9. [Google Scholar] [CrossRef]
- Yu, H.; Yang, X.; Tang, J.; Si, S.; Zhou, Z.; Lu, J.; Han, J.; Yuan, B.; Wu, Q.; Lu, Q.; et al. ALKBH5 Inhibited Cell Proliferation and Sensitized Bladder Cancer Cells to Cisplatin by m6A-CK2α-Mediated Glycolysis. Mol. Ther. Nucleic. Acids 2020, 23, 27–41. [Google Scholar] [CrossRef]
- Lu, S.; Han, L.; Hu, X.; Sun, T.; Xu, D.; Li, Y.; Chen, Q.; Yao, W.; He, M.; Wang, Z.; et al. N6-methyladenosine reader IMP2 stabilizes the ZFAS1/OLA1 axis and activates the Warburg effect: Implication in colorectal cancer. J. Hematol. Oncol. 2021, 14, 188. [Google Scholar] [CrossRef]
- Hu, C.; Liu, T.; Han, C.; Xuan, Y.; Jiang, D.; Sun, Y.; Zhang, X.; Zhang, W.; Xu, Y.; Liu, Y.; et al. HPV E6/E7 promotes aerobic glycolysis in cervical cancer by regulating IGF2BP2 to stabilize m6A-MYC expression. Int. J. Biol. Sci. 2022, 18, 507–521. [Google Scholar] [CrossRef]
- Luo, F.; Lin, K. N6-methyladenosine (m6A) reader IGF2BP1 accelerates gastric cancer aerobic glycolysis in c-Myc-dependent manner. Exp. Cell Res. 2022, 417, 113176. [Google Scholar] [CrossRef]
- Wang, Y.; Lu, J.-H.; Wu, Q.-N.; Jin, Y.; Wang, D.-S.; Chen, Y.-X.; Liu, J.; Luo, X.-J.; Meng, Q.; Pu, H.-Y.; et al. LncRNA LINRIS stabilizes IGF2BP2 and promotes the aerobic glycolysis in colorectal cancer. Mol. Cancer 2019, 18, 174. [Google Scholar] [CrossRef]
- Li, Z.; Peng, Y.; Li, J.; Chen, Z.; Chen, F.; Tu, J.; Lin, S.; Wang, H. N6-methyladenosine regulates glycolysis of cancer cells through PDK4. Nat. Commun. 2020, 11, 2578. [Google Scholar] [CrossRef]
- Huangfu, N.; Zheng, W.; Xu, Z.; Wang, S.; Wang, Y.; Cheng, J.; Li, Z.; Cheng, K.; Zhang, S.; Chen, X.; et al. RBM4 regulates M1 macrophages polarization through targeting STAT1-mediated glycolysis. Int. Immunopharmacol. 2020, 83, 106432. [Google Scholar] [CrossRef]
- Stincone, A.; Prigione, A.; Cramer, T.; Wamelink, M.M.C.; Campbell, K.; Cheung, E.; Olin-Sandoval, V.; Grüning, N.-M.; Krüger, A.; Tauqeer Alam, M.; et al. The return of metabolism: Biochemistry and physiology of the pentose phosphate pathway. Biol. Rev. 2015, 90, 927–963. [Google Scholar] [CrossRef]
- Clasquin, M.F.; Melamud, E.; Singer, A.; Gooding, J.R.; Xu, X.; Dong, A.; Cui, H.; Campagna, S.R.; Savchenko, A.; Yakunin, A.F.; et al. Riboneogenesis in Yeast. Cell 2011, 145, 969–980. [Google Scholar] [CrossRef]
- Wang, L.; Xie, J.; Schultz, P.G. Expanding the Genetic Code. Annu. Rev. Biophys. Biomol. Struct. 2006, 35, 225–249. [Google Scholar] [CrossRef]
- Hayes, J.D.; Dinkova-Kostova, A.T.; Tew, K.D. Oxidative Stress in Cancer. Cancer Cell 2020, 38, 167–197. [Google Scholar] [CrossRef]
- Liu, Z.; Chen, Y.; Wang, L.; Ji, S. ALKBH5 Promotes the Proliferation of Glioma Cells via Enhancing the mRNA Stability of G6PD. Neurochem. Res. 2021, 46, 3003–3011. [Google Scholar] [CrossRef]
- Sheng, H.; Li, Z.; Su, S.; Sun, W.; Zhang, X.; Li, L.; Li, J.; Liu, S.; Lu, B.; Zhang, S.; et al. YTH domain family 2 promotes lung cancer cell growth by facilitating 6-phosphogluconate dehydrogenase mRNA translation. Carcinog. 2019, 41, 541–550. [Google Scholar] [CrossRef]
- Chen, B.; Hong, Y.; Gui, R.; Zheng, H.; Tian, S.; Zhai, X.; Xie, X.; Chen, Q.; Qian, Q.; Ren, X.; et al. N6-methyladenosine modification of circ_0003215 suppresses the pentose phosphate pathway and malignancy of colorectal cancer through the miR-663b/DLG4/G6PD axis. Cell Death Dis. 2022, 13, 804. [Google Scholar] [CrossRef]
- Han, H.-S.; Kang, G.; Kim, J.S.; Choi, B.H.; Koo, S.-H. Regulation of glucose metabolism from a liver-centric perspective. Exp. Mol. Med. 2016, 48, e218. [Google Scholar] [CrossRef]
- Li, H.; Zhang, N.; Jiao, X.; Wang, C.; Sun, W.; He, Y.; Ren, G.; Huang, S.; Li, M.; Chang, Y.; et al. Downregulation of microRNA-6125 promotes colorectal cancer growth through YTHDF2-dependent recognition of N6-methyladenosine-modified GSK3β. Clin. Transl. Med. 2021, 11, e602. [Google Scholar] [CrossRef]
- Zhou, L.; Jiang, J.; Huang, Z.; Jin, P.; Peng, L.; Luo, M.; Zhang, Z.; Chen, Y.; Na Xie, N.; Gao, W.; et al. Hypoxia-induced lncRNA STEAP3-AS1 activates Wnt/β-catenin signaling to promote colorectal cancer progression by preventing m6A-mediated degradation of STEAP3 mRNA. Mol. Cancer 2022, 21, 168. [Google Scholar] [CrossRef]
- Zou, Y.; Zheng, S.; Xie, X.; Ye, F.; Hu, X.; Tian, Z.; Yan, S.-M.; Yang, L.; Kong, Y.; Tang, Y.; et al. N6-methyladenosine regulated FGFR4 attenuates ferroptotic cell death in recalcitrant HER2-positive breast cancer. Nat. Commun. 2022, 13, 2672. [Google Scholar] [CrossRef]
- Diao, M.-Y.; Zhu, Y.; Yang, J.; Xi, S.-S.; Wen, X.; Gu, Q.; Hu, W. Hypothermia protects neurons against ischemia/reperfusion-induced pyroptosis via m6A-mediated activation of PTEN and the PI3K/Akt/GSK-3β signaling pathway. Brain Res. Bull. 2020, 159, 25–31. [Google Scholar] [CrossRef]
- Gao, X.-Q.; Zhang, Y.-H.; Liu, F.; Ponnusamy, M.; Zhao, X.-M.; Zhou, L.-Y.; Zhai, M.; Liu, C.-Y.; Li, X.-M.; Wang, M.; et al. The piRNA CHAPIR regulates cardiac hypertrophy by controlling METTL3-dependent N6-methyladenosine methylation of Parp10 mRNA. Nature 2020, 22, 1319–1331. [Google Scholar] [CrossRef]
- Hatting, M.; Tavares, C.D.; Sharabi, K.; Rines, A.K.; Puigserver, P. Insulin regulation of gluconeogenesis. Ann. N. Y. Acad. Sci. 2017, 1411, 21–35. [Google Scholar] [CrossRef]
- Liu, J.; Luo, G.; Sun, J.; Men, L.; Ye, H.; He, C.; Ren, D. METTL14 is essential for β-cell survival and insulin secretion. Biochim. Biophys. Acta (BBA) Mol. Basis Dis. 2019, 1865, 2138–2148. [Google Scholar] [CrossRef]
- Zechner, R.; Zimmermann, R.; Eichmann, T.O.; Kohlwein, S.D.; Haemmerle, G.; Lass, A.; Madeo, F. FAT SIGNALS—Lipases and Lipolysis in Lipid Metabolism and Signaling. Cell Metab. 2012, 15, 279–291. [Google Scholar] [CrossRef]
- Alves-Bezerra, M.; Cohen, D.E. Triglyceride Metabolism in the Liver. Compr. Physiol. 2017, 8, 1–8. [Google Scholar]
- Kang, H.; Zhang, Z.; Yu, L.; Li, Y.; Liang, M.; Zhou, L. FTO reduces mitochondria and promotes hepatic fat accumulation through RNA demethylation. J. Cell. Biochem. 2018, 119, 5676–5685. [Google Scholar] [CrossRef]
- Wu, R.; Chen, Y.; Liu, Y.; Zhuang, L.; Chen, W.; Zeng, B.; Liao, X.; Guo, G.; Wang, Y.; Wang, X. m6A methylation promotes white-to-beige fat transition by facilitating Hif1a translation. EMBO Rep. 2021, 22, e52348. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Wu, R.; Liu, Y.; Zhao, Y.; Bi, Z.; Yao, Y.; Liu, Q.; Shi, H.; Wang, F.; Wang, Y. m6A mRNA methylation controls autophagy and adipogenesis by targeting Atg5 and Atg7. Autophagy 2019, 16, 1221–1235. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Song, C.; Na Wang, N.; Li, S.; Liu, Q.; Sun, Z.; Wang, K.; Yu, S.-C.; Yang, Q. NADP modulates RNA m6A methylation and adipogenesis via enhancing FTO activity. Nat. Chem. Biol. 2020, 16, 1394–1402. [Google Scholar] [CrossRef] [PubMed]
- Xie, W.; Ma, L.L.; Xu, Y.Q.; Wang, B.H.; Li, S.M. METTL3 inhibits hepatic insulin sensitivity via N6-methyladenosine modification of Fasn mRNA and promoting fatty acid metabolism. Biochem. Biophys. Res. Commun. 2019, 518, 120–126. [Google Scholar] [CrossRef] [PubMed]
- Sun, D.; Zhao, T.; Zhang, Q.; Wu, M.; Zhang, Z. Fat mass and obesity-associated protein regulates lipogenesis via m6A modification in fatty acid synthase mRNA. Cell Biol. Int. 2020, 45, 334–344. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, Q.; Cui, G.; Zhao, F.; Tian, X.; Sun, B.-F.; Yang, Y.; Li, W. m6A Regulates Liver Metabolic Disorders and Hepatogenous Diabetes. Genom. Proteom. Bioinform. 2020, 18, 371–383. [Google Scholar] [CrossRef]
- Zhou, B.; Liu, C.; Xu, L.; Yuan, Y.; Zhao, J.; Zhao, W.; Chen, Y.; Qiu, J.; Meng, M.; Zheng, Y.; et al. N6-Methyladenosine Reader Protein YT521-B Homology Domain-Containing 2 Suppresses Liver Steatosis by Regulation of mRNA Stability of Lipogenic Genes. Hepatology 2020, 73, 91–103. [Google Scholar] [CrossRef]
- Yu, Y.; Pan, Y.; Fan, Z.; Xu, S.; Gao, Z.; Ren, Z.; Yu, J.; Li, W.; Liu, F.; Gu, J.; et al. LuHui Derivative, A Novel Compound That Inhibits the Fat Mass and Obesity-Associated (FTO), Alleviates the Inflammatory Response and Injury in Hyperlipidemia-Induced Cardiomyopathy. Front. Cell Dev. Biol. 2021, 9, 3217. [Google Scholar] [CrossRef]
- Zong, X.; Zhao, J.; Wang, H.; Lu, Z.; Wang, F.; Du, H.; Wang, Y. Mettl3 Deficiency Sustains Long-Chain Fatty Acid Absorption through Suppressing Traf6-Dependent Inflammation Response. J. Immunol. 2019, 202, 567–578. [Google Scholar] [CrossRef]
- Takemoto, S.; Nakano, M.; Fukami, T.; Nakajima, M. m6A modification impacts hepatic drug and lipid metabolism properties by regulating carboxylesterase 2. Biochem. Pharmacol. 2021, 193, 114766. [Google Scholar] [CrossRef] [PubMed]
- Jia, Y.; Yan, Q.; Zheng, Y.; Li, L.; Zhang, B.; Chang, Z.; Wang, Z.; Tang, H.; Qin, Y.; Guan, X.-Y. Long non-coding RNA NEAT1 mediated RPRD1B stability facilitates fatty acid metabolism and lymph node metastasis via c-Jun/c-Fos/SREBP1 axis in gastric cancer. J. Exp. Clin. Cancer Res. 2022, 41, 287. [Google Scholar] [CrossRef]
- Liu, P.; Fan, B.; Othmane, B.; Hu, J.; Li, H.; Cui, Y.; Ou, Z.; Chen, J.; Zu, X. m6A-induced lncDBET promotes the malignant progression of bladder cancer through FABP5-mediated lipid metabolism. Theranostics 2022, 12, 6291–6307. [Google Scholar] [CrossRef] [PubMed]
- Espinosa, G.; López-Montero, I.; Monroy, F.; Langevin, D. Shear rheology of lipid monolayers and insights on membrane fluidity. Proc. Natl. Acad. Sci. USA 2011, 108, 6008–6013. [Google Scholar] [CrossRef]
- Brown, M.S.; Goldstein, J.L. The SREBP Pathway: Regulation of Cholesterol Metabolism by Proteolysis of a Membrane-Bound Transcription Factor. Cell 1997, 89, 331–340. [Google Scholar] [CrossRef] [PubMed]
- Zelcer, N.; Hong, C.; Boyadjian, R.; Tontonoz, P. LXR Regulates Cholesterol Uptake Through Idol-Dependent Ubiquitination of the LDL Receptor. Science 2009, 325, 100–104. [Google Scholar] [CrossRef] [PubMed]
- Gong, C.; Fan, Y.; Liu, J. METTL14 mediated m6A modification to LncRNA ZFAS1/RAB22A: A novel therapeutic target for atherosclerosis. Int. J. Cardiol. 2020, 328, 177. [Google Scholar] [CrossRef]
- Yang, Z.; Yu, G.-L.; Zhu, X.; Peng, T.-H.; Lv, Y.-C. Critical roles of FTO-mediated mRNA m6A demethylation in regulating adipogenesis and lipid metabolism: Implications in lipid metabolic disorders. Genes Dis. 2021, 9, 51–61. [Google Scholar] [CrossRef]
- Yang, Y.; Cai, J.; Yang, X.; Wang, K.; Sun, K.; Yang, Z.; Zhang, L.; Yang, L.; Gu, C.; Huang, X.; et al. Dysregulated m6A modification promotes lipogenesis and development of non-alcoholic fatty liver disease and hepatocellular carcinoma. Mol. Ther. 2022, 30, 2342–2353. [Google Scholar] [CrossRef]
- Fang, R.; Chen, X.; Zhang, S.; Shi, H.; Ye, Y.; Shi, H.; Zou, Z.; Li, P.; Guo, Q.; Ma, L.; et al. EGFR/SRC/ERK-stabilized YTHDF2 promotes cholesterol dysregulation and invasive growth of glioblastoma. Nat. Commun. 2021, 12, 177. [Google Scholar] [CrossRef]
- Borén, J.; Taskinen, M.-R.; Adiels, M. Kinetic studies to investigate lipoprotein metabolism. J. Intern. Med. 2012, 271, 166–173. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Lai, K.; Yong, X.; Yin, H.; Chen, Z.; Wang, H.; Chen, K.; Zheng, J. Silencing METTL3 Stabilizes Atherosclerotic Plaques by Regulating the Phenotypic Transformation of Vascular Smooth Muscle Cells via the miR-375-3p/PDK1 Axis. Cardiovasc. Drugs Ther. 2022, 1–16. [Google Scholar] [CrossRef]
- Liu, Y.; Luo, G.; Tang, Q.; Song, Y.; Liu, D.; Wang, H.; Ma, J. Methyltransferase-like 14 silencing relieves the development of atherosclerosis via m6A modification of p65 mRNA. Bioengineered 2022, 13, 11832–11843. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Xu, Q.; Huangfu, N.; Chen, X.; Zhu, J. Mettl3 promotes oxLDL-mediated inflammation through activating STAT1 signaling. J. Clin. Lab. Anal. 2021, 36, e24019. [Google Scholar] [CrossRef] [PubMed]
- Ye, D.-W.; Rong, X.-L.; Xu, A.-M.; Guo, J. Liver-adipose tissue crosstalk: A key player in the pathogenesis of glucolipid metabolic disease. Chin. J. Integr. Med. 2017, 23, 410–414. [Google Scholar] [CrossRef]
- Rønningen, T.; Dahl, M.B.; Valderhaug, T.G.; Cayir, A.; Keller, M.; Tönjes, A.; Blüher, M.; Böttcher, Y. m6A Regulators in Human Adipose Tissue-Depot-Specificity and Correlation with Obesity. Front. Endocrinol. 2021, 12, 1647. [Google Scholar] [CrossRef]
- Cecil, J.E.; Tavendale, R.; Watt, P.; Hetherington, M.M.; Palmer, C.N. An obesity-associated FTO gene variant and increased energy intake in children. N. Engl. J. Med. 2008, 359, 2558–2566. [Google Scholar] [CrossRef]
- Wardle, J.; Carnell, S.; Haworth, C.M.A.; Farooqi, I.S.; O’Rahilly, S.; Plomin, R. Obesity Associated Genetic Variation in FTO Is Associated with Diminished Satiety. J. Clin. Endocrinol. Metab. 2008, 93, 3640–3643. [Google Scholar] [CrossRef]
- Zhao, X.; Yang, Y.; Sun, B.-F.; Shi, Y.; Yang, X.; Xiao, W.; Hao, Y.-J.; Ping, X.-L.; Chen, Y.-S.; Wang, W.-J.; et al. FTO-dependent demethylation of N6-methyladenosine regulates mRNA splicing and is required for adipogenesis. Cell Res. 2014, 24, 1403–1419. [Google Scholar] [CrossRef]
- Shen, G.-S.; Zhou, H.-B.; Zhang, H.; Chen, B.; Liu, Z.-P.; Yuan, Y.; Zhou, X.-Z.; Xu, Y.-J. The GDF11-FTO-PPARγ axis controls the shift of osteoporotic MSC fate to adipocyte and inhibits bone formation during osteoporosis. Biochim. Biophys. Acta (BBA) Mol. Basis Dis. 2018, 1864, 3644–3654. [Google Scholar] [CrossRef]
- Zhou, X.; Chen, J.; Chen, J.; Wu, W.; Wang, X.; Wang, Y. The beneficial effects of betaine on dysfunctional adipose tissue and N6-methyladenosine mRNA methylation requires the AMP-activated protein kinase α1 subunit. J. Nutr. Biochem. 2015, 26, 1678–1684. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Q.; Sun, B.; Liu, Q.; Cai, M.; Wu, R.; Wang, F.; Yao, Y.; Wang, Y.; Wang, X. MTCH2 promotes adipogenesis in intramuscular preadipocytes via an m6A-YTHDF1-dependent mechanism. FASEB J. 2018, 33, 2971–2981. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Zhao, Y.; Wu, R.; Jiang, Q.; Cai, M.; Bi, Z.; Liu, Y.; Yao, Y.; Feng, J.; Wang, Y.; et al. ZFP217 regulates adipogenesis by controlling mitotic clonal expansion in a METTL3-m6A dependent manner. RNA Biol. 2019, 16, 1785–1793. [Google Scholar] [CrossRef]
- Gernapudi, R.; Wolfson, B.; Zhang, Y.; Yao, Y.; Yang, P.; Asahara, H.; Zhou, Q. MicroRNA 140 Promotes Expression of Long Noncoding RNA NEAT1 in Adipogenesis. Mol. Cell. Biol. 2016, 36, 30–38. [Google Scholar] [CrossRef]
- Bornaque, F.; Delannoy, C.P.; Courty, E.; Rabhi, N.; Carney, C.; Rolland, L.; Moreno, M.; Gromada, X.; Bourouh, C.; Petit, P.; et al. Glucose Regulates m6A Methylation of RNA in Pancreatic Islets. Cells 2022, 11, 291. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Shen, F.; Huang, W.; Qin, S.; Huang, J.-T.; Sergi, C.; Yuan, B.-F.; Liu, S.-M. Glucose Is Involved in the Dynamic Regulation of m6A in Patients with Type 2 Diabetes. J. Clin. Endocrinol. Metab. 2018, 104, 665–673. [Google Scholar] [CrossRef]
- Regué, L.; Zhao, L.; Ji, F.; Wang, H.; Avruch, J.; Dai, N. RNA m6A reader IMP2/IGF2BP2 promotes pancreatic β-cell proliferation and insulin secretion by enhancing PDX1 expression. Mol. Metab. 2021, 48, 101209. [Google Scholar] [CrossRef]
- Wang, Y.; Sun, J.; Lin, Z.; Zhang, W.; Wang, S.; Wang, W.; Wang, Q.; Ning, G. m(6)A mRNA Methylation Controls Functional Maturation in Neonatal Murine beta-Cells. Diabetes 2020, 69, 1708–1722. [Google Scholar] [CrossRef]
- Li, X.; Jiang, Y.; Sun, X.; Wu, Y.; Chen, Z. METTL3 is required for maintaining β-cell function. Metabolism 2021, 116, 154702. [Google Scholar] [CrossRef]
- Sun, J.; Liu, G.; Chen, R.; Zhou, J.; Chen, T.; Cheng, Y.; Lou, Q.; Wang, H. PARP1 Is Upregulated by Hyperglycemia Via N6-methyladenosine Modification and Promotes Diabetic Retinopathy. Discov. Med. 2022, 34, 115–129. [Google Scholar]
- Meng, L.; Lin, H.; Huang, X.; Weng, J.; Peng, F.; Wu, S. METTL14 suppresses pyroptosis and diabetic cardiomyopathy by downregulating TINCR lncRNA. Cell Death Dis. 2022, 13, 38. [Google Scholar] [CrossRef] [PubMed]
- Peng, T.; Liu, M.; Hu, L.; Guo, D.; Di Wang, D.; Qi, B.; Ren, G.; Hu, C.; Zhang, F.; Chun, H.J.; et al. LncRNA Airn alleviates diabetic cardiac fibrosis by inhibiting activation of cardiac fibroblasts via a m6A-IMP2-p53 axis. Biol. Direct 2022, 17, 32. [Google Scholar] [CrossRef] [PubMed]
- Cao, X.; Song, Y.; Huang, L.-L.; Tian, Y.-J.; Wang, X.-L.; Hua, L.-Y. mA transferase METTL3 regulates endothelial-mesenchymal transition in diabetic retinopathy via lncRNA SNHG7/KHSRP/MKL1 axis. Genomics 2022, 114, 110498. [Google Scholar] [CrossRef]
- Berglund, L.; Brunzell, J.D.; Goldberg, A.C.; Goldberg, I.J.; Sacks, F.; Murad, M.H.; Stalenhoef, A.F.H. Evaluation and Treatment of Hypertriglyceridemia: An Endocrine Society Clinical Practice Guideline. J. Clin. Endocrinol. Metab. 2012, 97, 2969–2989. [Google Scholar] [CrossRef] [PubMed]
- Jiao, Y.; Williams, A.; Wei, N. Quercetin ameliorated insulin resistance via regulating METTL3-mediated N6-methyladenosine modification of PRKD2 mRNA in skeletal muscle and C2C12 myocyte cell line. Nutr. Metab. Cardiovasc. Dis. 2022, 32, 2655–2668. [Google Scholar] [CrossRef]
- Dang, Y.; Xu, J.; Yang, Y.; Li, C.; Zhang, Q.; Zhou, W.; Zhang, L.; Ji, G. Ling-gui-zhu-gan decoction alleviates hepatic steatosis through SOCS2 modification by N6-methyladenosine. Biomed. Pharmacother. 2020, 127, 109976. [Google Scholar] [CrossRef]
- Park, M.H.; Jeong, E.; Choudhury, M. Mono-(2-Ethylhexyl)phthalate Regulates Cholesterol Efflux via MicroRNAs Regulated m6A RNA Methylation. Chem. Res. Toxicol. 2019, 33, 461–469. [Google Scholar] [CrossRef]
- Wu, J.; Li, Y.; Yu, J.; Gan, Z.; Wei, W.; Wang, C.; Zhang, L.; Wang, T.; Zhong, X. Resveratrol Attenuates High-Fat Diet Induced Hepatic Lipid Homeostasis Disorder and Decreases m6A RNA Methylation. Front. Pharmacol. 2020, 11, 568006. [Google Scholar] [CrossRef]
- Hardie, D.G.; Ross, F.A.; Hawley, S.A. AMPK: A nutrient and energy sensor that maintains energy homeostasis. Nat. Rev. Mol. Cell Biol. 2012, 13, 251–262. [Google Scholar] [CrossRef]
- Lin, L.; Hales, C.M.; Garber, K.; Jin, P. Fat mass and obesity-associated (FTO) protein interacts with CaMKII and modulates the activity of CREB signaling pathway. Hum. Mol. Genet. 2014, 23, 3299–3306. [Google Scholar] [CrossRef]
- Younossi, Z.M. Non-alcoholic fatty liver disease—A global public health perspective. J. Hepatol. 2019, 70, 531–544. [Google Scholar] [CrossRef] [PubMed]
- Neuschwander-Tetri, B.A. Hepatic lipotoxicity and the pathogenesis of nonalcoholic steatohepatitis: The central role of nontriglyceride fatty acid metabolites. Hepatology 2010, 52, 774–788. [Google Scholar] [CrossRef] [PubMed]
- Peng, Z.; Gong, Y.; Wang, X.; He, W.; Wu, L.; Zhang, L.; Xiong, L.; Huang, Y.; Su, L.; Shi, P.; et al. METTL3-m6A-Rubicon axis inhibits autophagy in nonalcoholic fatty liver disease. Mol. Ther. 2021, 30, 932–946. [Google Scholar] [CrossRef]
- Hu, Y.; Feng, Y.; Zhang, L.; Jia, Y.; Cai, D.; Qian, S.-B.; Du, M.; Zhao, R. GR-mediated FTO transactivation induces lipid accumulation in hepatocytes via demethylation of m6A on lipogenic mRNAs. RNA Biol. 2020, 17, 930–942. [Google Scholar] [CrossRef] [PubMed]
- Gan, X.; Dai, Z.; Ge, C.; Yin, H.; Wang, Y.; Tan, J.; Sun, S.; Zhou, W.; Yuan, S.; Yang, F. FTO promotes liver inflammation by suppressing m6A mRNA methylation of IL-17RA. Front. Oncol. 2022, 12, 989353. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Wang, Y.; Lai, S.; Zhao, L.; Liu, W.; Liu, S.; Chen, H.; Wang, J.; Du, G.; Tang, B. LINC01468 drives NAFLD-HCC progression through CUL4A-linked degradation of SHIP2. Cell Death Discov. 2022, 8, 449. [Google Scholar] [CrossRef]
- Sun, R.; Tian, X.; Li, Y.; Zhao, Y.; Wang, Z.; Hu, Y.; Zhang, L.; Wang, Y.; Gao, D.; Zheng, S.; et al. The m6A reader YTHDF3-mediated PRDX3 translation alleviates liver fibrosis. Redox Biol. 2022, 54, 102378. [Google Scholar] [CrossRef]
- Yang, J.-J.; Wang, J.; Yang, Y.; Li, J.; Lu, D.; Lu, C. ALKBH5 ameliorated liver fibrosis and suppressed HSCs activation via triggering PTCH1 activation in an m6A dependent manner. Eur. J. Pharmacol. 2022, 922, 174900. [Google Scholar] [CrossRef]
- Cheng, W.; Li, M.; Zhang, L.; Zhou, C.; Yu, S.; Peng, X.; Zhang, W.; Zhang, W. New roles of N6-methyladenosine methylation system regulating the occurrence of non-alcoholic fatty liver disease with N6-methyladenosine-modified MYC. Front. Pharmacol. 2022, 13, 973116. [Google Scholar] [CrossRef]
- Luo, Y.; Zhang, Z.; Xiang, L.; Zhou, B.; Wang, X.; Lin, Y.; Ding, X.; Liu, F.; Lu, Y.; Peng, Y. Analysis of N6-Methyladenosine Methylation Modification in Fructose-Induced Non-Alcoholic Fatty Liver Disease. Front. Endocrinol. 2021, 12, 780617. [Google Scholar] [CrossRef]
- Lionakis, N. Hypertension in the elderly. World J. Cardiol. 2012, 4, 135–147. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Yuan, X.; Han, R.; Zhang, H.; Xiu, R. Epitranscriptomic mechanisms of N6-methyladenosine methylation regulating mammalian hypertension development by determined spontaneously hypertensive rats pericytes. Epigenomics 2019, 11, 1359–1370. [Google Scholar] [CrossRef]
- Mo, X.-B.; Lei, S.-F.; Zhang, Y.-H.; Zhang, H. Examination of the associations between m6A-associated single-nucleotide polymorphisms and blood pressure. Hypertens. Res. 2019, 42, 1582–1589. [Google Scholar] [CrossRef] [PubMed]
- Marcadenti, A.; Fuchs, F.D.; Matte, U.; Sperb, F.; Moreira, L.B.; Fuchs, S.C. Effects of FTO RS9939906 and MC4R RS17782313 on obesity, type 2 diabetes mellitus and blood pressure in patients with hypertension. Cardiovasc. Diabetol. 2013, 12, 103–108. [Google Scholar] [CrossRef] [PubMed]
- Mouton, A.J.; Li, X.; Hall, M.E.; Hall, J.E. Obesity, Hypertension, and Cardiac Dysfunction. Circ. Res. 2020, 126, 789–806. [Google Scholar] [CrossRef] [PubMed]
- Fu, Z.; Zhou, E.; Wang, X.; Tian, M.; Kong, J.; Li, J.; Ji, L.; Niu, C.; Shen, H.; Dong, S.; et al. Oxidized low-density lipoprotein-induced microparticles promote endothelial monocyte adhesion via intercellular adhesion molecule 1. Am. J. Physiol. Physiol. 2017, 313, C567–C574. [Google Scholar] [CrossRef]
- Zhou, Y.; Jiang, R.; Jiang, Y.; Fu, Y.; Manafhan, Y.; Zhu, J.; Jia, E. Exploration of N6-Methyladenosine Profiles of mRNAs and the Function of METTL3 in Atherosclerosis. Cells 2022, 11, 2980. [Google Scholar] [CrossRef]
- Chien, C.-S.; Li, J.Y.-S.; Chien, Y.; Wang, M.-L.; Yarmishyn, A.A.; Tsai, P.-H.; Juan, C.-C.; Nguyen, P.; Cheng, H.-M.; Huo, T.-I.; et al. METTL3-dependent N6-methyladenosine RNA modification mediates the atherogenic inflammatory cascades in vascular endothelium. Proc. Natl. Acad. Sci. USA 2021, 118, e2025070118. [Google Scholar] [CrossRef]
- Dong, G.; Yu, J.; Shan, G.; Su, L.; Yu, N.; Yang, S. N6-Methyladenosine Methyltransferase METTL3 Promotes Angiogenesis and Atherosclerosis by Upregulating the JAK2/STAT3 Pathway via m6A Reader IGF2BP1. Front. Cell Dev. Biol. 2021, 9, 731810. [Google Scholar] [CrossRef]
- Jian, D.; Wang, Y.; Jian, L.; Tang, H.; Rao, L.; Chen, K.; Jia, Z.; Zhang, W.; Liu, Y.; Chen, X.; et al. METTL14 aggravates endothelial inflammation and atherosclerosis by increasing FOXO1 N6-methyladeosine modifications. Theranostics 2020, 10, 8939–8956. [Google Scholar] [CrossRef]
- Kumari, R.; Dutta, R.; Ranjan, P.; Suleiman, Z.G.; Goswami, S.K.; Li, J.; Pal, H.C.; Verma, S.K. ALKBH5 Regulates SPHK1-Dependent Endothelial Cell Angiogenesis Following Ischemic Stress. Front. Cardiovasc. Med. 2022, 8, 2160. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.-Y.; Han, L.; Tang, Y.-F.; Zhang, G.-X.; Fan, X.-L.; Zhang, J.-J.; Xue, Q.; Xu, Z.-Y. METTL14 regulates M6A methylation-modified primary miR-19a to promote cardiovascular endothelial cell proliferation and invasion. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 7015–7023. [Google Scholar] [PubMed]
- Zheng, Y.; Li, Y.; Ran, X.; Wang, D.; Zheng, X.; Zhang, M.; Yu, B.; Sun, Y.; Wu, J. Mettl14 mediates the inflammatory response of macrophages in atherosclerosis through the NF-κB/IL-6 signaling pathway. Cell. Mol. Life Sci. 2022, 79, 311. [Google Scholar] [CrossRef]
- Guo, M.; Yan, R.; Ji, Q.; Yao, H.; Sun, M.; Duan, L.; Xue, Z.; Jia, Y. IFN regulatory Factor-1 induced macrophage pyroptosis by modulating m6A modification of circ_0029589 in patients with acute coronary syndrome. Int. Immunopharmacol. 2020, 86, 106800. [Google Scholar] [CrossRef]
- Cheng, M.-L.; Shiao, M.-S.; Chiu, D.T.-Y.; Weng, S.-F.; Tang, H.-Y.; Ho, H.-Y. Biochemical disorders associated with antiproliferative effect of dehydroepiandrosterone in hepatoma cells as revealed by LC-based metabolomics. Biochem. Pharmacol. 2011, 82, 1549–1561. [Google Scholar] [CrossRef]
- Lu, S.; Ke, S.; Wang, C.; Xu, Y.; Li, Z.; Song, K.; Bai, M.; Zhou, M.; Yu, H.; Yin, B.; et al. NNMT promotes the progression of intrahepatic cholangiocarcinoma by regulating aerobic glycolysis via the EGFR-STAT3 axis. Oncogenesis 2022, 11, 39. [Google Scholar] [CrossRef]
- Rome, F.I.; Hughey, C.C. Disrupted liver oxidative metabolism in glycine N-methyltransferase-deficient mice is mitigated by dietary methionine restriction. Mol. Metab. 2022, 58, 101452. [Google Scholar] [CrossRef]
- E Christensen, K.; Mikael, L.G.; Leung, K.-Y.; Lévesque, N.; Deng, L.; Wu, Q.; Malysheva, O.V.; Best, A.; A Caudill, M.; DE Greene, N.; et al. High folic acid consumption leads to pseudo-MTHFR deficiency, altered lipid metabolism, and liver injury in mice. Am. J. Clin. Nutr. 2015, 101, 646–658. [Google Scholar] [CrossRef]


| Type | Regulator | Biological Function | References |
|---|---|---|---|
| m6A writers | METTL3 | Catalyzes m6A modification | [16] |
| METTL14 | Assists METTL3 to recognize the specific subtract and enhances the stability of MTC structure | [16] | |
| METTL16 | Catalyzes m6A modification and regulates SAM homeostasis | [17] | |
| WTAP | Promotes the cellular m6A deposition | [18] | |
| VIRMA (KIAA1429) | Guides the core component of MTC to specific RNA region and interacts with cleavage and polyadenylation specific factor 5/6 (CPSF5/6) | [18] | |
| RBM15/15B | Recruits METTL3-METTL14 heterodimer to specific RNA sites | [19] | |
| ZC3H13 | Contributes to the nuclear localization of MTC | [20] | |
| IME4 | Mediates m6A modification of yeast mRNA | [21] | |
| MUM2 | Mediates m6A modification of yeast mRNA | [21] | |
| m6A erasers | FTO | Removes m6A modification to promote mRNA splicing and translation | [22] |
| ALKBH5 | Removes m6A modification to promote mRNA splicing, mRNA nuclear output, and long 3′-UTR mRNA production | [23] | |
| m6A readers | YTHDF1 | Promotes mRNA translation and protein synthesis | [24] |
| YTHDF2 | Reduces mRNA stability and regulates mRNA localization | [24] | |
| YTHDF3 | Interacts with YTHDF1 to promote mRNA translation or assists YTHDF2-mediated RNA degradation | [24] | |
| YTHDC1 | Contributes to RNA splicing, export, and transcriptional silencing | [25] | |
| YTHDC2 | Promotes the translation of target RNA but reduces their abundance | [26] | |
| eIF3 | Promotes mRNA translation | [27] | |
| IGF2BPs | Enhances the stability and translation of target RNA | [28] | |
| HNRNPA2B1 | Regulates alternative splicing and primary microRNA processing | [29] | |
| HNRNPC/G | Mediates mRNA splicing | [29] |
| Disease | Regulator | Target | Function | References |
|---|---|---|---|---|
| Obesity | FTO | SRSF2, RUNX1T1 | FTO depletion induces preadipocyte differentiation by regulating the RNA binding ability of SRSF2 and exonic splicing of RUNX1T1 | [99] |
| FTO | PPARG | FTO promotes the differentiation of BMSCs into adipocytes by increasing the expression of PPARG | [100] | |
| YTHDF1 | MTCH2 | YTHDF1 facilitates MTCH2 mRNA translation and adipogenesis | [102] | |
| METTL3/YTHDF2 | CCND1 | METTL3/YTHDF2 mediated CCND1 degradation inhibits cell-cycle progression and adipogenesis | [103] | |
| DM | FTO | FOXO1, G6PC, DGAT2 | FTO promotes the expression of FOXO1, G6PC and DGAT2, which results in hyperglycemic phenotype | [106] |
| METTL3 | FASN | METTL3 facilitates fatty acid metabolism and insulin resistance by increasing the expression of FASN | [75] | |
| YTHDC2 | FASN, ACC, SREBP-1c, SCD-1 | YTHDC2 suppresses liver steatosis by decreasing of mRNA stability of lipogenic genes | [78] | |
| IMP2/IGF2BP2 | PDX1 | IMP2/IGF2BP2 promotes pancreatic β-cell proliferation and insulin secretion by enhancing PDX1 expression | [107] | |
| METTL3/ METTL14 | Pdx1, MafA, Nkx6.1, GLUT2 | METTL3/14 promotes β-cell proliferation and functional maturation by increasing the expression of Pdx1, MafA, Nkx6.1, and GLUT2 | [108,109] | |
| METTL14/ YTHDF2 | LncRNA TINCR | METTL14/YTHDF2 suppresses pyroptosis and diabetic cardiomyopathy by mediating the degradation of lncRNA TINCR | [111] | |
| IMP2 | p53 | LncRNA Airn prevents the development of cardiac fibrosis in the diabetic heart via the IMP2-p53 axis | [112] | |
| METTL3 | LncRNA SNHG7 | METTL3 regulates endothelial-mesenchymal transition in diabetic retinopathy by enhancing the stability of lncRNA SNHG7 | [113] | |
| Hyperlipemia | METTL3 | PRKD2 | METTL3 inhibits PRKD2 expression and the activity of GLUT4/IRS-1/AKT signaling, which results in high levels of glucose and TG | [115] |
| FTO | CREB, AMPK | FTO modulates the activity of the CREB signaling pathway and AMPK pathway | [119,120] | |
| NAFLD | METTL3/YTHDF1 | Rubicon | METTL3/YTHDF1 promotes Rubicon expression and hepatic lipid deposition | [123] |
| FTO | SREBF1, SCD1 | FTO induces lipid accumulation by increasing the expression of SREBF1 and SCD1 | [124] | |
| FTO | IL-17 | FTO promotes IL-17 expression and chronic liver inflammation | [125] | |
| ALKBH5 | LINC01468 | ALKBH5 drives lipogenesis and NAFLD-HCC progression by increasing the RNA stability of LINC01468 | [126] | |
| YTHDF3 | PRDX3 | YTHDF3-mediated PRDX3 translation alleviates liver fibrosis | [127] | |
| ALKBH5 | PTCH1 | ALKBH5 ameliorated liver fibrosis and suppressed HSCs activation via triggering PTCH1 activation | [128] | |
| Hypertension | FTO rs9939609 | MC4R rs17782313 | FTO rs9939609 is positively correlated with MC4R rs17782313, which is inversely correlated with diastolic blood pressure in male patients | [134] |
| AS | METTL14 | lncRNA ZFAS1 | METTL14 limits cholesterol efflux by mediating the m6A modification of lncRNA ZFAS1 | [87] |
| METTL3 | PI3K/AKT pathway | METTL3 facilitates HCASMC proliferation and migration by activating PI3K/AKT pathway | [137] | |
| METTL3/IGF2BP1 | JAK2/STAT3 pathway | METTL3/IGF2BP1 promoted HUVEC proliferation, migration, and angiogenesis by positively regulating JAK2/STAT3 pathway | [139] | |
| METTL14 | FOXO1 | METTL14 aggravates endothelial inflammation and AS by increasing the expression of FOXO1 | [140] | |
| ALKBH5 | SPHK1 | ALKBH5 helps in the maintenance of angiogenesis in endothelial cells following acute ischemic stress via increased SPHK1 expression | [141] | |
| METTL14 | miR-19a | METTL14 enhances the proliferation and invasion of ASVEC by promoting the processing of mature miR-19a | [142] | |
| METTL14 | NF-κB/IL-6 pathway | Mettl14 mediates the inflammatory response of macrophages through the NF-κB/IL-6 pathway | [143] | |
| METTL3 | circ_0029589 | METTL3 induces macrophage pyroptosis and inflammation by inhibiting circ_0029589 | [144] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, H.; Li, Y.; Huang, L.; Fang, M.; Xu, S. The Epigenetic Regulation of RNA N6-Methyladenosine Methylation in Glycolipid Metabolism. Biomolecules 2023, 13, 273. https://doi.org/10.3390/biom13020273
Yang H, Li Y, Huang L, Fang M, Xu S. The Epigenetic Regulation of RNA N6-Methyladenosine Methylation in Glycolipid Metabolism. Biomolecules. 2023; 13(2):273. https://doi.org/10.3390/biom13020273
Chicago/Turabian StyleYang, Haiqing, Yuting Li, Linying Huang, Miaochun Fang, and Shun Xu. 2023. "The Epigenetic Regulation of RNA N6-Methyladenosine Methylation in Glycolipid Metabolism" Biomolecules 13, no. 2: 273. https://doi.org/10.3390/biom13020273
APA StyleYang, H., Li, Y., Huang, L., Fang, M., & Xu, S. (2023). The Epigenetic Regulation of RNA N6-Methyladenosine Methylation in Glycolipid Metabolism. Biomolecules, 13(2), 273. https://doi.org/10.3390/biom13020273






