Emergent Role of IFITM1/3 towards Splicing Factor (SRSF1) and Antigen-Presenting Molecule (HLA-B) in Cervical Cancer
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Western Blotting
2.3. Antibodies
2.4. Trichloroacetic Acid (TCA) Precipitation
2.5. Immunofluorescence
2.6. RT-qPCR
2.7. Proximity Ligation Assay (PLA)
2.8. RNA In Situ Hybridization-PLA (rISH-PLA)
2.9. Bioinformatic Analysis of the RNA Sequencing
2.10. Sucrose Gradient Sedimentation
2.11. Cloning, Transfection and Affinity Purification of IFITM1 from Heavy Isotope Labeled Cells
2.12. Peptide Generation Using FASP
2.13. LC-MS/MS Analysis of SILAC Labeled Samples
2.14. Database Searching and Analysis
3. Results
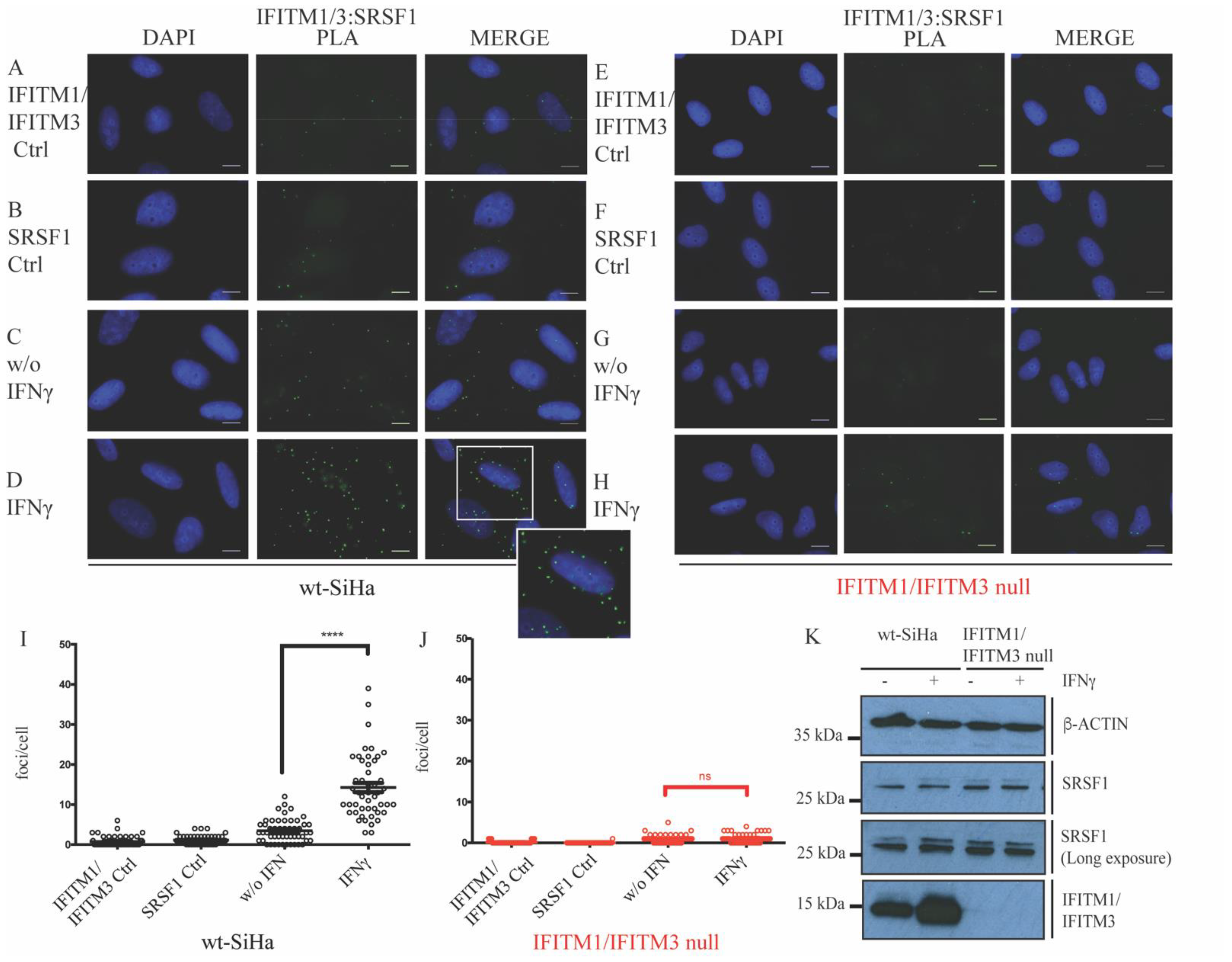
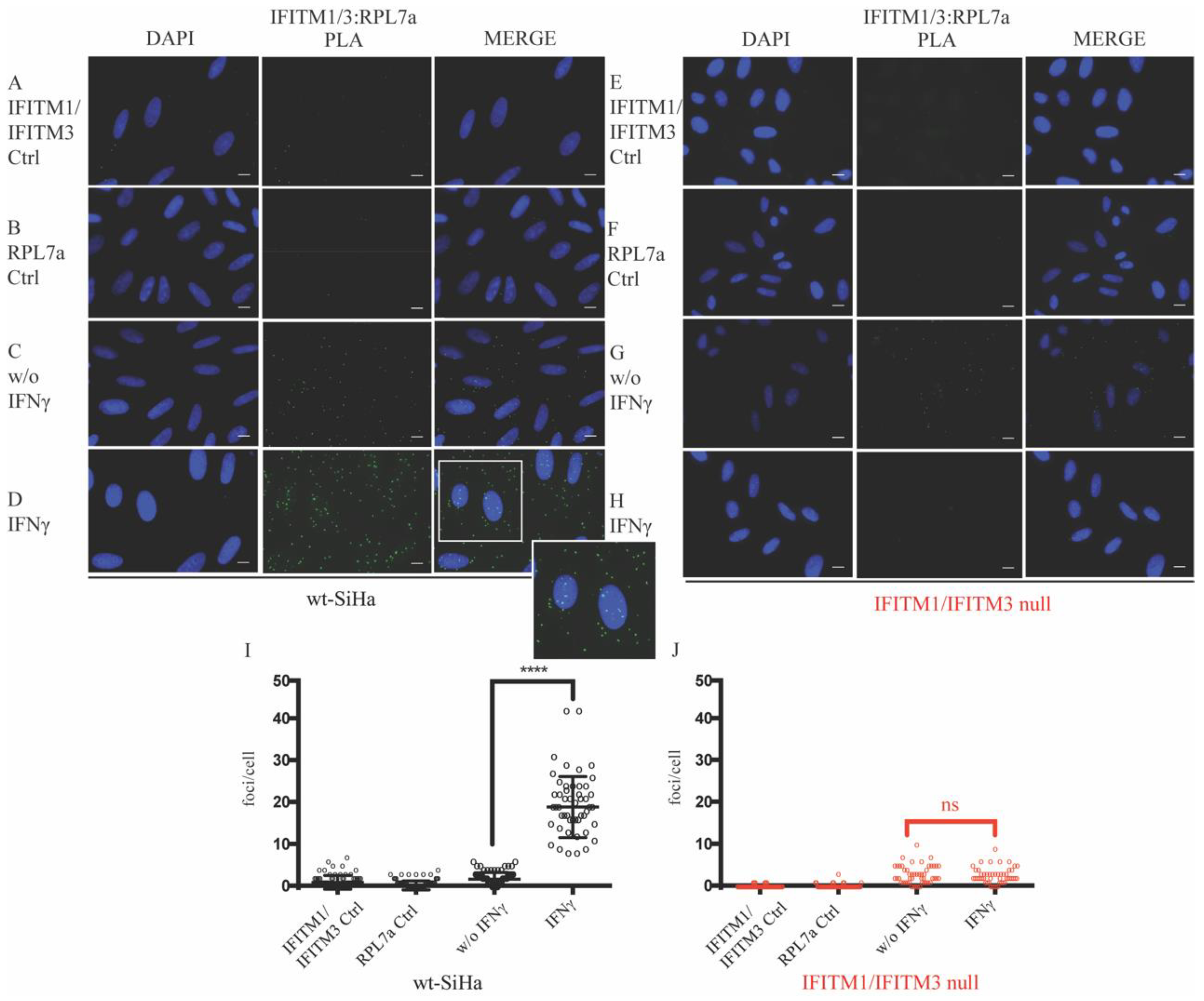
3.1. Defining New Protein–Protein Association with IFITM Proteins
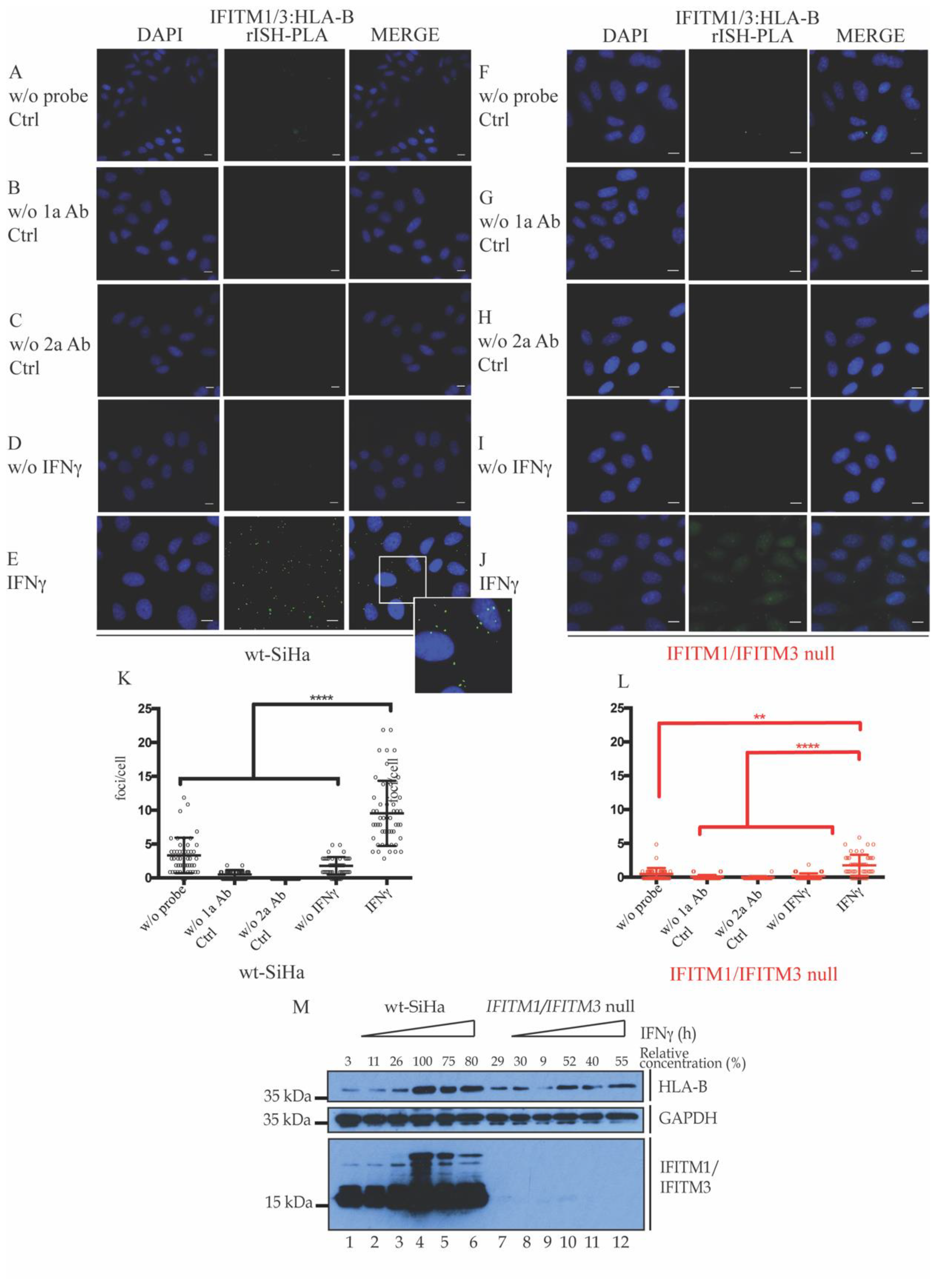
3.2. Association of IFITM1/3 Proteins with HLA-B RNA
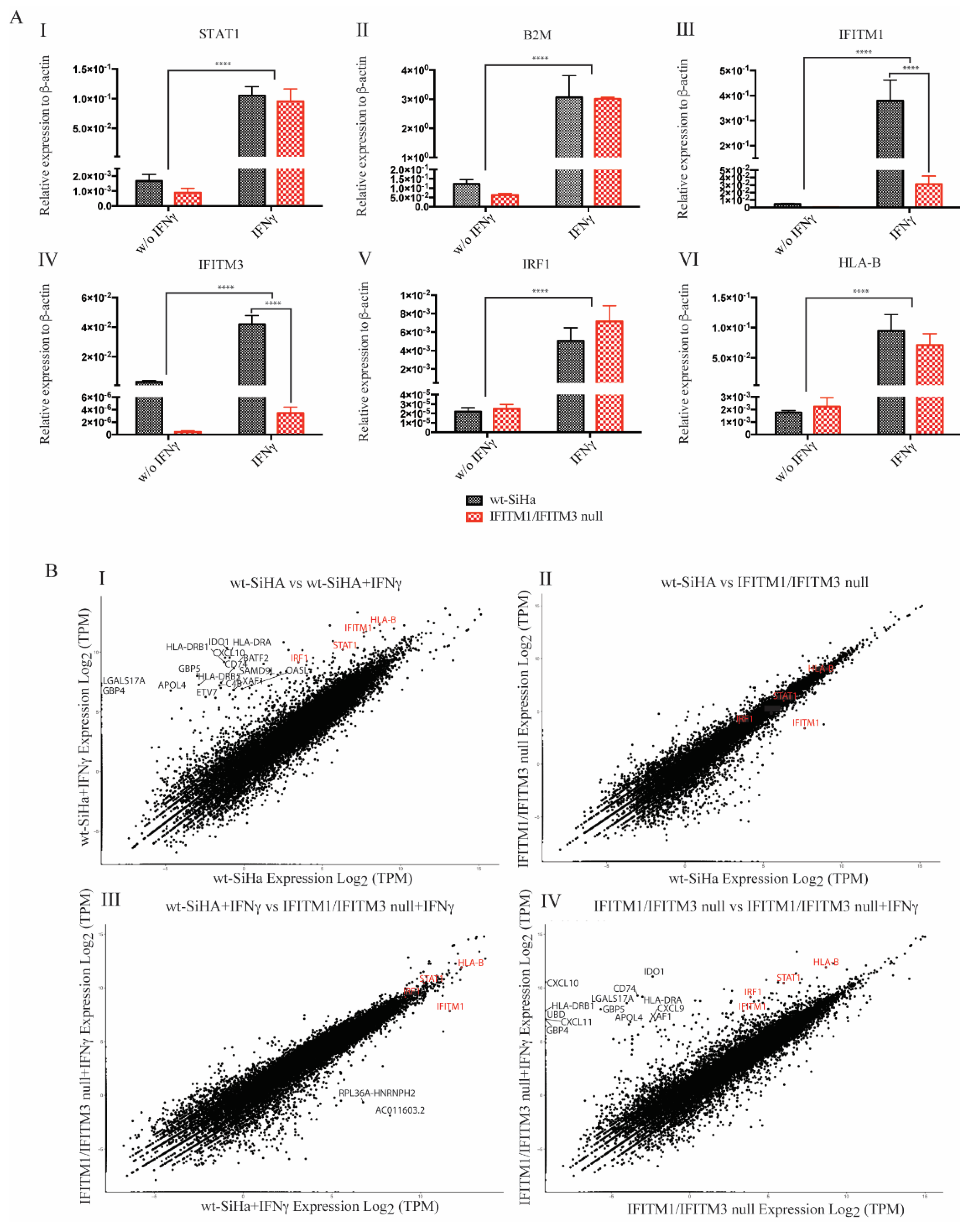
3.3. Exploring RNA Transcripts in wt-SiHA and IFITM1/IFITM3 Null Cells
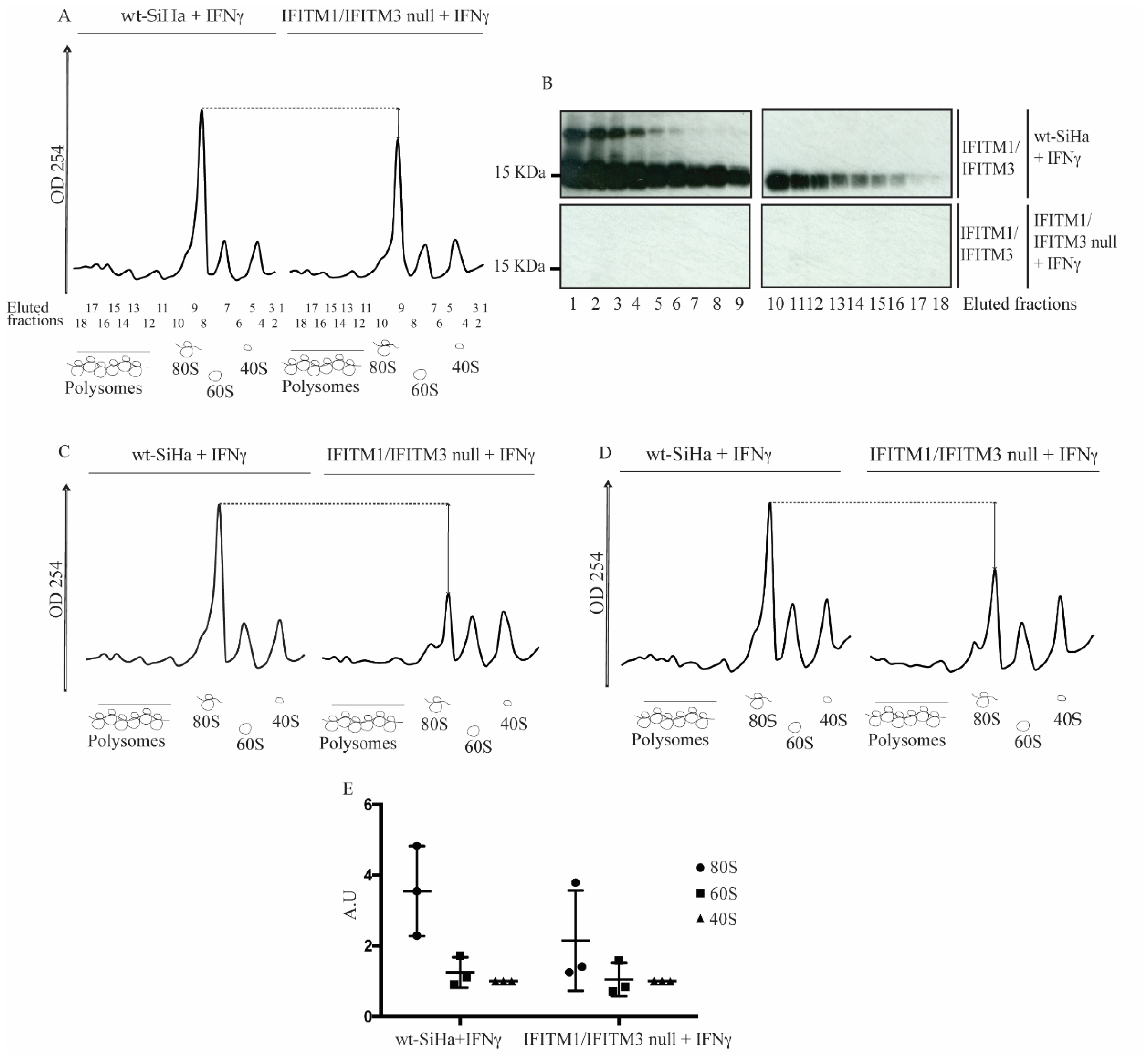
3.4. Reduction in 80S Ribosomal RNA Levels in the IFITM1/IFITM3 Null Cells Using Sucrose Gradient Sedimentation
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Robichaud, N.; Sonenberg, N.; Ruggero, D.; Schneider, R.J. Translational Control in Cancer. Cold Spring Harb. Perspect. Biol. 2019, 11, a032896. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, Y.; Ruggero, D. The Role of Translation Control in Tumorigenesis and Its Therapeutic Implications. Annu. Rev. Cancer Biol. 2020, 4, 437–457. [Google Scholar] [CrossRef] [Green Version]
- Platanias, L.C. Mechanisms of type-I- and type-II-interferon-mediated signalling. Nat. Rev. Immunol. 2005, 5, 375–386. [Google Scholar] [CrossRef] [PubMed]
- Marth, C.; Windbichler, G.; Hausmaninger, H.; Petru, E.; Estermann, K.; Pelzer, A.; Mueller-Holzner, E. Interferon-gamma in combination with carboplatin and paclitaxel as a safe and effective first-line treatment option for advanced ovarian cancer: Results of a phase I/II study. Int. J. Gynecol. Cancer 2006, 16, 1522–1528. [Google Scholar] [CrossRef]
- Pujade-Lauraine, E.; Guastalla, J.P.; Colombo, N.; DeVillier, P.; François, E.; Fumoleau, P.; Monnier, A.; Nooy, M.; Mignot, L.; Bugat, R.; et al. Intraperitoneal recombinant interferon gamma in ovarian cancer patients with residual disease at second-look laparotomy. J. Clin. Oncol. 1996, 14, 343–350. [Google Scholar] [CrossRef] [PubMed]
- Windbichler, G.H.; Hausmaninger, H.; Stummvoll, W.; Graf, A.H.; Kainz, C.; Lahodny, J.; Denison, U.; Müller-Holzner, E.; Marth, C. Interferon-gamma in the first-line therapy of ovarian cancer: A randomized phase III trial. Br. J. Cancer 2000, 82, 1138–1144. [Google Scholar] [CrossRef] [Green Version]
- Alberts, D.S.; Marth, C.; Alvarez, R.D.; Johnson, G.; Bidzinski, M.; Kardatzke, D.R.; Bradford, W.Z.; Loutit, J.; Kirn, D.H.; Clouser, M.C.; et al. Randomized phase 3 trial of interferon gamma-1b plus standard carboplatin/paclitaxel versus carboplatin/paclitaxel alone for first-line treatment of advanced ovarian and primary peritoneal carcinomas: Results from a prospectively designed analysis of progression-free survival. Gynecol. Oncol. 2008, 109, 174–181. [Google Scholar] [CrossRef]
- Tamura, K.; Makino, S.; Araki, Y.; Imamura, T.; Seita, M. Recombinant interferon beta and gamma in the treatment of adult T-cell leukemia. Cancer 1987, 59, 1059–1062. [Google Scholar] [CrossRef]
- Van der Kooij, M.K.; Verdegaal, E.M.E.; Visser, M.; Visser, M.; de Bruin, L.; van der Minne, C.E.; Meij, P.M.; Roozen, I.C.F.M.; Jonker, M.A.; van den Bosch, S.; et al. Phase I/II study protocol to assess safety and efficacy of adoptive cell therapy with anti-PD-1 plus low-dose pegylated-interferon-alpha in patients with metastatic melanoma refractory to standard of care treatments: The ACTME trial. BMJ Open 2020, 10, e044036. [Google Scholar] [CrossRef]
- Ives, N.J.; Suciu, S.; Eggermont, A.M.M.; Kirkwood, J.; Lorigan, P.; Markovic, S.N.; Garbe, C.; Wheatley, K.; Bufalino, R.; Cameron, D.; et al. Adjuvant interferon-α for the treatment of high-risk melanoma: An individual patient data meta-analysis. Eur. J. Cancer 2017, 82, 171–183. [Google Scholar] [CrossRef]
- Parker, B.S.; Rautela, J.; Hertzog, P.J. Antitumour actions of interferons: Implications for cancer therapy. Nat. Rev. Cancer 2016, 16, 131–144. [Google Scholar] [CrossRef]
- Budhwani, M.; Mazzieri, R.; Dolcetti, R. Plasticity of Type I Interferon-Mediated Responses in Cancer Therapy: From Anti-tumor Immunity to Resistance. Front. Oncol. 2018, 8, 322. [Google Scholar] [CrossRef] [Green Version]
- Green, D.S.; Nunes, A.T.; Annunziata, C.M.; Zoon, K.C. Monocyte and interferon based therapy for the treatment of ovarian cancer. Cytokine Growth Factor Rev. 2016, 29, 109–115. [Google Scholar] [CrossRef] [Green Version]
- Booy, S.; Hofland, L.; van Eijck, C. Potentials of Interferon Therapy in the Treatment of Pancreatic Cancer. J. Interf. Cytokine Res. 2015, 35, 327–339. [Google Scholar] [CrossRef]
- Wang, B.X.; Rahbar, R.; Fish, E.N. Interferon: Current Status and Future Prospects in Cancer Therapy. J. Interf. Cytokine Res. 2011, 31, 545–552. [Google Scholar] [CrossRef] [PubMed]
- Kolosenko, I.; Fryknäs, M.; Forsberg, S.; Johnsson, P.; Cheon, H.; Holvey-Bates, E.G.; Edsbäcker, E.; Pellegrini, P.; Rassoolzadeh, H.; Brnjic, S.; et al. Cell crowding induces interferon regulatory factor 9, which confers resistance to chemotherapeutic drugs. Int. J. Cancer 2015, 136, E51–E61. [Google Scholar] [CrossRef] [PubMed]
- Wallace, T.A.; Martin, D.N.; Ambs, S. Interactions among genes, tumor biology and the environment in cancer health disparities: Examining the evidence on a national and global scale. Carcinogenesis 2011, 32, 1107–1121. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weichselbaum, R.R.; Ishwaran, H.; Yoon, T.; Nuyten, D.S.A.; Baker, S.W.; Khodarev, N.; Su, A.W.; Shaikh, A.Y.; Roach, P.; Kreike, B.; et al. An interferon-related gene signature for DNA damage resistance is a predictive marker for chemotherapy and radiation for breast cancer. Proc. Natl. Acad. Sci. USA 2008, 105, 18490–18495. [Google Scholar] [CrossRef] [Green Version]
- Cheon, H.; Borden, E.C.; Stark, G.R. Interferons and Their Stimulated Genes in the Tumor Microenvironment. Semin. Oncol. 2014, 41, 156–173. [Google Scholar] [CrossRef] [Green Version]
- Jorgovanovic, D.; Song, M.; Wang, L.; Zhang, Y. Roles of IFN-γ in tumor progression and regression: A review. Biomark. Res. 2020, 8, 49. [Google Scholar] [CrossRef]
- Bekisz, J.; Schmeisser, H.; Hernandez, J.; Goldman, N.D.; Zoon, K.C. Mini ReviewHuman Interferons Alpha, Beta and Omega. Growth Factors 2004, 22, 243–251. [Google Scholar] [CrossRef] [Green Version]
- Borden, E.C.; Sen, G.C.; Uze, G.; Silverman, R.H.; Ransohoff, R.M.; Foster, G.R.; Stark, G.R. Interferons at age 50: Past, current and future impact on biomedicine. Nat. Rev. Drug Discov. 2007, 6, 975–990. [Google Scholar] [CrossRef] [PubMed]
- Darnell, J.E., Jr.; Kerr, I.M.; Stark, G.R. Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science 1994, 264, 1415–1421. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Williams, B.R.G. Transcriptional regulation of interferon-stimulated genes. JBIC J. Biol. Inorg. Chem. 1991, 200, 111–121. [Google Scholar] [CrossRef]
- Bailey, C.C.; Zhong, G.; Huang, I.-C.; Farzan, M. IFITM-Family Proteins: The Cell's First Line of Antiviral Defense. Annu. Rev. Virol. 2014, 1, 261–283. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jia, R.; Ding, S.; Pan, Q.; Liu, S.-L.; Qiao, W.; Liang, C. The C-Terminal Sequence of IFITM1 Regulates Its Anti-HIV-1 Activity. PLoS ONE 2015, 10, e0118794. [Google Scholar] [CrossRef] [Green Version]
- Weston, S.; Czieso, S.; White, I.J.; Smith, S.; Kellam, P.; Marsh, M. A Membrane Topology Model for Human Interferon Inducible Transmembrane Protein 1. PLoS ONE 2014, 9, e104341. [Google Scholar] [CrossRef]
- Amini-Bavil-Olyaee, S.; Choi, Y.J.; Lee, J.H.; Shi, M.; Huang, I.-C.; Farzan, M.; Jung, J.U. The Antiviral Effector IFITM3 Disrupts Intracellular Cholesterol Homeostasis to Block Viral Entry. Cell Host Microbe 2013, 13, 452–464. [Google Scholar] [CrossRef] [Green Version]
- Yu, F.; Xie, D.; Ng, S.S.; Lum, C.T.; Cai, M.-Y.; Cheung, W.K.; Kung, H.-F.; Lin, G.; Wang, X.; Lin, M.C. IFITM1 promotes the metastasis of human colorectal cancer via CAV-1. Cancer Lett. 2015, 368, 135–143. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Yang, G.; Hu, G. Binding of IFITM1 enhances the inhibiting effect of caveolin-1 on ERK activation. Acta Biochim. Biophys. Sin. 2009, 41, 488–494. [Google Scholar] [CrossRef] [Green Version]
- Narayana, S.K.; Helbig, K.J.; McCartney, E.M.; Eyre, N.S.; Bull, R.A.; Eltahla, A.; Lloyd, A.R.; Beard, M.R. The Interferon-induced Transmembrane Proteins, IFITM1, IFITM2, and IFITM3 Inhibit Hepatitis C Virus Entry. J. Biol. Chem. 2015, 290, 25946–25959. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, X.; Li, J.; Winkler, C.A.; An, P.; Guo, J.-T. IFITM Genes, Variants, and Their Roles in the Control and Pathogenesis of Viral Infections. Front. Microbiol. 2019, 9, 3228. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, T.H.; Schreiber, K.; Arina, A.; Khodarev, N.N.; Efimova, E.V.; Rowley, D.A.; Weichselbaum, R.R.; Schreiber, H. Progression of cancer from indolent to aggressive despite antigen retention and increased expression of interferon-gamma inducible genes—PubMed. Cancer Immunol. 2011, 11, 2. [Google Scholar]
- Borg, D.; Hedner, C.; Gaber, A.; Nodin, B.; Fristedt, R.; Jirström, K.; Eberhard, J.; Johnsson, A. Expression of IFITM1 as a prognostic biomarker in resected gastric and esophageal adenocarcinoma. Biomark. Res. 2016, 4, 10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Györffy, B.; Dietel, M.; Fekete, T.; Lage, H. A snapshot of microarray-generated gene expression signatures associated with ovarian carcinoma. Int. J. Gynecol. Cancer 2008, 18, 1215–1233. [Google Scholar] [CrossRef] [PubMed]
- Sari, I.N.; Yang, Y.-G.; Phi, L.; Kim, H.; Baek, M.J.; Jeong, D.; Kwon, H.Y. Interferon-Induced Transmembrane Protein 1 (IFITM1) Is Required for the Progression of Colorectal Cancer. Oncotarget 2016, 7, 86039. [Google Scholar] [CrossRef] [Green Version]
- Fan, J.; Peng, Z.; Zhou, C.; Qiu, G.; Tang, H.; Sun, Y.; Wang, X.; Li, Q.; Le, X.; Xie, K. Gene-expression profiling in Chinese patients with colon cancer by coupling experimental and bioinformatic genomewide gene-expression analyses. Cancer 2008, 113, 266–275. [Google Scholar] [CrossRef]
- Liu, Y.; Lu, R.; Cui, W.; Pang, Y.; Liu, C.; Cui, L.; Qian, T.; Quan, L.; Dai, Y.; Jiao, Y.; et al. High IFITM3 expression predicts adverse prognosis in acute myeloid leukemia. Cancer Gene Ther. 2020, 27, 38–44. [Google Scholar] [CrossRef]
- Wang, H.; Tang, F.; Bian, E.; Zhang, Y.; Ji, X.; Yang, Z.; Zhao, B. IFITM3/STAT3 axis promotes glioma cells invasion and is modulated by TGF-β. Mol. Biol. Rep. 2020, 47, 433–441. [Google Scholar] [CrossRef]
- Ogony, J.; Choi, H.J.; Lui, A.; Cristofanilli, M.; Lewis-Wambi, J. Interferon-induced transmembrane protein 1 (IFITM1) overexpression enhances the aggressive phenotype of SUM149 inflammatory breast cancer cells in a signal transducer and activator of transcription 2 (STAT2)-dependent manner. Breast Cancer Res. 2016, 18, 25. [Google Scholar] [CrossRef] [Green Version]
- Khodarev, N.N.; Roizman, B.; Weichselbaum, R.R. Molecular Pathways: Interferon/Stat1 Pathway: Role in the Tumor Resistance to Genotoxic Stress and Aggressive Growth. Clin. Cancer Res. 2012, 18, 3015–3021. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, W.-Y.J.; Fu, R.M.; Liang, C.; Sloan, R.D. IFITM proteins inhibit HIV-1 protein synthesis. Sci. Rep. 2018, 8, 14551. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gómez-Herranz, M.; Nekulova, M.; Faktor, J.; Hernychova, L.; Kote, S.; Sinclair, E.H.; Nenutil, R.; Vojtesek, B.; Ball, K.L.; Hupp, T.R. The effects of IFITM1 and IFITM3 gene deletion on IFNγ stimulated protein synthesis. Cell. Signal. 2019, 60, 39–56. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Sanford, J.R.; Gray, N.K.; Beckmann, K.; Cáceres, J.F. A novel role for shuttling SR proteins in mRNA translation. Genes Dev. 2004, 18, 755–768. [Google Scholar] [CrossRef] [Green Version]
- Wiśniewski, J.R.; Zougman, A.; Nagaraj, N.; Mann, M. Universal sample preparation method for proteome analysis. Nat. Methods 2009, 6, 359–362. [Google Scholar] [CrossRef]
- Pan, Z.; Chen, S.; Pan, X.; Wang, Z.; Han, H.; Zheng, W.; Wang, X.; Li, F.; Qu, S.; Shao, R. Differential gene expression identified in Uigur women cervical squamous cell carcinoma by suppression subtractive hybridization. Neoplasma 2010, 57, 123–128. [Google Scholar] [CrossRef] [Green Version]
- Kim, T.-J.; Choi, J.; Kim, W.Y.; Choi, C.H.; Lee, J.-W.; Bae, D.-S.; Son, D.-S.; Kim, J.; Park, B.K.; Ahn, G.; et al. Gene expression profiling for the prediction of lymph node metastasis in patients with cervical cancer. Cancer Sci. 2008, 99, 31–38. [Google Scholar] [CrossRef] [PubMed]
- Siegrist, F.; Ebeling, M.; Certa, U. The Small Interferon-Induced Transmembrane Genes and Proteins. J. Interf. Cytokine Res. 2011, 31, 183–197. [Google Scholar] [CrossRef] [PubMed]
- Winkler, M.; Wrensch, F.; Bosch, P.; Knoth, M.; Schindler, M.; Gärtner, S.; Pöhlmann, S. Analysis of IFITM-IFITM Interactions by a Flow Cytometry-Based FRET Assay. Int. J. Mol. Sci. 2019, 20, 3859. [Google Scholar] [CrossRef] [Green Version]
- Rahman, K.; A Coomer, C.; Majdoul, S.; Ding, S.Y.; Padilla-Parra, S.; A Compton, A. Homology-guided identification of a conserved motif linking the antiviral functions of IFITM3 to its oligomeric state. eLife 2020, 9, e58537. [Google Scholar] [CrossRef] [PubMed]
- Seyfried, N.T.; Huysentruyt, L.C.; Atwood, J.A.; Xia, Q.; Seyfried, T.N.; Orlando, R. Up-regulation of NG2 proteoglycan and interferon-induced transmembrane proteins 1 and 3 in mouse astrocytoma: A membrane proteomics approach. Cancer Lett. 2008, 263, 243–252. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Keefe, A.D.; Wilson, D.S.; Seelig, B.; Szostak, J.W. One-Step Purification of Recombinant Proteins Using a Nanomolar-Affinity Streptavidin-Binding Peptide, the SBP-Tag. Protein Expr. Purif. 2001, 23, 440–446. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haward, F.; Maslon, M.M.; Yeyati, P.L.; Bellora, N.; Hansen, J.N.; Aitken, S.; Lawson, J.; von Kriegsheim, A.; Wachten, D.; Mill, P.; et al. Nucleo-cytoplasmic shuttling of splicing factor SRSF1 is required for development and cilia function. eLife 2021, 10, e65104. [Google Scholar] [CrossRef] [PubMed]
- Jeong, S. SR Proteins: Binders, Regulators, and Connectors of RNA. Mol. Cells 2017, 40, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Maslon, M.; Heras, S.R.; Bellora, N.; Eyras, E.; Cáceres, J.F. The translational landscape of the splicing factor SRSF1 and its role in mitosis. eLife 2014, 3, e02028. [Google Scholar] [CrossRef] [Green Version]
- Cáceres, J.F.; Screaton, G.R.; Krainer, A.R. A specific subset of SR proteins shuttles continuously between the nucleus and the cytoplasm. Genes Dev. 1998, 12, 55–66. [Google Scholar] [CrossRef] [Green Version]
- Zheng, X.; Peng, Q.; Wang, L.; Zhang, X.; Huang, L.; Wang, J.; Qin, Z. Serine/arginine-rich splicing factors: The bridge linking alternative splicing and cancer. Int. J. Biol. Sci. 2020, 16, 2442–2453. [Google Scholar] [CrossRef]
- Weibrecht, I.; Leuchowius, K.-J.; Clausson, C.-M.; Conze, T.; Jarvius, M.; Howell, W.M.; Kamali-Moghaddam, M.; Söderberg, O. Proximity ligation assays: A recent addition to the proteomics toolbox. Expert Rev. Proteom. 2010, 7, 401–409. [Google Scholar] [CrossRef]
- Roussis, I.M.; Myers, F.A.; Scarlett, G.P. RNA Whole-Mount In Situ Hybridization Proximity Ligation Assay (rISH-PLA), an Assay for Detecting RNA-Protein Complexes in Intact Cells. Curr. Protoc. Cell Biol. 2017, 74, 17.20.1–17.20.10. [Google Scholar] [CrossRef]
- Tuladhar, R.; Yeu, Y.; Piazza, J.T.; Tan, Z.; Clemenceau, J.R.; Wu, X.; Barrett, Q.; Herbert, J.; Mathews, D.H.; Kim, J.; et al. CRISPR-Cas9-based mutagenesis frequently provokes on-target mRNA misregulation. Nat. Commun. 2019, 10, 4056. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feng, J.; Cao, Z.; Wang, L.; Wan, Y.; Peng, N.; Wang, Q.; Chen, X.; Zhou, Y.; Zhu, Y. Inducible GBP5 Mediates the Antiviral Response via Interferon-Related Pathways during Influenza A Virus Infection. J. Innate Immun. 2017, 9, 419–435. [Google Scholar] [CrossRef] [PubMed]
- Mach, B.; Steimle, V.; Martinez-Soria, E.; Reith, W. Regulation of MHC class II genes: Lessons from a Disease. Annu. Rev. Immunol. 1996, 14, 301–331. [Google Scholar] [CrossRef] [PubMed]
- Soejima, K.; Rollins, B.J. A Functional IFN-γ-Inducible Protein-10/CXCL10-Specific Receptor Expressed by Epithelial and Endothelial Cells That Is Neither CXCR3 Nor Glycosaminoglycan. J. Immunol. 2001, 167, 6576–6582. [Google Scholar] [CrossRef] [Green Version]
- Erdal, E.; Haider, S.; Rehwinkel, J.; Harris, A.L.; McHugh, P.J. A prosurvival DNA damage-induced cytoplasmic interferon response is mediated by end resection factors and is limited by Trex1. Genes Dev. 2017, 31, 353–369. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Robledo, S.; Idol, R.A.; Crimmins, D.L.; Ladenson, J.H.; Mason, P.J.; Bessler, M. The role of human ribosomal proteins in the maturation of rRNA and ribosome production. RNA 2008, 14, 1918–1929. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, W.-C.; Liu, H.-W.; Lin, A. Human ribosomal protein L7 displays an ER binding property and is involved in ribosome-ER association. FEBS Lett. 2007, 581, 651–657. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, J.; Robinson, M.E.; Ma, N.; Artadji, D.; Ahmed, M.A.; Xiao, G.; Sadras, T.; Deb, G.; Winchester, J.; Cosgun, K.N.; et al. IFITM3 functions as a PIP3 scaffold to amplify PI3K signalling in B cells. Nature 2020, 588, 491–497. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Shen, Z.; Man, D.; Ruan, H.; Huang, S. miR-152-3p Affects the Progression of Colon Cancer via the KLF4/IFITM3 Axis. Comput. Math. Methods Med. 2020, 2020, 8209504. [Google Scholar] [CrossRef]
- Yang, J.; Li, L.; Xi, Y.; Sun, R.; Wang, H.; Ren, Y.; Zhao, L.; Wang, X.; Li, X. Combination of IFITM1 knockdown and radiotherapy inhibits the growth of oral cancer. Cancer Sci. 2018, 109, 3115–3128. [Google Scholar] [CrossRef] [Green Version]
- Yang, G.; Xu, Y.; Chen, X.; Hu, G. IFITM1 plays an essential role in the antiproliferative action of interferon-γ. Oncogene 2007, 26, 594–603. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, W.; Zhao, Z.; Yi, X.; Zuo, Q.; Li, H.; Guo, X.; Li, D.; He, H.; Pan, Z.; Fan, P.; et al. Down-regulation of IFITM1 and its growth inhibitory role in cervical squamous cell carcinoma. Cancer Cell Int. 2017, 17, 88. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Das, S.; Krainer, A.R. Emerging Functions of SRSF1, Splicing Factor and Oncoprotein, in RNA Metabolism and Cancer. Mol. Cancer Res. 2014, 12, 1195–1204. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Neumann, F.; Krawinkel, U. Constitutive Expression of Human Ribosomal Protein L7 Arrests the Cell Cycle in G1and Induces Apoptosis in Jurkat T-Lymphoma Cells. Exp. Cell Res. 1997, 230, 252–261. [Google Scholar] [CrossRef]
- Neumann, F.; Hemmerich, P.; Von Mikecz, A.; Peter, H.-H.; Krawinkel, U. Human ribosomal protein L7 inhibits cell-free translation in reticulocyte lysates and affects the expression of nuclear proteins upon stable transfection into Jurkat T-lymphoma cells. Nucleic Acids Res. 1995, 23, 195–202. [Google Scholar] [CrossRef]
- Connor, M.E.; Stern, P.L. Loss of MHC class-I expression in cervical carcinomas. Int. J. Cancer 1990, 46, 1029–1034. [Google Scholar] [CrossRef]
- Cromme, F.V.; Meijer, C.J.; Snijders, P.J.; Uyterlinde, A.M.; Kenemans, P.; Helmerhorst, T.J.M.; Stern, P.L.; Brule, A.J.V.D.; Walboomers, J.M. Analysis of MHC class I and II expression in relation to presence of HPV genotypes in premalignant and malignant cervical lesions. Br. J. Cancer 1993, 67, 1372–1380. [Google Scholar] [CrossRef] [Green Version]
- Honma, S.; Tsukada, S.; Honda, S.; Nakamura, M.; Takakuwa, K.; Maruhashi, T.; Kodama, S.; Kanazawa, K.; Takahashi, T.; Tanaka, K. Biological-clinical significance of selective loss of HLA-class-I allelic product expression in squamous-cell carcinoma of the uterine cervix. Int. J. Cancer 1994, 57, 650–655. [Google Scholar] [CrossRef]
- Torres, L.M.; Cabrera, T.; Concha, A.; Oliva, M.R.; Ruiz-Cabello, F.; Garrido, F. HLA class I expression and HPV-16 sequences in premalignant and malignant lesions of the cervix. Tissue Antigens 1993, 41, 65–71. [Google Scholar] [CrossRef]
- Koopman, L.A.; Corver, W.E.; van der Slik, A.R.; Giphart, M.J.; Fleuren, G.J. Multiple Genetic Alterations Cause Frequent and Heterogeneous Human Histocompatibility Leukocyte Antigen Class I Loss in Cervical Cancer. J. Exp. Med. 2000, 191, 961–976. [Google Scholar] [CrossRef]
- Ferns, D.M.; Heeren, A.M.; Samuels, S.; Bleeker, M.C.G.; de Gruijl, T.D.; Kenter, G.G.; Jordanova, E.S. Classical and non-classical HLA class I aberrations in primary cervical squamous- and adenocarcinomas and paired lymph node metastases. J. Immunother. Cancer 2016, 4, 78. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cromme, F.V.; Airey, J.; Heemels, M.T.; Ploegh, H.L.; Keating, P.J.; Stern, P.L.; Meijer, C.J.; Walboomers, J.M. Loss of transporter protein, encoded by the TAP-1 gene, is highly correlated with loss of HLA expression in cervical carcinomas. J. Exp. Med. 1994, 179, 335–340. [Google Scholar] [CrossRef] [PubMed] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gómez-Herranz, M.; Faktor, J.; Yébenes Mayordomo, M.; Pilch, M.; Nekulova, M.; Hernychova, L.; Ball, K.L.; Vojtesek, B.; Hupp, T.R.; Kote, S. Emergent Role of IFITM1/3 towards Splicing Factor (SRSF1) and Antigen-Presenting Molecule (HLA-B) in Cervical Cancer. Biomolecules 2022, 12, 1090. https://doi.org/10.3390/biom12081090
Gómez-Herranz M, Faktor J, Yébenes Mayordomo M, Pilch M, Nekulova M, Hernychova L, Ball KL, Vojtesek B, Hupp TR, Kote S. Emergent Role of IFITM1/3 towards Splicing Factor (SRSF1) and Antigen-Presenting Molecule (HLA-B) in Cervical Cancer. Biomolecules. 2022; 12(8):1090. https://doi.org/10.3390/biom12081090
Chicago/Turabian StyleGómez-Herranz, Maria, Jakub Faktor, Marcos Yébenes Mayordomo, Magdalena Pilch, Marta Nekulova, Lenka Hernychova, Kathryn L. Ball, Borivoj Vojtesek, Ted R. Hupp, and Sachin Kote. 2022. "Emergent Role of IFITM1/3 towards Splicing Factor (SRSF1) and Antigen-Presenting Molecule (HLA-B) in Cervical Cancer" Biomolecules 12, no. 8: 1090. https://doi.org/10.3390/biom12081090
APA StyleGómez-Herranz, M., Faktor, J., Yébenes Mayordomo, M., Pilch, M., Nekulova, M., Hernychova, L., Ball, K. L., Vojtesek, B., Hupp, T. R., & Kote, S. (2022). Emergent Role of IFITM1/3 towards Splicing Factor (SRSF1) and Antigen-Presenting Molecule (HLA-B) in Cervical Cancer. Biomolecules, 12(8), 1090. https://doi.org/10.3390/biom12081090






