Keeping Cell Death Alive: An Introduction into the French Cell Death Research Network
Abstract
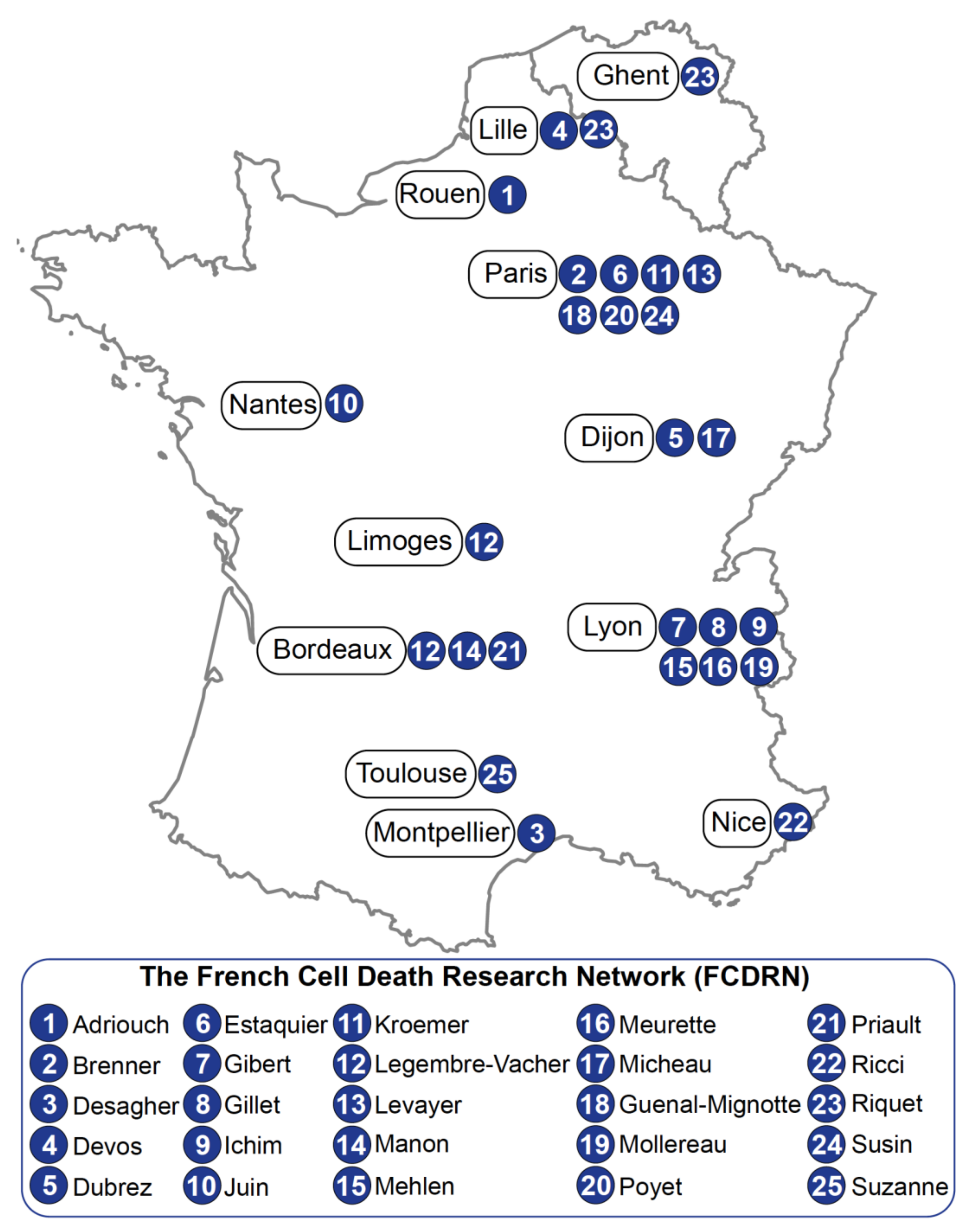
1. General Introduction
2. Team Work
2.1. Team Adriouch
2.2. Team Brenner
2.3. Team Desagher
2.4. Team Devos
2.5. Team Dubrez
2.6. Team Estaquier
2.7. Team Gibert
2.8. Team Gillet
2.9. Team Guenal-Mignotte
2.10. Team Ichim
2.11. Team Juin
2.12. Team Kroemer
2.13. Team Legembre-Vacher
2.14. Team Levayer
2.15. Team Manon
2.16. Team Mehlen
2.17. Team Meurette
2.18. Team Micheau
2.19. Team Mollereau
2.20. Team Priault
2.21. Team Ricci
2.22. Team Poyet
2.23. Team Riquet
2.24. Team Suzanne
2.25. Team Susin
3. Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Green, D.R. A Matter of Life and Death. Cold Spring Harb. Perspect. Biol. 2022, 14, a041004. [Google Scholar] [CrossRef] [PubMed]
- Ichim, G.; Tait, S.W.G. A fate worse than death: Apoptosis as an oncogenic process. Nat. Cancer 2016, 16, 539–548. [Google Scholar] [CrossRef] [PubMed]
- Lalier, L.; Vallette, F.; Manon, S. Bcl-2 Family Members and the Mitochondrial Import Machineries: The Roads to Death. Biomolecules 2022, 12, 162. [Google Scholar] [CrossRef] [PubMed]
- Zadoroznyj, A.; Dubrez, L. Cytoplasmic and Nuclear Functions of cIAP1. Biomolecules 2022, 12, 322. [Google Scholar] [CrossRef]
- Green, D.R. The Death Receptor Pathway of Apoptosis. Cold Spring Harb. Perspect. Biol. 2022, 14, a041053. [Google Scholar] [CrossRef]
- Bock, F.J.; Tait, S.W.G. Mitochondria as multifaceted regulators of cell death. Nat. Rev. Mol. Cell Biol. 2019, 21, 85–100. [Google Scholar] [CrossRef]
- Bahatyrevich-Kharitonik, B.; Medina-Guzman, R.; Flores-Cortes, A.; García-Cruzado, M.; Kavanagh, E.; Burguillos, M.A. Cell Death Related Proteins Beyond Apoptosis in the CNS. Front. Cell Dev. Biol. 2021, 9, 825747. [Google Scholar] [CrossRef]
- Juanez, K.; Ghose, P. Repurposing the Killing Machine: Non-canonical Roles of the Cell Death Apparatus in Caenorhabditis elegans Neurons. Front. Cell Dev. Biol. 2022, 10, 825124. [Google Scholar] [CrossRef]
- Colon-Plaza, S.; Su, T.T. Non-Apoptotic Role of Apoptotic Caspases in the Drosophila Nervous System. Front. Cell Dev. Biol. 2022, 10, 839358. [Google Scholar] [CrossRef]
- Green, D.R. Caspases and Their Substrates. Cold Spring Harb. Perspect. Biol. 2022, 14, a041012. [Google Scholar] [CrossRef]
- Ferrer, C.C.; Berthenet, K.; Ichim, G. Apoptosis—Fueling the oncogenic fire. FEBS J. 2020, 288, 4445–4463. [Google Scholar] [CrossRef]
- Ketelut-Carneiro, N.; Fitzgerald, K.A. Apoptosis, Pyroptosis, and Necroptosis—Oh My! The Many Ways a Cell Can Die. J. Mol. Biol. 2021, 434, 167378. [Google Scholar] [CrossRef]
- Zhang, L.; Jia, R.; Li, H.; Yu, H.; Ren, K.; Jia, S.; Li, Y.; Wang, Q. Insight into the Double-Edged Role of Ferroptosis in Disease. Biomolecules 2021, 11, 1790. [Google Scholar] [CrossRef]
- Gibert, B.; Mehlen, P. Dependence Receptors and Cancer: Addiction to Trophic Ligands. Cancer Res. 2015, 75, 5171–5175. [Google Scholar] [CrossRef]
- Newton, K.; Dixit, V.M.; Kayagaki, N. Dying cells fan the flames of inflammation. Science 2021, 374, 1076–1080. [Google Scholar] [CrossRef]
- André, S.; Picard, M.; Cezar, R.; Roux-Dalvai, F.; Alleaume-Butaux, A.; Soundaramourty, C.; Cruz, A.S.; Mendes-Frias, A.; Gotti, C.; Leclercq, M.; et al. T cell apoptosis characterizes severe Covid-19 disease. Cell Death Differ. 2022, 1–14. [Google Scholar] [CrossRef]
- LaForge, M.; Silvestre, R.; Rodrigues, V.; Garibal, J.; Campillo-Gimenez, L.; Mouhamad, S.; Monceaux, V.; Cumont, M.-C.; Rabezanahary, H.; Pruvost, A.; et al. The anti-caspase inhibitor Q-VD-OPH prevents AIDS disease progression in SIV-infected rhesus macaques. J. Clin. Investig. 2018, 128, 1627–1640. [Google Scholar] [CrossRef]
- Casares, N.; Pequignot, M.O.; Tesniere, A.; Ghiringhelli, F.; Roux, S.; Chaput, N.; Schmitt, E.; Hamai, A.; Hervas-Stubbs, S.; Obeid, M.; et al. Caspase-dependent immunogenicity of doxorubicin-induced tumor cell death. J. Exp. Med. 2005, 202, 1691–1701. [Google Scholar] [CrossRef]
- Galluzzi, L.; Buqué, A.; Kepp, O.; Zitvogel, L.; Kroemer, G. Immunogenic cell death in cancer and infectious disease. Nat. Rev. Immunol. 2017, 17, 97–111. [Google Scholar] [CrossRef]
- Kepp, O.; Tesniere, A.; Schlemmer, F.; Michaud, M.; Senovilla, L.; Zitvogel, L.; Kroemer, G. Immunogenic cell death modalities and their impact on cancer treatment. Apoptosis 2009, 14, 364–375. [Google Scholar] [CrossRef]
- Scarpitta, A.; Hacker, U.T.; Büning, H.; Boyer, O.; Adriouch, S. Pyroptotic and Necroptotic Cell Death in the Tumor Microenvironment and Their Potential to Stimulate Anti-Tumor Immune Responses. Front. Oncol. 2021, 11, 731598. [Google Scholar] [CrossRef]
- Shi, J.; Zhao, Y.; Wang, K.; Shi, X.; Wang, Y.; Huang, H.; Zhuang, Y.; Cai, T.; Wang, F.; Shao, F. Cleavage of GSDMD by inflammatory caspases determines pyroptotic cell death. Nature 2015, 526, 660–665. [Google Scholar] [CrossRef]
- Liu, X.; Zhang, Z.; Ruan, J.; Pan, Y.; Magupalli, V.G.; Wu, H.; Lieberman, J. Inflammasome-activated gasdermin D causes pyroptosis by forming membrane pores. Nature 2016, 535, 153–158. [Google Scholar] [CrossRef]
- Zhang, J.; Yang, Y.; He, W.; Sun, L. Necrosome core machinery: MLKL. Cell. Mol. Life Sci. 2016, 73, 2153–2163. [Google Scholar] [CrossRef]
- Man, S.M.; Kanneganti, T.-D. Gasdermin D: The long-awaited executioner of pyroptosis. Cell Res. 2015, 25, 1183–1184. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef]
- Hanahan, D. Hallmarks of Cancer: New Dimensions. Cancer Discov. 2022, 12, 31–46. [Google Scholar] [CrossRef]
- Hahn, V.S.; Lenihan, D.J.; Ky, B. Cancer Therapy–Induced Cardiotoxicity: Basic Mechanisms and Potential Cardioprotective Therapies. JAHA 2014, 3, e000665. [Google Scholar] [CrossRef] [PubMed]
- Mulrooney, D.A.; Yeazel, M.W.; Robison, L.L.; Kawashima, T.; Mertens, A.C.; Mitby, P.; Stovall, M.; Donaldson, S.S.; Green, D.M.; Sklar, C.A.; et al. Cardiac outcomes in a cohort of adult survivors of childhood and adolescent cancer: Retrospective analysis of the Childhood Cancer Survivor Study cohort. BMJ 2009, 339, b4606. [Google Scholar] [CrossRef]
- Liu, D.; Peyre, F.; Loissell-Baltazar, Y.A.; Courilleau, D.; Lacas-Gervais, S.; Nicolas, V.; Jacquet, E.; Dokudovskaya, S.; Taran, F.; Cintrat, J.-C.; et al. Identification of Small Molecules Inhibiting Cardiomyocyte Necrosis and Apoptosis by Autophagy Induction and Metabolism Reprogramming. Cells 2022, 11, 474. [Google Scholar] [CrossRef]
- Basu-Shrivastava, M.; Kozoriz, A.; Desagher, S.; Lassot, I. To Ubiquitinate or Not to Ubiquitinate: TRIM17 in Cell Life and Death. Cells 2021, 10, 1235. [Google Scholar] [CrossRef] [PubMed]
- Abbas, R.; Larisch, S. Killing by Degradation: Regulation of Apoptosis by the Ubiquitin-Proteasome-System. Cells 2021, 10, 3465. [Google Scholar] [CrossRef] [PubMed]
- Hollville, E.; Romero, S.E.; Deshmukh, M. Apoptotic cell death regulation in neurons. FEBS J. 2019, 286, 3276–3298. [Google Scholar] [CrossRef] [PubMed]
- Li, K.; van Delft, M.F.; Dewson, G. Too much death can kill you: Inhibiting intrinsic apoptosis to treat disease. EMBO J. 2021, 40, e107341. [Google Scholar] [CrossRef]
- Schmidt, M.F.; Gan, Z.Y.; Komander, D.; Dewson, G. Ubiquitin signalling in neurodegeneration: Mechanisms and therapeutic opportunities. Cell Death Differ. 2021, 28, 570–590. [Google Scholar] [CrossRef]
- D’Amico, F.; Mukhopadhyay, R.; Ovaa, H.; Mulder, M.P.C. Targeting TRIM Proteins: A Quest towards Drugging an Emerging Protein Class. ChemBioChem 2021, 22, 2011–2031. [Google Scholar] [CrossRef]
- Desagher, S.; Severac, D.; Lipkin, A.; Bernis, C.; Ritchie, W.; Le Digarcher, A.; Journot, L. Genes Regulated in Neurons Undergoing Transcription-dependent Apoptosis Belong to Signaling Pathways Rather than the Apoptotic Machinery. J. Biol. Chem. 2005, 280, 5693–5702. [Google Scholar] [CrossRef]
- Lassot, I.; Robbins, I.; Kristiansen, M.; Rahmeh, R.; Jaudon, F.; Magiera, M.M.; Mora, S.; Vanhille, L.; Lipkin, A.; Pettmann, B.; et al. Trim17, a novel E3 ubiquitin-ligase, initiates neuronal apoptosis. Cell Death Differ. 2010, 17, 1928–1941. [Google Scholar] [CrossRef]
- Mojsa, B.; Lassot, I.; Desagher, S. Mcl-1 Ubiquitination: Unique Regulation of an Essential Survival Protein. Cells 2014, 3, 418–437. [Google Scholar] [CrossRef]
- Magiera, M.M.; Mora, S.; Mojsa, B.; Robbins, I.; Lassot, I.; Desagher, S. Trim17-mediated ubiquitination and degradation of Mcl-1 initiate apoptosis in neurons. Cell Death Differ. 2013, 20, 281–292. [Google Scholar] [CrossRef]
- Lionnard, L.; Duc, P.; Brennan, M.S.; Kueh, A.J.; Pal, M.; Guardia, F.; Mojsa, B.; Damiano, M.-A.; Mora, S.; Lassot, I.; et al. TRIM17 and TRIM28 antagonistically regulate the ubiquitination and anti-apoptotic activity of BCL2A1. Cell Death Differ. 2019, 26, 902–917. [Google Scholar] [CrossRef]
- Basu-Shrivastava, M.; Mojsa, B.; Mora, S.; Robbins, I.; Bossis, G.; Lassot, I.; Desagher, S. Trim39 regulates neuronal apoptosis by acting as a SUMO-targeted E3 ubiquitin-ligase for the transcription factor NFATc3. Cell Death Differ. 2022, 1–16. [Google Scholar] [CrossRef]
- Lassot, I.; Mora, S.; Lesage, S.; Zieba, B.A.; Coque, E.; Condroyer, C.; Bossowski, J.P.; Mojsa, B.; Marelli, C.; Soulet, C.; et al. The E3 Ubiquitin Ligases TRIM17 and TRIM41 Modulate α-Synuclein Expression by Regulating ZSCAN21. Cell Rep. 2018, 25, 2484–2496.e9. [Google Scholar] [CrossRef]
- Mojsa, B.; Mora, S.; Bossowski, J.P.; Lassot, I.; Desagher, S. Control of neuronal apoptosis by reciprocal regulation of NFATc3 and Trim17. Cell Death Differ. 2015, 22, 274–286. [Google Scholar] [CrossRef]
- Devos, D.; Hirsch, E.; Wyse, R. Seven Solutions for Neuroprotection in Parkinson’s Disease. Mov. Disord. 2021, 36, 306–316. [Google Scholar] [CrossRef]
- Mahoney-Sánchez, L.; Bouchaoui, H.; Ayton, S.; Devos, D.; Duce, J.A.; Devedjian, J.-C. Ferroptosis and its potential role in the physiopathology of Parkinson’s Disease. Prog. Neurobiol. 2021, 196, 101890. [Google Scholar] [CrossRef]
- Dixon, S.J.; Lemberg, K.M.; Lamprecht, M.R.; Skouta, R.; Zaitsev, E.M.; Gleason, C.E.; Patel, D.N.; Bauer, A.J.; Cantley, A.M.; Yang, W.S.; et al. Ferroptosis: An Iron-Dependent Form of Nonapoptotic Cell Death. Cell 2012, 149, 1060–1072. [Google Scholar] [CrossRef]
- Masaldan, S.; Bush, A.I.; Devos, D.; Rolland, A.S.; Moreau, C. Striking while the iron is hot: Iron metabolism and ferroptosis in neurodegeneration. Free. Radic. Biol. Med. 2019, 133, 221–233. [Google Scholar] [CrossRef]
- Moreau, C.; Duce, J.A.; Rascol, O.; Devedjian, J.-C.; Berg, D.; Dexter, D.; Cabantchik, Z.I.; Bush, A.I.; Devos, D.; the FAIRPARK-II Study Group. Iron as a therapeutic target for Parkinson’s Disease: Iron and Parkinson’s disease. Mov. Disord. 2018, 33, 568–574. [Google Scholar] [CrossRef]
- Hopes, L.; Grolez, G.; Moreau, C.; Lopes, R.; Ryckewaert, G.; Carrière, N.; Auger, F.; Laloux, C.; Petrault, M.; Devedjian, J.-C.; et al. Magnetic Resonance Imaging Features of the Nigrostriatal System: Biomarkers of Parkinson’s Disease Stages? PLoS ONE 2016, 11, e0147947. [Google Scholar] [CrossRef]
- Grolez, G.; Moreau, C.; Sablonnière, B.; Garçon, G.; Devedjian, J.-C.; Meguig, S.; Gelé, P.; Delmaire, C.; Bordet, R.; Defebvre, L.; et al. Ceruloplasmin activity and iron chelation treatment of patients with Parkinson’s disease. BMC Neurol. 2015, 15, 74. [Google Scholar] [CrossRef]
- Cabantchik, Z.I.; Munnich, A.; Youdim, M.B.; Devos, D. Regional siderosis: A new challenge for iron chelation therapy. Front. Pharmacol. 2013, 4, 167. [Google Scholar] [CrossRef]
- Duce, J.A.; Wong, B.; Durham, H.; Devedjian, J.-C.; Smith, D.P.; Devos, D. Post translational changes to α-synuclein control iron and dopamine trafficking; a concept for neuron vulnerability in Parkinson’s disease. Mol. Neurodegener. 2017, 12, 45. [Google Scholar] [CrossRef]
- Do Van, B.; Gouel, F.; Jonneaux, A.; Timmerman, K.; Gelé, P.; Pétrault, M.; Bastide, M.; Laloux, C.; Moreau, C.; Bordet, R.; et al. Ferroptosis, a newly characterized form of cell death in Parkinson’s disease that is regulated by PKC. Neurobiol. Dis. 2016, 94, 169–178. [Google Scholar] [CrossRef]
- Devos, D.; Moreau, C.; Devedjian, J.C.; Kluza, J.; Petrault, M.; Laloux, C.; Jonneaux, A.; Ryckewaert, G.; Garcon, G.; Rouaix, N.; et al. Targeting Chelatable Iron as a Therapeutic Modality in Parkinson’s Disease. Antioxid. Redox Signal. 2014, 21, 195–210. [Google Scholar] [CrossRef]
- Sordet, O.; Rébé, C.; Plenchette, S.; Zermati, Y.; Hermine, O.; Vainchenker, W.; Garrido, C.; Solary, E.; Dubrez-Daloz, L. Specific involvement of caspases in the differentiation of monocytes into macrophages. Blood 2002, 100, 4446–4453. [Google Scholar] [CrossRef]
- Plenchette, S.; Cathelin, S.; Rébé, C.; Launay, S.; Ladoire, S.; Sordet, O.; Ponnelle, T.; Debili, N.; Phan, T.-H.; Padua, R.-A.; et al. Translocation of the inhibitor of apoptosis protein c-IAP1 from the nucleus to the Golgi in hematopoietic cells undergoing differentiation: A nuclear export signal-mediated event. Blood 2004, 104, 2035–2043. [Google Scholar] [CrossRef]
- Orme, M.; Meier, P. Inhibitor of apoptosis proteins in Drosophila: Gatekeepers of death. Apoptosis 2009, 14, 950–960. [Google Scholar] [CrossRef]
- Deveraux, Q.L.; Takahashi, R.; Salvesen, G.S.; Reed, J.C. X-Linked IAP Is a Direct Inhibitor of Cell-Death Proteases. Nature 1997, 388, 300–304. [Google Scholar] [CrossRef]
- Chai, J.; Shiozaki, E.; Srinivasula, S.M.; Wu, Q.; Dataa, P.; Alnemri, E.S.; Shi, Y. Structural Basis of Caspase-7 Inhibition by XIAP. Cell 2001, 104, 769–780. [Google Scholar] [CrossRef]
- Eckelman, B.P.; Salvesen, G.S.; Scott, F.L. Human inhibitor of apoptosis proteins: Why XIAP is the black sheep of the family. EMBO Rep. 2006, 7, 988–994. [Google Scholar] [CrossRef] [PubMed]
- Cartier, J.; Berthelet, J.; Marivin, A.; Gemble, S.; Edmond, V.; Plenchette, S.; Lagrange, B.; Hammann, A.; Dupoux, A.; Delva, L.; et al. Cellular Inhibitor of Apoptosis Protein-1 (cIAP1) Can Regulate E2F1 Transcription Factor-mediated Control of Cyclin Transcription. J. Biol. Chem. 2011, 286, 26406–26417. [Google Scholar] [CrossRef] [PubMed]
- Cirulli, V.; Yebra, M. Netrins: Beyond the brain. Nat. Rev. Mol. Cell Biol. 2007, 8, 296–306. [Google Scholar] [CrossRef] [PubMed]
- Barnault, R.; Verzeroli, C.; Fournier, C.; Michelet, M.; Redavid, A.R.; Chicherova, I.; Plissonnier, M.; Adrait, A.; Khomich, O.; Chapus, F.; et al. Hepatic inflammation elicits production of proinflammatory netrin-1 through exclusive activation of translation. Hepatology 2022, 32446. [Google Scholar] [CrossRef]
- Paradisi, A.; Creveaux, M.; Gibert, B.; Devailly, G.; Redoulez, E.; Neves, D.; Cleyssac, E.; Treilleux, I.; Klein, C.; Niederfellner, G.; et al. Combining chemotherapeutic agents and netrin-1 interference potentiates cancer cell death. EMBO Mol. Med. 2013, 5, 1821–1834. [Google Scholar] [CrossRef]
- Cassier, P.; Eberst, L.; Garin, G.; Courbebaisse, Y.; Terret, C.; Robert, M.; Frenel, J.-S.; Depil, S.; Delord, J.-P.; Perol, D.; et al. A first in human, phase I trial of NP137, a first-in-class antibody targeting netrin-1, in patients with advanced refractory solid tumors. Ann. Oncol. 2019, 30, v159. [Google Scholar] [CrossRef]
- Reuten, R.; Patel, T.R.; McDougall, M.; Rama, N.; Nikodemus, D.; Gibert, B.; Delcros, J.-G.; Prein, C.; Meier, M.; Metzger, S.; et al. Structural decoding of netrin-4 reveals a regulatory function towards mature basement membranes. Nat. Commun. 2016, 7, 13515. [Google Scholar] [CrossRef]
- Wang, H.; Copeland, N.G.; Gilbert, D.J.; Jenkins, N.A.; Tessier-Lavigne, M. Netrin-3, a Mouse Homolog of Human NTN2L, Is Highly Expressed in Sensory Ganglia and Shows Differential Binding to Netrin Receptors. J. Neurosci. 1999, 19, 4938–4947. [Google Scholar] [CrossRef]
- Jiang, S.; Richaud, M.; Vieugué, P.; Rama, N.; Delcros, J.; Siouda, M.; Sanada, M.; Redavid, A.; Ducarouge, B.; Hervieu, M.; et al. Targeting netrin-3 in small cell lung cancer and neuroblastoma. EMBO Mol. Med. 2021, 13, e12878. [Google Scholar] [CrossRef]
- Kalifi, M.; Walter, T.; Milot, L.; Hervieu, V.; Millot, I.; Gibert, B.; Roche, C.; Forestier, J.; Lombard-Bohas, C.; Pasquer, A.; et al. Unifocal versus Multiple Ileal Neuroendocrine Tumors Location: An Embryological Origin. Neuroendocrinology 2021, 111, 786–793. [Google Scholar] [CrossRef]
- Gerard, L.; Garcia, J.; Gauthier, A.; Lopez, J.; Durand, A.; Hervieu, V.; Lemelin, A.; Chardon, L.; Landel, V.; Gibert, B.; et al. ctDNA in Neuroendocrine Carcinoma of Gastroenteropancreatic Origin or of Unknown Primary: The CIRCAN-NEC Pilot Study. Neuroendocrinology 2020, 111, 951–964. [Google Scholar] [CrossRef]
- Gillet, G.; Guerin, M.; Trembleau, A.; Brun, G. A Bcl-2-related gene is activated in avian cells transformed by the Rous sarcoma virus. EMBO J. 1995, 14, 1372–1381. [Google Scholar] [CrossRef]
- Popgeorgiev, N.; Bonneau, B.; Ferri, K.F.; Prudent, J.; Thibaut, J.; Gillet, G. The Apoptotic Regulator Nrz Controls Cytoskeletal Dynamics via the Regulation of Ca2+ Trafficking in the Zebrafish Blastula. Dev. Cell 2011, 20, 663–676. [Google Scholar] [CrossRef]
- Prudent, J.; Popgeorgiev, N.; Bonneau, B.; Thibaut, J.; Gadet, R.; Lopez, J.; Gonzalo, P.; Rimokh, R.; Manon, S.; Houart, C.; et al. Bcl-wav and the mitochondrial calcium uniporter drive gastrula morphogenesis in zebrafish. Nat. Commun. 2013, 4, 2330. [Google Scholar] [CrossRef]
- Nougarede, A.; Popgeorgiev, N.; Kassem, L.; Omarjee, S.; Borel, S.; Mikaelian, I.; Lopez, J.; Gadet, R.; Marcillat, O.; Treilleux, I.; et al. Breast Cancer Targeting through Inhibition of the Endoplasmic Reticulum-Based Apoptosis Regulator Nrh/BCL2L10. Cancer Res. 2018, 78, 1404–1417. [Google Scholar] [CrossRef]
- Popgeorgiev, N.; Sa, J.D.; Jabbour, L.; Banjara, S.; Nguyen, T.T.M.; Akhavan-E-Sabet, A.; Gadet, R.; Ralchev, N.; Manon, S.; Hinds, M.G.; et al. Ancient and conserved functional interplay between Bcl-2 family proteins in the mitochondrial pathway of apoptosis. Sci. Adv. 2020, 6, eabc4149. [Google Scholar] [CrossRef]
- Vayssiere, J.L.; Petit, P.X.; Risler, Y.; Mignotte, B. Commitment to apoptosis is associated with changes in mitochondrial biogenesis and activity in cell lines conditionally immortalized with simian virus 40. Proc. Natl. Acad. Sci. USA 1994, 91, 11752–11756. [Google Scholar] [CrossRef]
- Seervi, M.; Xue, D. Mitochondrial Cell Death Pathways in Caenorhabiditis Elegans. In Current Topics in Developmental Biology; Elsevier: Amsterdam, The Netherlands, 2015; Volume 114, pp. 43–65. ISBN 978-0-12-410425-9. [Google Scholar]
- Sandu, C.; Ryoo, H.D.; Steller, H. Drosophila IAP antagonists form multimeric complexes to promote cell death. J. Cell Biol. 2010, 190, 1039–1052. [Google Scholar] [CrossRef]
- Clavier, A.; Rincheval-Arnold, A.; Colin, J.; Mignotte, B.; Guénal, I. Apoptosis in Drosophila: Which role for mitochondria? Apoptosis 2016, 21, 239–251. [Google Scholar] [CrossRef]
- Clavier, A.; Ruby, V.; Rincheval-Arnold, A.; Mignotte, B.; Guénal, I. The Drosophila retinoblastoma protein, Rbf1, induces a debcl and drp1-dependent mitochondrial apoptosis. J. Cell Sci. 2015, 128, 169896. [Google Scholar] [CrossRef]
- Clavier, A.; Rincheval-Arnold, A.; Baillet, A.; Mignotte, B.; Guénal, I. Two different specific JNK activators are required to trigger apoptosis or compensatory proliferation in response to Rbf1 in Drosophila. Cell Cycle 2016, 15, 283–294. [Google Scholar] [CrossRef][Green Version]
- Berthenet, K.; Ferrer, C.C.; Fanfone, D.; Popgeorgiev, N.; Neves, D.; Bertolino, P.; Gibert, B.; Hernandez-Vargas, H.; Ichim, G. Failed Apoptosis Enhances Melanoma Cancer Cell Aggressiveness. Cell Rep. 2020, 31, 107731. [Google Scholar] [CrossRef]
- Gyrd-Hansen, M.; Meier, P. IAPs: From caspase inhibitors to modulators of NF-κB, inflammation and cancer. Nat. Cancer 2010, 10, 561–574. [Google Scholar] [CrossRef]
- Jäger, R.; Zwacka, R.M. The Enigmatic Roles of Caspases in Tumor Development. Cancers 2010, 2, 1952–1979. [Google Scholar] [CrossRef]
- Gensbittel, V.; Kräter, M.; Harlepp, S.; Busnelli, I.; Guck, J.; Goetz, J.G. Mechanical Adaptability of Tumor Cells in Metastasis. Dev. Cell 2021, 56, 164–179. [Google Scholar] [CrossRef]
- Shen, M.; Kang, Y. Stresses in the metastatic cascade: Molecular mechanisms and therapeutic opportunities. Genes Dev. 2020, 34, 1577–1598. [Google Scholar] [CrossRef]
- Fanfone, D.; Wu, Z.; Mammi, J.; Berthenet, K.; Neves, D.; Weber, K.; Halaburkova, A.; Virard, F.; Bunel, F.; Jamard, C.; et al. Confined migration promotes cancer metastasis through resistance to anoikis and increased invasiveness. eLife 2022, 11, e73150. [Google Scholar] [CrossRef]
- Charles, E.M.; Rehm, M. Key regulators of apoptosis execution as biomarker candidates in melanoma. Mol. Cell. Oncol. 2014, 1, e964037. [Google Scholar] [CrossRef]
- Fink, D.; Schlagbauer-Wadl, H.; Selzer, E.; Lucas, T.; Wolff, K.; Pehamberger, H.; Eichler, H.-G.; Jansen, B. Elevated procaspase levels in human melanoma. Melanoma Res. 2001, 11, 385–393. [Google Scholar] [CrossRef]
- Lohard, S.; Bourgeois, N.; Maillet, L.; Gautier, F.; Fétiveau, A.; Lasla, H.; Nguyen, F.; Vuillier, C.; Dumont, A.; Moreau-Aubry, A.; et al. STING-dependent paracriny shapes apoptotic priming of breast tumors in response to anti-mitotic treatment. Nat. Commun. 2020, 11, 259. [Google Scholar] [CrossRef]
- Louault, K.; Bonneaud, T.L.; Séveno, C.; Gomez-Bougie, P.; Nguyen, F.; Gautier, F.; Bourgeois, N.; Loussouarn, D.; Kerdraon, O.; Barille-Nion, S.; et al. Interactions between cancer-associated fibroblasts and tumor cells promote MCL-1 dependency in estrogen receptor-positive breast cancers. Oncogene 2019, 38, 3261–3273. [Google Scholar] [CrossRef] [PubMed]
- Vuillier, C.; Lohard, S.; Fétiveau, A.; Allègre, J.; Kayaci, C.; King, L.E.; Braun, F.; Barillé-Nion, S.; Gautier, F.; Dubrez, L.; et al. E2F1 interacts with BCL - xL and regulates its subcellular localization dynamics to trigger cell death. EMBO Rep. 2017, 19, 234–243. [Google Scholar] [CrossRef] [PubMed]
- De Carné Trécesson, S.; Souazé, F.; Basseville, A.; Bernard, A.-C.; Pécot, J.; Lopez, J.; Bessou, M.; Sarosiek, K.; Letai, A.; Barille-Nion, S.; et al. BCL-XL directly modulates RAS signalling to favour cancer cell stemness. Nat. Commun. 2017, 8, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Pécot, J.; Maillet, L.; Le Pen, J.; Vuillier, C.; de Carné Trécesson, S.; Fétiveau, A.; Sarosiek, K.; Bock, F.J.; Braun, F.; Letai, A.; et al. Tight Sequestration of BH3 Proteins by BCL-xL at Subcellular Membranes Contributes to Apoptotic Resistance. Cell Rep. 2016, 17, 3347–3358. [Google Scholar] [CrossRef]
- Le Pen, J.; Laurent, M.; Sarosiek, K.; Vuillier, C.; Gautier, F.; Montessuit, S.; Martinou, J.-C.; Letaï, A.; Braun, F.; Juin, P.P. Constitutive p53 heightens mitochondrial apoptotic priming and favors cell death induction by BH3 mimetic inhibitors of BCL-xL. Cell Death Dis. 2016, 7, e2083. [Google Scholar] [CrossRef]
- Vacchelli, E.; Ma, Y.; Baracco, E.E.; Sistigu, A.; Enot, D.P.; Pietrocola, F.; Yang, H.; Adjemian, S.; Chaba, K.; Semeraro, M.; et al. Chemotherapy-induced antitumor immunity requires formyl peptide receptor 1. Science 2015, 350, 972–978. [Google Scholar] [CrossRef]
- Apetoh, L.; Ghiringhelli, F.; Tesniere, A.; Obeid, M.; Ortiz, C.; Criollo, A.; Mignot, G.; Maiuri, M.C.; Ullrich, E.; Saulnier, P.; et al. Toll-like receptor 4–dependent contribution of the immune system to anticancer chemotherapy and radiotherapy. Nat. Med. 2007, 13, 1050–1059. [Google Scholar] [CrossRef]
- Ma, Y.; Adjemian, S.; Mattarollo, S.R.; Yamazaki, T.; Aymeric, L.; Yang, H.; Catani, J.P.P.; Hannani, D.; Duret, H.; Steegh, K.; et al. Anticancer Chemotherapy-Induced Intratumoral Recruitment and Differentiation of Antigen-Presenting Cells. Immunity 2013, 38, 729–741. [Google Scholar] [CrossRef]
- Pietrocola, F.; Pol, J.; Vacchelli, E.; Rao, S.; Enot, D.P.; Baracco, E.E.; Levesque, S.; Castoldi, F.; Jacquelot, N.; Yamazaki, T.; et al. Caloric Restriction Mimetics Enhance Anticancer Immunosurveillance. Cancer Cell 2016, 30, 147–160. [Google Scholar] [CrossRef]
- Obeid, M.; Tesniere, A.; Ghiringhelli, F.; Fimia, G.M.; Apetoh, L.; Perfettini, J.-L.; Castedo, M.; Mignot, G.; Panaretakis, T.; Casares, N.; et al. Calreticulin exposure dictates the immunogenicity of cancer cell death. Nat. Med. 2007, 13, 54–61. [Google Scholar] [CrossRef]
- Humeau, J.; Sauvat, A.; Cerrato, G.; Xie, W.; Loos, F.; Iannantuoni, F.; Bezu, L.; Lévesque, S.; Paillet, J.; Pol, J.; et al. Inhibition of transcription by dactinomycin reveals a new characteristic of immunogenic cell stress. EMBO Mol. Med. 2020, 12, e11622. [Google Scholar] [CrossRef]
- Sistigu, A.; Yamazaki, T.; Vacchelli, E.; Chaba, K.; Enot, D.P.; Adam, J.; Vitale, I.; Goubar, A.; Baracco, E.E.; Remédios, C.; et al. Cancer cell–autonomous contribution of type I interferon signaling to the efficacy of chemotherapy. Nat. Med. 2014, 20, 1301–1309. [Google Scholar] [CrossRef]
- Liu, P.; Zhao, L.; Loos, F.; Marty, C.; Xie, W.; Martins, I.; Lachkar, S.; Qu, B.; Waeckel-Énée, E.; Plo, I.; et al. Immunosuppression by Mutated Calreticulin Released from Malignant Cells. Mol. Cell 2020, 77, 748–760.e9. [Google Scholar] [CrossRef]
- Le Naour, J.; Liu, P.; Zhao, L.; Adjemian, S.; Sztupinszki, Z.; Taieb, J.; Mulot, C.; Silvin, A.; Dutertre, C.-A.; Ginhoux, F.; et al. A TLR3 Ligand Reestablishes Chemotherapeutic Responses in the Context of FPR1 Deficiency. Cancer Discov. 2021, 11, 408–423. [Google Scholar] [CrossRef]
- Malleter, M.; Tauzin, S.; Bessede, A.; Castellano, R.; Goubard, A.; Godey, F.; Levêque, J.; Jézéquel, P.; Campion, L.; Campone, M.; et al. CD95L Cell Surface Cleavage Triggers a Prometastatic Signaling Pathway in Triple-Negative Breast Cancer. Cancer Res. 2013, 73, 6711–6721. [Google Scholar] [CrossRef]
- Poissonnier, A.; Sanséau, D.; Le Gallo, M.; Malleter, M.; Levoin, N.; Viel, R.; Morere, L.; Penna, A.; Blanco, P.; Dupuy, A.; et al. CD95-Mediated Calcium Signaling Promotes T Helper 17 Trafficking to Inflamed Organs in Lupus-Prone Mice. Immunity 2016, 45, 209–223. [Google Scholar] [CrossRef]
- Guégan, J.-P.; Pollet, J.; Ginestier, C.; Charafe-Jauffret, E.; Peter, M.E.; Legembre, P. CD95/Fas suppresses NF-κB activation through recruitment of KPC2 in a CD95L/FasL-independent mechanism. iScience 2021, 24, 103538. [Google Scholar] [CrossRef]
- Qadir, A.S.; Guégan, J.P.; Ginestier, C.; Chaibi, A.; Bessede, A.; Charafe-Jauffret, E.; Macario, M.; Lavoué, V.; de la Motte Rouge, T.; Law, C.; et al. CD95/Fas protects triple negative breast cancer from anti-tumor activity of NK cells. iScience 2021, 24, 103348. [Google Scholar] [CrossRef]
- Khadra, N.; Bresson-Bepoldin, L.; Penna, A.; Chaigne-Delalande, B.; Ségui, B.; Levade, T.; Vacher, A.-M.; Reiffers, J.; Ducret, T.; Moreau, J.-F.; et al. CD95 triggers Orai1-mediated localized Ca 2+ entry, regulates recruitment of protein kinase C (PKC) β2, and prevents death-inducing signaling complex formation. Proc. Natl. Acad. Sci. USA 2011, 108, 19072–19077. [Google Scholar] [CrossRef]
- Domingo-Fernández, R.; Coll, R.C.; Kearney, J.; Breit, S.; O’Neill, L.A. The intracellular chloride channel proteins CLIC1 and CLIC4 induce IL-1β transcription and activate the NLRP3 inflammasome. J. Biol. Chem. 2017, 292, 12077–12087. [Google Scholar] [CrossRef]
- Murakami, T.; Ockinger, J.; Yu, J.; Byles, V.; McColl, A.; Hofer, A.M.; Horng, T. Critical role for calcium mobilization in activation of the NLRP3 inflammasome. Proc. Natl. Acad. Sci. USA 2012, 109, 11282–11287. [Google Scholar] [CrossRef]
- Muñoz-Planillo, R.; Kuffa, P.; Martínez-Colón, G.; Smith, B.L.; Rajendiran, T.M.; Núñez, G. K+ Efflux Is the Common Trigger of NLRP3 Inflammasome Activation by Bacterial Toxins and Particulate Matter. Immunity 2013, 38, 1142–1153. [Google Scholar] [CrossRef]
- Clavería, C.; Torres, M. Cell Competition: Mechanisms and Physiological Roles. Annu. Rev. Cell Dev. Biol. 2016, 32, 411–439. [Google Scholar] [CrossRef]
- Levayer, R.; Dupont, C.; Moreno, E. Tissue Crowding Induces Caspase-Dependent Competition for Space. Curr. Biol. 2016, 26, 670–677. [Google Scholar] [CrossRef]
- Wagstaff, L.; Goschorska, M.; Kozyrska, K.; Duclos, G.; Kucinski, I.; Chessel, A.; Hampton-O’Neil, L.; Bradshaw, C.R.; Allen, G.E.; Rawlins, E.L.; et al. Mechanical cell competition kills cells via induction of lethal p53 levels. Nat. Commun. 2016, 7, 11373. [Google Scholar] [CrossRef]
- Matamoro-Vidal, A.; Levayer, R. Multiple Influences of Mechanical Forces on Cell Competition. Curr. Biol. 2019, 29, R762–R774. [Google Scholar] [CrossRef]
- Moreno, E.; Valon, L.; Levillayer, F.; Levayer, R. Competition for Space Induces Cell Elimination through Compaction-Driven ERK Downregulation. Curr. Biol. 2019, 29, 23–34.e8. [Google Scholar] [CrossRef]
- Valon, L.; Davidović, A.; Levillayer, F.; Villars, A.; Chouly, M.; Cerqueira-Campos, F.; Levayer, R. Robustness of epithelial sealing is an emerging property of local ERK feedback driven by cell elimination. Dev. Cell 2021, 56, 1700–1711.e8. [Google Scholar] [CrossRef]
- Bock, F.J.; Sedov, E.; Koren, E.; Koessinger, A.L.; Cloix, C.; Zerbst, D.; Athineos, D.; Anand, J.; Campbell, K.J.; Blyth, K.; et al. Apoptotic stress-induced FGF signalling promotes non-cell autonomous resistance to cell death. Nat. Commun. 2021, 12, 6572. [Google Scholar] [CrossRef]
- Villars, A.; Levayer, R. Collective effects in epithelial cell death and cell extrusion. Curr. Opin. Genet. Dev. 2022, 72, 8–14. [Google Scholar] [CrossRef]
- Sato, T.; Hanada, M.; Bodrug, S.; Irie, S.; Iwama, N.; Boise, L.H.; Thompson, C.B.; Golemis, E.; Fong, L.; Wang, H.G. Interactions among members of the Bcl-2 protein family analyzed with a yeast two-hybrid system. Proc. Natl. Acad. Sci. USA 1994, 91, 9238–9242. [Google Scholar] [CrossRef] [PubMed]
- Renault, T.T.; Dejean, L.M.; Manon, S. A brewing understanding of the regulation of Bax function by Bcl-xL and Bcl-2. Mech. Ageing Dev. 2017, 161, 201–210. [Google Scholar] [CrossRef] [PubMed]
- Bellot, G.; Cartron, P.-F.; Er, E.; Oliver, L.; Juin, P.; Armstrong, L.C.; Bornstein, P.; Mihara, K.; Manon, S.; Vallette, F. TOM22, a core component of the mitochondria outer membrane protein translocation pore, is a mitochondrial receptor for the proapoptotic protein Bax. Cell Death Differ. 2007, 14, 785–794. [Google Scholar] [CrossRef] [PubMed]
- Simonyan, L.; Renault, T.T.; da Costa Novais, M.J.; Sousa, M.J.; Côrte-Real, M.; Camougrand, N.; Gonzalez, C.; Manon, S. Regulation of Bax/mitochondria interaction by AKT. FEBS Lett. 2015, 590, 13–21. [Google Scholar] [CrossRef]
- Légiot, A.; Céré, C.; Dupoiron, T.; Kaabouni, M.; Camougrand, N.; Manon, S. Mitochondria-Associated Membranes (MAMs) are involved in Bax mitochondrial localization and cytochrome c release. Microb. Cell 2019, 6, 257–266. [Google Scholar] [CrossRef]
- Thibert, C.; Teillet, M.-A.; Lapointe, F.; Mazelin, L.; Le Douarin, N.M.; Mehlen, P. Inhibition of Neuroepithelial Patched-Induced Apoptosis by Sonic Hedgehog. Science 2003, 301, 843–846. [Google Scholar] [CrossRef]
- Castets, M.; Broutier, L.; Molin, Y.; Brevet, M.; Chazot, G.; Gadot, N.; Paquet, A.; Mazelin, L.; Jarrosson-Wuilleme, L.; Scoazec, J.-Y.; et al. DCC constrains tumour progression via its dependence receptor activity. Nature 2012, 482, 534–537. [Google Scholar] [CrossRef]
- Mehlen, P.; Delloye-Bourgeois, C.; Chedotal, A. Novel roles for Slits and netrins: Axon guidance cues as anticancer targets? Nat. Cancer 2011, 11, 188–197. [Google Scholar] [CrossRef]
- Fitamant, J.; Guenebeaud, C.; Coissieux, M.-M.; Guix, C.; Treilleux, I.; Scoazec, J.-Y.; Bachelot, T.; Bernet, A.; Mehlen, P. Netrin-1 expression confers a selective advantage for tumor cell survival in metastatic breast cancer. Proc. Natl. Acad. Sci. USA 2008, 105, 4850–4855. [Google Scholar] [CrossRef]
- Grandin, M.; Meier, M.; Delcros, J.G.; Nikodemus, D.; Reuten, R.; Patel, T.; Goldschneider, D.; Orriss, G.; Krahn, N.; Boussouar, A.; et al. Structural Decoding of the Netrin-1/UNC5 Interaction and its Therapeutical Implications in Cancers. Cancer Cell 2016, 29, 173–185. [Google Scholar] [CrossRef]
- Bedoui, S.; Herold, M.; Strasser, A. Emerging connectivity of programmed cell death pathways and its physiological implications. Nat. Rev. Mol. Cell Biol. 2020, 21, 678–695. [Google Scholar] [CrossRef]
- Napoletano, F.; Baron, O.; Vandenabeele, P.; Mollereau, B.; Fanto, M. Intersections between Regulated Cell Death and Autophagy. Trends Cell Biol. 2019, 29, 323–338. [Google Scholar] [CrossRef]
- Bata, N.; Cosford, N.D.P. Cell Survival and Cell Death at the Intersection of Autophagy and Apoptosis: Implications for Current and Future Cancer Therapeutics. ACS Pharmacol. Transl. Sci. 2021, 4, 1728–1746. [Google Scholar] [CrossRef]
- Causeret, F.; Sumia, I.; Pierani, A. Kremen1 and Dickkopf1 control cell survival in a Wnt-independent manner. Cell Death Differ. 2016, 23, 323–332. [Google Scholar] [CrossRef]
- Dufour, F.; Rattier, T.; Shirley, S.; Picarda, G.; Constantinescu, A.A.; Morlé, A.; Zakaria, A.B.; Marcion, G.; Causse, S.; Szegezdi, E.; et al. N-glycosylation of mouse TRAIL-R and human TRAIL-R1 enhances TRAIL-induced death. Cell Death Differ. 2017, 24, 500–510. [Google Scholar] [CrossRef]
- Holler, N.; Zaru, R.; Micheau, O.; Thome, M.; Attinger, A.; Valitutti, S.; Bodmer, J.-L.; Schneider, P.; Seed, B.; Tschopp, J. Fas triggers an alternative, caspase-8–independent cell death pathway using the kinase RIP as effector molecule. Nat. Immunol. 2000, 1, 489–495. [Google Scholar] [CrossRef]
- Micheau, O.; Rizzi, M.; Smulski, C.R. Editorial: TNFR Superfamily Oligomerization and Signaling. Front. Cell Dev. Biol. 2021, 9, 682472. [Google Scholar] [CrossRef]
- Micheau, O.; Tschopp, J. Induction of TNF Receptor I-Mediated Apoptosis via Two Sequential Signaling Complexes. Cell 2003, 114, 181–190. [Google Scholar] [CrossRef]
- Kischkel, F.C.; Hellbardt, S.; Behrmann, I.; Germer, M.; Pawlita, M.; Krammer, P.H.; Peter, M.E. Cytotoxicity-dependent APO-1 (Fas/CD95)-associated proteins form a death-inducing signaling complex (DISC) with the receptor. EMBO J. 1995, 14, 5579–5588. [Google Scholar] [CrossRef]
- Micheau, O.; Lens, S.; Gaide, O.; Alevizopoulos, K.; Tschopp, J. NF-κB Signals Induce the Expression of c-FLIP. Mol. Cell. Biol. 2001, 21, 5299–5305. [Google Scholar] [CrossRef]
- Irmler, M.; Thome, M.; Hahne, M.; Schneider, P.; Hofmann, K.; Steiner, V.; Bodmer, J.-L.; Schröter, M.; Burns, K.; Mattmann, C.; et al. Inhibition of Death Receptor Signals by Cellular FLIP. Nature 1997, 388, 190–195. [Google Scholar] [CrossRef]
- Azijli, K.; Weyhenmeyer, B.; Peters, G.J.; De Jong, S.; Kruyt, F.A.E. Non-canonical kinase signaling by the death ligand TRAIL in cancer cells: Discord in the death receptor family. Cell Death Differ. 2013, 20, 858–868.e9. [Google Scholar] [CrossRef]
- Jin, Z.; El-Deiry, W.S. Distinct Signaling Pathways in TRAIL- versus Tumor Necrosis Factor-Induced Apoptosis. Mol. Cell. Biol. 2006, 26, 8136–8148. [Google Scholar] [CrossRef]
- Takeda, K.; Hayakawa, Y.; Smyth, M.; Kayagaki, N.; Yamaguchi, N.; Kakuta, S.; Iwakura, Y.; Yagita, H.; Okumura, K. Involvement of tumor necrosis factor-related apoptosis-inducing ligand in surveillance of tumor metastasis by liver natural killer cells. Nat. Med. 2001, 7, 94–100. [Google Scholar] [CrossRef]
- Takeda, K.; Smyth, M.; Cretney, E.; Hayakawa, Y.; Kayagaki, N.; Yagita, H.; Okumura, K. Critical Role for Tumor Necrosis Factor–related Apoptosis-inducing Ligand in Immune Surveillance Against Tumor Development. J. Exp. Med. 2002, 195, 161–169. [Google Scholar] [CrossRef]
- French, L.; Tschopp, J. The TRAIL to selective tumor death. Nat. Med. 1999, 5, 146–147. [Google Scholar] [CrossRef]
- Von Karstedt, S.; Montinaro, A.; Walczak, H. Exploring the TRAILs less travelled: TRAIL in cancer biology and therapy. Nat. Cancer 2017, 17, 352–366. [Google Scholar] [CrossRef]
- Micheau, O.; Shirley, S.; Dufour, F. Death receptors as targets in cancer: TRAIL Clinical Trials. Br. J. Pharmacol. 2013, 169, 1723–1744. [Google Scholar] [CrossRef]
- Hengartner, M.; Horvitz, H.R. Programmed cell death in Caenorhabditis elegans. Curr. Opin. Genet. Dev. 1994, 4, 581–586. [Google Scholar] [CrossRef]
- Saleem, S. Apoptosis, Autophagy, Necrosis and their multi galore crosstalk in neurodegeneration. Neuroscience 2021, 469, 162–174. [Google Scholar] [CrossRef]
- Mollereau, B. Cell death: What can we learn from flies? Editorial for the special review issue on Drosophila apoptosis. Apoptosis 2009, 14, 929–934. [Google Scholar] [CrossRef] [PubMed]
- Marcel, V.; Dichtel-Danjoy, M.-L.; Sagne, C.; Hafsi, H.; Ma, D.; Ortiz-Cuaran, S.; Olivier, M.; Hall, J.; Mollereau, B.; Hainaut, P.; et al. Biological functions of p53 isoforms through evolution: Lessons from animal and cellular models. Cell Death Differ. 2011, 18, 1815–1824. [Google Scholar] [CrossRef] [PubMed]
- Mollereau, B.; Ma, D. The p53 control of apoptosis and proliferation: Lessons from Drosophila. Apoptosis 2014, 19, 1421–1429. [Google Scholar] [CrossRef] [PubMed]
- Mollereau, B.; Perez-Garijo, A.; Bergmann, A.; Miura, M.; Gerlitz, O.; Ryoo, H.D.; Steller, H.; Morata, G. Compensatory proliferation and apoptosis-induced proliferation: A need for clarification. Cell Death Differ. 2013, 20, 181. [Google Scholar] [CrossRef] [PubMed]
- Dichtel-Danjoy, M.-L.; Ma, D.; Dourlen, P.; Chatelain, G.; Napoletano, F.; Robin, M.; Corbet, M.; Levet, C.; Hafsi, H.; Hainaut, P.; et al. Drosophila p53 isoforms differentially regulate apoptosis and apoptosis-induced proliferation. Cell Death Differ. 2013, 20, 108–116. [Google Scholar] [CrossRef] [PubMed]
- Mollereau, B. Establishing Links between Endoplasmic Reticulum-Mediated Hormesis and Cancer. Mol. Cell. Biol. 2013, 33, 2372–2374. [Google Scholar] [CrossRef]
- Mendes, C.S.; Levet, C.; Chatelain, G.; Dourlen, P.; Fouillet, A.; Dichtel-Danjoy, M.-L.; Gambis, A.; Ryoo, H.D.; Steller, H.; Mollereau, B. ER stress protects from retinal degeneration. EMBO J. 2009, 28, 1296–1307. [Google Scholar] [CrossRef]
- Fouillet, A.; Levet, C.; Virgone, A.; Robin, M.; Dourlen, P.; Rieusset, J.; Belaidi, E.; Ovize, M.; Touret, M.; Nataf, S.; et al. ER stress inhibits neuronal death by promoting autophagy. Autophagy 2012, 8, 915–926. [Google Scholar] [CrossRef]
- Youle, R.J.; Narendra, D.P. Mechanisms of mitophagy. Nat. Rev. Mol. Cell Biol. 2011, 12, 9–14. [Google Scholar] [CrossRef]
- Yang, D.-S.; Kumar, A.; Stavrides, P.; Peterson, J.; Peterhoff, C.M.; Pawlik, M.; Levy, E.; Cataldo, A.M.; Nixon, R.A. Neuronal Apoptosis and Autophagy Cross Talk in Aging PS/APP Mice, a Model of Alzheimer’s Disease. Am. J. Pathol. 2008, 173, 665–681. [Google Scholar] [CrossRef]
- Robin, M.; Issa, A.R.; Santos, C.C.; Napoletano, F.; Petitgas, C.; Chatelain, G.; Ruby, M.; Walter, L.; Birman, S.; Domingos, P.; et al. Drosophila p53 integrates the antagonism between autophagy and apoptosis in response to stress. Autophagy 2019, 15, 771–784. [Google Scholar] [CrossRef]
- Napoletano, F.; Gibert, B.; Yacobi-Sharon, K.; Vincent, S.; Favrot, C.; Mehlen, P.; Girard, V.; Teil, M.; Chatelain, G.; Walter, L.; et al. p53-dependent programmed necrosis controls germ cell homeostasis during spermatogenesis. PLoS Genet. 2017, 13, e1007024. [Google Scholar] [CrossRef]
- Yacobi-Sharon, K.; Namdar, Y.; Arama, E. Alternative Germ Cell Death Pathway in Drosophila Involves HtrA2/Omi, Lysosomes, and a Caspase-9 Counterpart. Dev. Cell 2013, 25, 29–42. [Google Scholar] [CrossRef]
- Priault, M.; Hue, E.; Marhuenda, F.; Pilet, P.; Oliver, L.; Vallette, F.M. Differential Dependence on Beclin 1 for the Regulation of Pro-Survival Autophagy by Bcl-2 and Bcl-xL in HCT116 Colorectal Cancer Cells. PLoS ONE 2010, 5, e8755. [Google Scholar] [CrossRef]
- Beaumatin, F.; El Dhaybi, M.; Lasserre, J.-P.; Salin, B.; Moyer, M.P.; Verdier, M.; Manon, S.; Priault, M. N52 monodeamidated Bcl-xL shows impaired oncogenic properties in vivo and in vitro. Oncotarget 2016, 7, 17129–17143. [Google Scholar] [CrossRef]
- Bobo, C.; Céré, C.; Dufossée, M.; Dautant, A.; Moreau, V.; Manon, S.; Beaumatin, F.; Priault, M. Improved Electrophoretic Separation to Assist the Monitoring of Bcl-xL Post-Translational Modifications. Int. J. Mol. Sci. 2019, 20, 5571. [Google Scholar] [CrossRef]
- Boudier-Lemosquet, A.; Mahler, A.; Bobo, C.; Moreau, V.; Priault, M. 1D continuous gel electrophoresis composition for the separation of deamidated proteins. Methods 2022, 200, 23–30. [Google Scholar] [CrossRef]
- Boudier-Lemosquet, A.; Mahler, A.; Bobo, C.; Dufossée, M.; Priault, M. Introducing protein deamidation: Landmark discoveries, societal outreach, and tentative priming workflow to address deamidation. Methods 2022, 200, 3–14. [Google Scholar] [CrossRef]
- McCarthy, N.J.; Whyte, M.K.; Gilbert, C.S.; Evan, G.I. Inhibition of Ced-3/ICE-related Proteases Does Not Prevent Cell Death Induced by Oncogenes, DNA Damage, or the Bcl-2 Homologue Bak. J. Cell Biol. 1997, 136, 215–227. [Google Scholar] [CrossRef]
- Haraguchi, M.; Torii, S.; Matsuzawa, S.I.; Xie, Z.; Kitada, S.; Krajewski, S.; Yoshida, H.; Mak, T.W.; Reed, J.C. Apoptotic Protease Activating Factor 1 (Apaf-1)–Independent Cell Death Suppression by Bcl-2. J. Exp. Med. 2000, 191, 1709–1720. [Google Scholar] [CrossRef]
- Tait, S.W.G.; Green, D.R. Caspase-independent cell death: Leaving the set without the final cut. Oncogene 2008, 27, 6452–6461. [Google Scholar] [CrossRef]
- Zunino, B.; Rubio-Patiño, C.; Villa, E.; Meynet, O.; Proics, E.; Cornille, A.; Pommier, S.; Mondragón, L.; Chiche, J.; Bereder, J.-M.; et al. Hyperthermic intraperitoneal chemotherapy leads to an anticancer immune response via exposure of cell surface heat shock protein 90. Oncogene 2016, 35, 261–268. [Google Scholar] [CrossRef]
- Meynet, O.; Bénéteau, M.; Jacquin, M.A.; Pradelli, L.A.; Cornille, A.; Carles, M.; Ricci, J.E. Glycolysis inhibition targets Mcl-1 to restore sensitivity of lymphoma cells to ABT-737-induced apoptosis. Leukemia 2012, 26, 1145–1147. [Google Scholar] [CrossRef][Green Version]
- Meynet, O.; Zunino, B.; Happo, L.; Pradelli, L.A.; Chiche, J.; Jacquin, M.A.; Mondragón, L.; Tanti, J.-F.; Taillan, B.; Garnier, G.; et al. Caloric restriction modulates Mcl-1 expression and sensitizes lymphomas to BH3 mimetic in mice. Blood 2013, 122, 2402–2411. [Google Scholar] [CrossRef]
- Lavallard, V.J.; Pradelli, L.A.; Paul, A.; Bénéteau, M.; Jacquel, A.; Auberger, P.; Ricci, J.E. Modulation of Caspase-Independent Cell Death Leads to Resensitization of Imatinib Mesylate–Resistant Cells. Cancer Res. 2009, 69, 3013–3020. [Google Scholar] [CrossRef]
- Villa, E.; Paul, R.; Meynet, O.; Volturo, S.; Pinna, G.; Ricci, J.-E. The E3 ligase UBR2 regulates cell death under caspase deficiency via Erk/MAPK pathway. Cell Death Dis. 2020, 11, 1041. [Google Scholar] [CrossRef]
- Chiche, J.; Pommier, S.; Bénéteau, M.; Mondragón, L.; Meynet, O.; Zunino, B.; Mouchotte, A.; Verhoeyen, E.; Guyot, M.; Pagès, G.; et al. GAPDH enhances the aggressiveness and the vascularization of non-Hodgkin’s B lymphomas via NF-κB-dependent induction of HIF-1α. Leukemia 2015, 29, 1163–1176. [Google Scholar] [CrossRef]
- Chiche, J.; Reverso-Meinietti, J.; Mouchotte, A.; Rubio-Patiño, C.; Mhaidly, R.; Villa, E.; Bossowski, J.P.; Proics, E.; Grima-Reyes, M.; Paquet, A.; et al. GAPDH Expression Predicts the Response to R-CHOP, the Tumor Metabolic Status, and the Response of DLBCL Patients to Metabolic Inhibitors. Cell Metab. 2019, 29, 1243–1257.e10. [Google Scholar] [CrossRef]
- Mondragón, L.; Mhaidly, R.; De Donatis, G.M.; Tosolini, M.; Dao, P.; Martin, A.R.; Pons, C.; Chiche, J.; Jacquin, M.; Imbert, V.; et al. GAPDH Overexpression in the T Cell Lineage Promotes Angioimmunoblastic T Cell Lymphoma through an NF-κB-Dependent Mechanism. Cancer Cell 2019, 36, 268–287.e10. [Google Scholar] [CrossRef]
- Villa, E.; Marchetti, S.; Ricci, J.-E. No Parkin Zone: Mitophagy without Parkin. Trends Cell Biol. 2018, 28, 882–895. [Google Scholar] [CrossRef]
- Villa, E.; Proïcs, E.; Rubio-Patiño, C.; Obba, S.; Zunino, B.; Bossowski, J.P.; Rozier, R.M.; Chiche, J.; Mondragón, L.; Riley, J.S.; et al. Parkin-Independent Mitophagy Controls Chemotherapeutic Response in Cancer Cells. Cell Rep. 2017, 20, 2846–2859. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Wu, W.; Liu, D.; Lv, Y.; Deng, H.; Gao, S.; Gu, Y.; Huang, M.; Guo, X.; Liu, B.; et al. Evolution and Structure of API5 and Its Roles in Anti-Apoptosis. Protein Pept. Lett. 2021, 28, 612–622. [Google Scholar] [CrossRef] [PubMed]
- Cho, H.; Chung, J.-Y.; Song, K.-H.; Noh, K.H.; Kim, B.W.; Chung, E.J.; Ylaya, K.; Kim, J.H.; Kim, T.W.; Hewitt, S.M.; et al. Apoptosis inhibitor-5 overexpression is associated with tumor progression and poor prognosis in patients with cervical cancer. BMC Cancer 2014, 14, 545. [Google Scholar] [CrossRef] [PubMed]
- Faye, A.; Poyet, J.-L. Targeting AAC-11 in cancer therapy. Expert Opin. Ther. Targets 2010, 14, 57–65. [Google Scholar] [CrossRef]
- Jang, H.S.; Woo, S.R.; Song, K.-H.; Cho, H.; Chay, D.B.; Hong, S.-O.; Lee, H.-J.; Oh, S.J.; Chung, J.-Y.; Kim, J.-H.; et al. API5 induces cisplatin resistance through FGFR signaling in human cancer cells. Exp. Mol. Med. 2017, 49, e374. [Google Scholar] [CrossRef]
- Kim, J.W.; Cho, H.S.; Kim, J.H.; Hur, S.Y.; Kim, T.E.; Lee, J.M.; Kim, I.-K.; Namkoong, S.E. AAC-11 Overexpression Induces Invasion and Protects Cervical Cancer Cells from Apoptosis. Lab. Investig. 2000, 80, 587–594. [Google Scholar] [CrossRef][Green Version]
- Rigou, P.; Piddubnyak, V.; Faye, A.; Rain, J.-C.; Michel, L.; Calvo, F.; Poyet, J.-L. The antiapoptotic protein AAC-11 interacts with and regulates Acinus-mediated DNA fragmentation. EMBO J. 2009, 28, 1576–1588. [Google Scholar] [CrossRef]
- Song, K.-H.; Kim, S.-H.; Noh, K.H.; Bae, H.C.; Kim, J.H.; Lee, H.-J.; Song, J.; Kang, T.H.; Kim, D.-W.; Oh, S.-J.; et al. Apoptosis inhibitor 5 increases metastasis via Erk-mediated MMP expression. BMB Rep. 2015, 48, 330–335. [Google Scholar] [CrossRef]
- Noh, K.H.; Kim, S.-H.; Kim, J.H.; Song, K.-H.; Lee, Y.-H.; Kang, T.H.; Han, H.D.; Sood, A.K.; Ng, J.; Kim, K.; et al. API5 Confers Tumoral Immune Escape through FGF2-Dependent Cell Survival Pathway. Cancer Res. 2014, 74, 3556–3566. [Google Scholar] [CrossRef]
- Bousquet, G.; Feugeas, J.-P.; Gu, Y.; Leboeuf, C.; El Bouchtaoui, M.; Lu, H.; Espié, M.; Janin, A.; Di Benedetto, M. High expression of apoptosis protein (Api-5) in chemoresistant triple-negative breast cancers: An innovative target. Oncotarget 2019, 10, 6577–6588. [Google Scholar] [CrossRef][Green Version]
- Habault, J.; Fraser, C.; Pasquereau-Kotula, E.; Born-Bony, M.; Marie-Cardine, A.; Poyet, J.-L. Efficient Therapeutic Delivery by a Novel Cell-Penetrating Peptide Derived from Acinus. Cancers 2020, 12, 1858. [Google Scholar] [CrossRef]
- Habault, J.; Kaci, A.; Pasquereau-Kotula, E.; Fraser, C.; Chomienne, C.; Dombret, H.; Braun, T.; Pla, M.; Poyet, J.-L. Prophylactic and therapeutic antileukemic effects induced by the AAC-11-derived Peptide RT53. OncoImmunology 2020, 9, 1728871. [Google Scholar] [CrossRef]
- Jagot-Lacoussiere, L.; Kotula, E.; Villoutreix, B.O.; Bruzzoni-Giovanelli, H.; Poyet, J.-L. A Cell-Penetrating Peptide Targeting AAC-11 Specifically Induces Cancer Cells Death. Cancer Res. 2016, 76, 5479–5490. [Google Scholar] [CrossRef]
- Pasquereau-Kotula, E.; Habault, J.; Kroemer, G.; Poyet, J.-L. The anticancer peptide RT53 induces immunogenic cell death. PLoS ONE 2018, 13, e0201220. [Google Scholar] [CrossRef]
- Levine, J.H.; Lin, Y.; Elowitz, M.B. Functional Roles of Pulsing in Genetic Circuits. Science 2013, 342, 1193–1200. [Google Scholar] [CrossRef]
- Purvis, J.E.; Lahav, G. Encoding and Decoding Cellular Information through Signaling Dynamics. Cell 2013, 152, 945–956. [Google Scholar] [CrossRef]
- Albeck, J.G.; Mills, G.B.; Brugge, J.S. Frequency-Modulated Pulses of ERK Activity Transmit Quantitative Proliferation Signals. Mol. Cell 2013, 49, 249–261. [Google Scholar] [CrossRef]
- Aoki, K.; Kumagai, Y.; Sakurai, A.; Komatsu, N.; Fujita, Y.; Shionyu, C.; Matsuda, M. Stochastic ERK Activation Induced by Noise and Cell-to-Cell Propagation Regulates Cell Density-Dependent Proliferation. Mol. Cell 2013, 52, 529–540. [Google Scholar] [CrossRef]
- Ryu, H.; Chung, M.; Dobrzyński, M.; Fey, D.; Blum, Y.; Lee, S.S.; Peter, M.; Kholodenko, B.; Jeon, N.L.; Pertz, O. Frequency modulation of ERK activation dynamics rewires cell fate. Mol. Syst. Biol. 2016, 12, 866. [Google Scholar] [CrossRef]
- Pasparakis, M.; Vandenabeele, P. Necroptosis and its role in inflammation. Nature 2015, 517, 311–320. [Google Scholar] [CrossRef]
- Wallach, D.; Kovalenko, A.; Kang, T.-B. ‘Necrosome’-induced inflammation: Must cells die for it? Trends Immunol. 2011, 32, 505–509. [Google Scholar] [CrossRef]
- Jouan-Lanhouet, S.; Riquet, F.; Duprez, L.; Berghe, T.V.; Takahashi, N.; Vandenabeele, P. Necroptosis, in vivo detection in experimental disease models. Semin. Cell Dev. Biol. 2014, 35, 2–13. [Google Scholar] [CrossRef] [PubMed]
- Mifflin, L.; Ofengeim, D.; Yuan, J. Receptor-interacting protein kinase 1 (RIPK1) as a therapeutic target. Nat. Rev. Drug Discov. 2020, 19, 553–571. [Google Scholar] [CrossRef] [PubMed]
- Wallach, D.; Kang, T.-B.; Dillon, C.P.; Green, D.R. Programmed necrosis in inflammation: Toward identification of the effector molecules. Science 2016, 352, aaf2154. [Google Scholar] [CrossRef] [PubMed]
- Najjar, M.; Saleh, D.; Zelic, M.; Nogusa, S.; Shah, S.; Tai, A.; Finger, J.N.; Polykratis, A.; Gough, P.J.; Bertin, J.; et al. RIPK1 and RIPK3 Kinases Promote Cell-Death-Independent Inflammation by Toll-like Receptor 4. Immunity 2016, 45, 46–59. [Google Scholar] [CrossRef] [PubMed]
- Sipieter, F.; Cappe, B.; Leray, A.; De Schutter, E.; Bridelance, J.; Hulpiau, P.; Van Camp, G.; Declercq, W.; Héliot, L.; Vincent, P.; et al. Characteristic ERK1/2 signaling dynamics distinguishes necroptosis from apoptosis. iScience 2021, 24, 103074. [Google Scholar] [CrossRef] [PubMed]
- Montero, J.A.; Hurlé, J.M. Sculpturing digit shape by cell death. Apoptosis 2010, 15, 365–375. [Google Scholar] [CrossRef]
- Toyama, Y.; Peralta, X.G.; Wells, A.R.; Kiehart, D.P.; Edwards, G.S. Apoptotic Force and Tissue Dynamics During Drosophila Embryogenesis. Science 2008, 321, 1683–1686. [Google Scholar] [CrossRef]
- Suzanne, M.; Petzoldt, A.G.; Spéder, P.; Coutelis, J.-B.; Steller, H.; Noselli, S. Coupling of Apoptosis and L/R Patterning Controls Stepwise Organ Looping. Curr. Biol. 2010, 20, 1773–1778. [Google Scholar] [CrossRef]
- Kuranaga, E.; Matsunuma, T.; Kanuka, H.; Takemoto, K.; Koto, A.; Kimura, K.-I.; Miura, M. Apoptosis controls the speed of looping morphogenesis in Drosophila male terminalia. Development 2011, 138, 1493–1499. [Google Scholar] [CrossRef]
- Monier, B.; Gettings, M.; Gay, G.; Mangeat, T.; Schott, S.; Guarner, A.; Suzanne, M. Apico-basal forces exerted by apoptotic cells drive epithelium folding. Nature 2015, 518, 245–248. [Google Scholar] [CrossRef]
- Ambrosini, A.; Rayer, M.; Monier, B.; Suzanne, M. Mechanical Function of the Nucleus in Force Generation during Epithelial Morphogenesis. Dev. Cell 2019, 50, 197–211.e5. [Google Scholar] [CrossRef]
- Roellig, D.; Theis, S.; Proag, A.; Allio, G.; Bénazéraf, B.; Gros, J.; Suzanne, M. Force-generating apoptotic cells orchestrate avian neural tube bending. Dev. Cell 2022, 57, 707–718. [Google Scholar] [CrossRef]
- Eisenhoffer, G.T.; Loftus, P.D.; Yoshigi, M.; Otsuna, H.; Chien, C.-B.; Morcos, P.A.; Rosenblatt, J. Crowding induces live cell extrusion to maintain homeostatic cell numbers in epithelia. Nature 2012, 484, 546–549. [Google Scholar] [CrossRef]
- Gracia, M.; Theis, S.; Proag, A.; Gay, G.; Benassayag, C.; Suzanne, M. Mechanical impact of epithelial−mesenchymal transition on epithelial morphogenesis in Drosophila. Nat. Commun. 2019, 10, 2951. [Google Scholar] [CrossRef]
- Hallek, M.; Cheson, B.D.; Catovsky, D.; Caligaris-Cappio, F.; Dighiero, G.; Döhner, H.; Hillmen, P.; Keating, M.; Montserrat, E.; Chiorazzi, N.; et al. iwCLL guidelines for diagnosis, indications for treatment, response assessment, and supportive management of CLL. Blood 2018, 131, 2745–2760. [Google Scholar] [CrossRef]
- Chapiro, E.; Leporrier, N.; Radford-Weiss, I.; Bastard, C.; Mossafa, H.; Leroux, D.; Tigaud, I.; De Braekeleer, M.; Terré, C.; Brizard, F.; et al. Gain of the short arm of chromosome 2 (2p) is a frequent recurring chromosome aberration in untreated chronic lymphocytic leukemia (CLL) at advanced stages. Leuk. Res. 2010, 34, 63–68. [Google Scholar] [CrossRef]
- Cosson, A.; Chapiro, E.; Bougacha, N.; Lambert, J.; Herbi, L.; Cung, H.-A.; Algrin, C.; Keren, B.; Damm, F.; Gabillaud, C.; et al. Gain in the short arm of chromosome 2 (2p+) induces gene overexpression and drug resistance in chronic lymphocytic leukemia: Analysis of the central role of XPO1. Leukemia 2017, 31, 1625–1629. [Google Scholar] [CrossRef]
- Pramil, E.; Herbi Bastian, L.; Denèfle, T.; Nemati, F.; Xiao, M.; Lardé, E.; Maloum, K.; Roos-Weil, D.; Chapiro, E.; Le Garff-Tavernier, M.; et al. Targeting chronic lymphocytic leukemia with N-methylated thrombospondin-1–derived peptides overcomes drug resistance. Blood Adv. 2019, 3, 2920–2933. [Google Scholar] [CrossRef]
- Martinez-Torres, A.-C.; Quiney, C.; Attout, T.; Boullet, H.; Herbi, L.; Vela, L.; Barbier, S.; Chateau, D.; Chapiro, E.; Nguyen-Khac, F.; et al. CD47 Agonist Peptides Induce Programmed Cell Death in Refractory Chronic Lymphocytic Leukemia B Cells via PLCγ1 Activation: Evidence from Mice and Humans. PLOS Med. 2015, 12, e1001796. [Google Scholar] [CrossRef]
- Barbier, S.; Chatre, L.; Bras, M.; Sancho, P.; Roué, G.; Virely, C.; Yuste, V.J.; Baudet, S.; Rubio, M.; Esquerda, J.E.; et al. Caspase-independent type III programmed cell death in chronic lymphocytic leukemia: The key role of the F-actin cytoskeleton. Haematologica 2009, 94, 507–517. [Google Scholar] [CrossRef]
- Sarfati, M.; Fortin, G.; Raymond, M.; Susin, S. CD47 in the Immune Response: Role of Thrombospondin and SIRP-α Reverse Signaling. Curr. Drug Targets 2008, 9, 842–850. [Google Scholar] [CrossRef]
- Mathieu, M.; Martin-Jaular, L.; Lavieu, G.; Théry, C. Specificities of secretion and uptake of exosomes and other extracellular vesicles for cell-to-cell communication. Nat. Cell Biol. 2019, 21, 9–17. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ichim, G.; Gibert, B.; Adriouch, S.; Brenner, C.; Davoust, N.; Desagher, S.; Devos, D.; Dokudovskaya, S.; Dubrez, L.; Estaquier, J.; et al. Keeping Cell Death Alive: An Introduction into the French Cell Death Research Network. Biomolecules 2022, 12, 901. https://doi.org/10.3390/biom12070901
Ichim G, Gibert B, Adriouch S, Brenner C, Davoust N, Desagher S, Devos D, Dokudovskaya S, Dubrez L, Estaquier J, et al. Keeping Cell Death Alive: An Introduction into the French Cell Death Research Network. Biomolecules. 2022; 12(7):901. https://doi.org/10.3390/biom12070901
Chicago/Turabian StyleIchim, Gabriel, Benjamin Gibert, Sahil Adriouch, Catherine Brenner, Nathalie Davoust, Solange Desagher, David Devos, Svetlana Dokudovskaya, Laurence Dubrez, Jérôme Estaquier, and et al. 2022. "Keeping Cell Death Alive: An Introduction into the French Cell Death Research Network" Biomolecules 12, no. 7: 901. https://doi.org/10.3390/biom12070901
APA StyleIchim, G., Gibert, B., Adriouch, S., Brenner, C., Davoust, N., Desagher, S., Devos, D., Dokudovskaya, S., Dubrez, L., Estaquier, J., Gillet, G., Guénal, I., Juin, P. P., Kroemer, G., Legembre, P., Levayer, R., Manon, S., Mehlen, P., Meurette, O., ... Mollereau, B. (2022). Keeping Cell Death Alive: An Introduction into the French Cell Death Research Network. Biomolecules, 12(7), 901. https://doi.org/10.3390/biom12070901












