Influence of Carbon Sources on the Phenolic Compound Production by Euglena gracilis Using an Untargeted Metabolomic Approach
Abstract
:1. Introduction
2. Materials and Methods
2.1. Algal Growth and Biomass Harvesting
2.2. Antioxidant Activity Assay
2.3. Phenolic Compounds Extraction and HPLC Separation
2.4. Mass Spectrometry Analysis
2.5. Statistical Analysis
3. Results
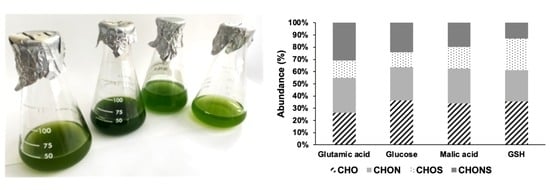
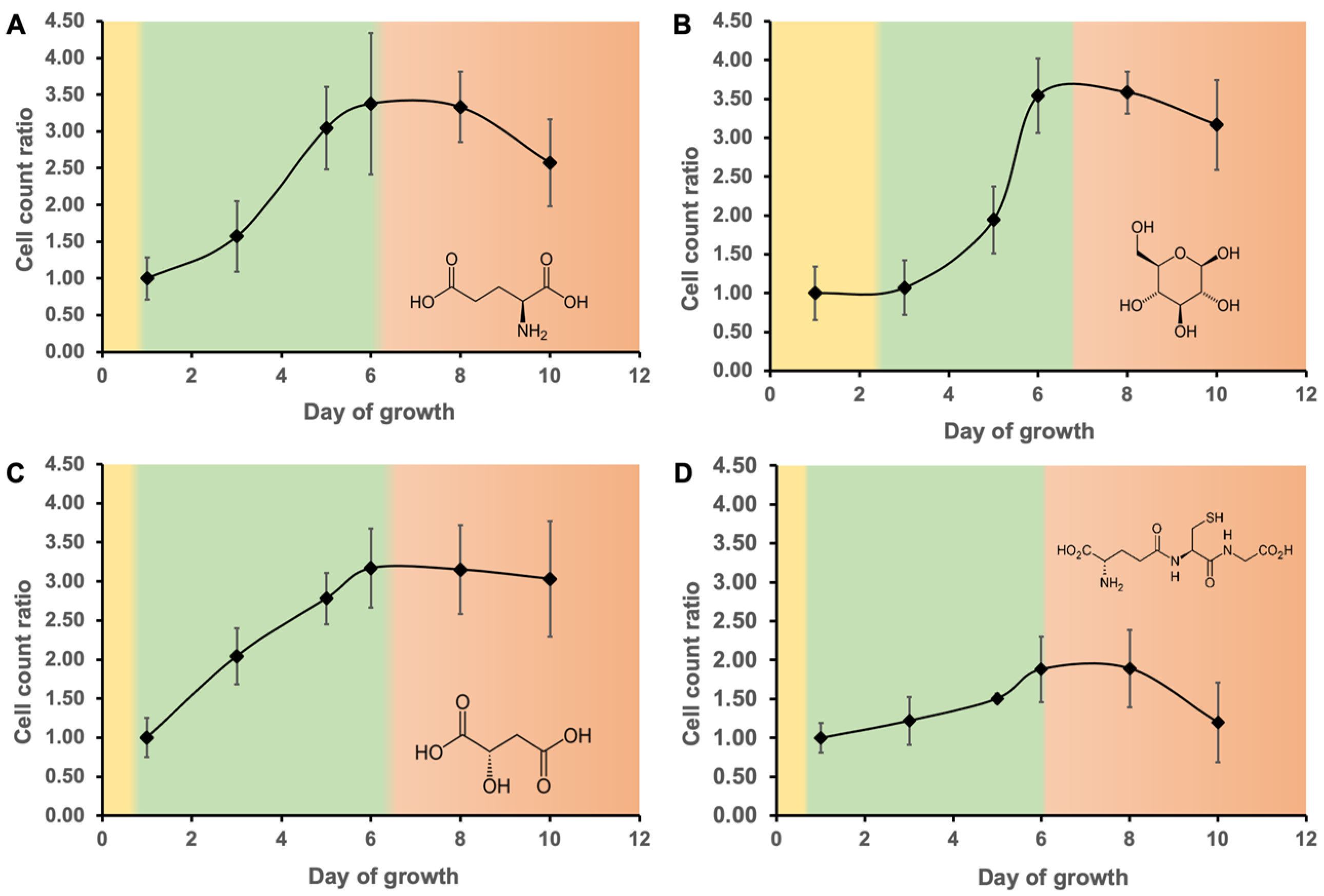
3.1. Growth Curve and Carbon Sources
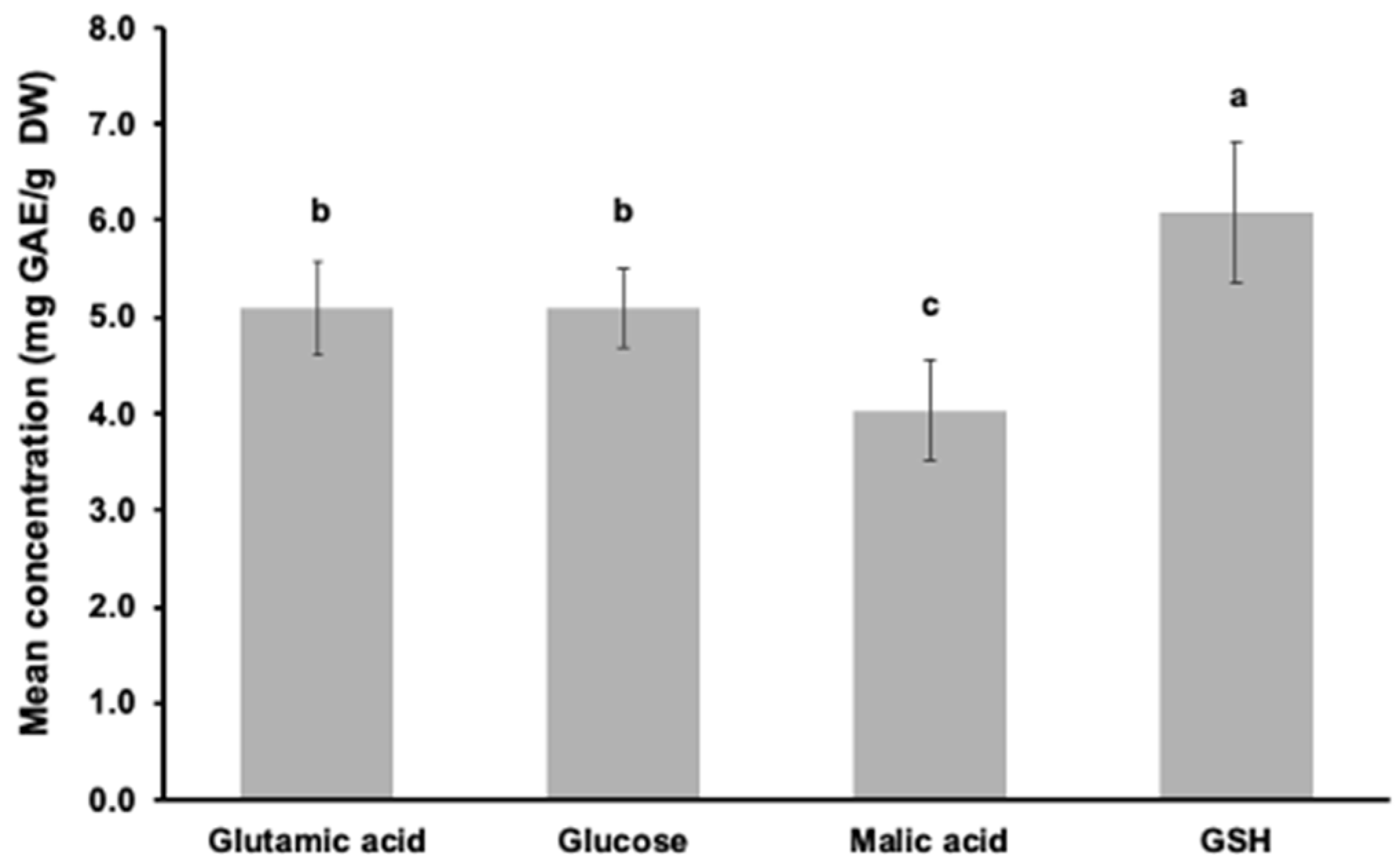
3.2. Phenolic Concentration Based on the Folin–Ciocalteu Assay
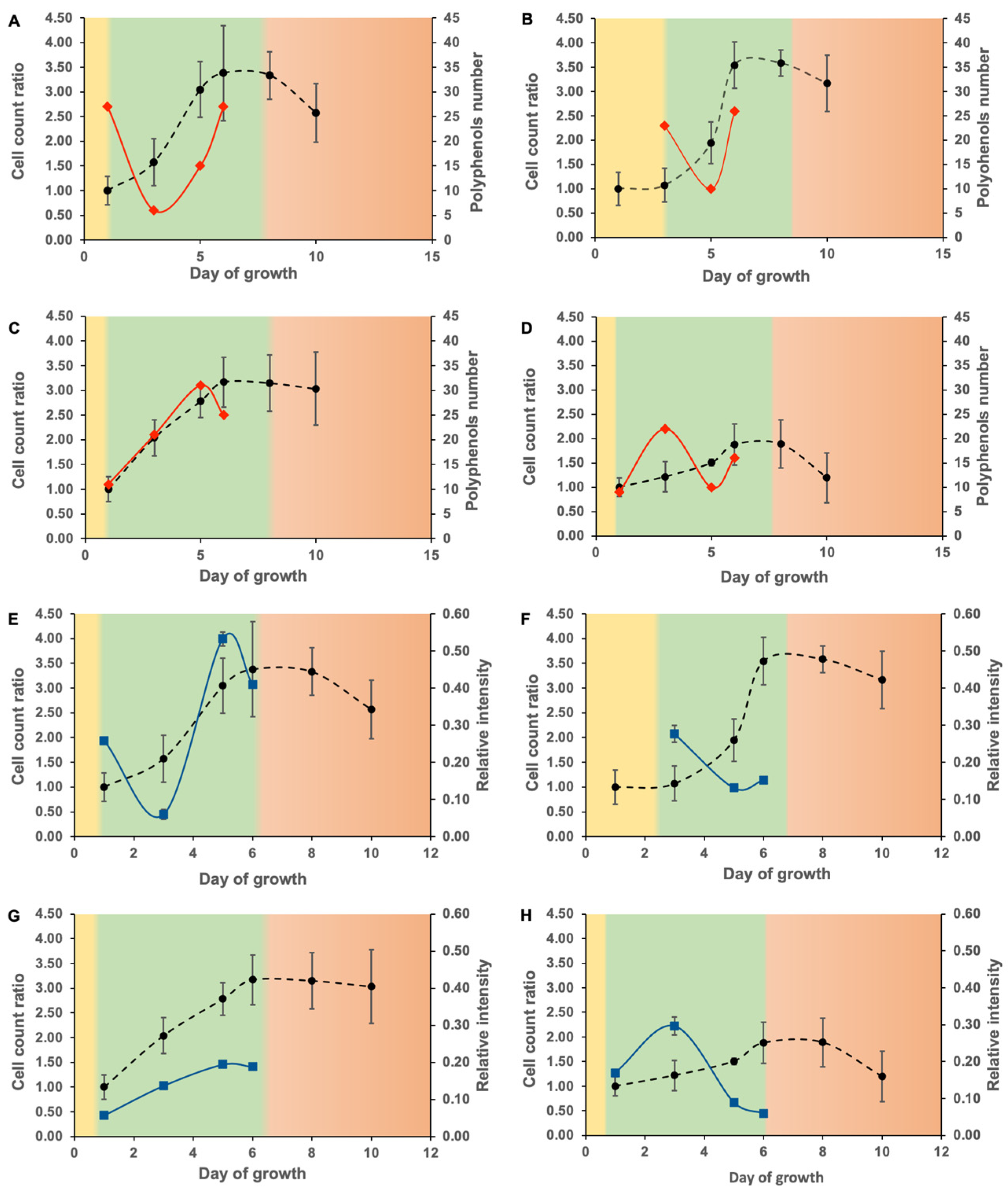
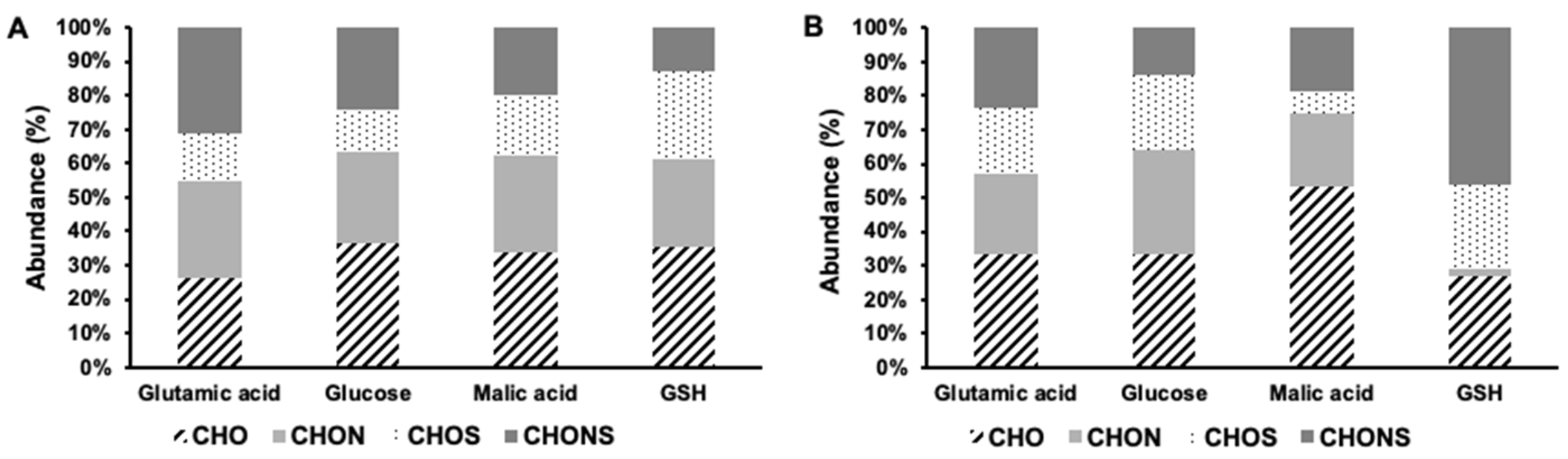
3.3. Phenolic Compound Molecular Characterization
4. Discussion
4.1. Impacts on Phenolic Production
4.2. Impacts on Phenolic Molecular Composition
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Joseph, L.; Jun, B.M.; Flora, J.R.V.; Park, C.M.; Yoon, Y. Removal of heavy metals from water sources in the developing world using low cost materials: A review. Chemosphere 2019, 229, 142–159. [Google Scholar] [CrossRef]
- Zhou, Q.; Yang, N.; Li, Y.; Ren, B.; Ding, X.; Bian, H.; Yao, X. Total concentrations and sources of heavy metal pollution in global river and lake water bodies from 1972 to 2017. Glob. Ecol. Conserv. 2020, 22, e00925. [Google Scholar] [CrossRef]
- Briffa, J.; Sinagra, E.; Blundell, R. Heavy metal pollution in the environment and their toxicological effects on humans. Heliyon 2020, 6, E04691. [Google Scholar] [CrossRef]
- Hedbavna, P.; Rolfe, S.A.; Huang, W.E.; Thornton, S.F. Biodegradation of phenolic compounds and their metabolites in contaminated groundwater using microbial fuel cells. Bioresour. Technol. 2016, 200, 426–434. [Google Scholar] [CrossRef]
- Bulgariu, L.; Bulgariu, D. Bioremediation of toxic heavy metals using marine algae biomasse. In Green Materials for Wastewater Treatment; Lichtfouse, E., Schwarzbauer, J., Robert, D., Eds.; Springer: Berlin/Heidelberg, Germany, 2005; pp. 69–98. [Google Scholar] [CrossRef]
- Ogawa, T.; Nakamoto, M.; Tanaka, Y.; Sato, K.; Okazawa, A.; Kanaya, S.; Ohta, D. Exploration and characterization of chemical stimulators to maximize the wax ester production by Euglena gracilis. J. Biosci. Bioeng. 2022, 133, 243–249. [Google Scholar] [CrossRef]
- Barsanti, L.; Bastianini, A.; Passarelli, V.; Tredici, M.R.; Gualtieri, P. Fatty Acid Content in Wild Type and WZSL Mutant of Euglena gracilis—Effects of Carbon Source and Growth Conditions. J. Appl. Phycol. 2000, 12, 515–520. [Google Scholar] [CrossRef]
- Lewis, A.; Guéguen, C. How growth conditions of Euglena gracilis cells influence cellular composition as evidenced by Fourier transform infrared and direct infusion high resolution mass spectrometry. J. Appl. Phycol. 2019, 32, 153–163. [Google Scholar] [CrossRef]
- Tolivia, A.; Conforti, V.; Córdoba, O.; Flores, L. Chemical constituents and biological activity of Euglena gracilis extracts. J. Pharm. Res. 2013, 7, 209–214. [Google Scholar] [CrossRef]
- Moreno-Sánchez, R.; Rodríguez-Enríquez, S.; Jasso-Chávez, R.; Saavedra, E.; García-García, J.D. Biochemistry and physiology of heavy metal resistance and accumulation in Euglena. In Advances in Experimental Medicine and Biology; Schwatzbach, S.D., Shigeoka, S., Eds.; Springer: Cham, Switzerland, 2017; Volume 979, pp. 91–121. [Google Scholar] [CrossRef]
- Santiago-Martinez, M.G.; Lira-Silva, E.; Encalada, R.; Pineda, E.; Gallardo-Perez, J.C.; Zepeda-Rodriguez, A.; Moreno-Sanchez, R.; Saavedra, E.; Jasso-Chavez, R. Cadmium removal by Euglena gracilis is enhanced under anaerobic growth conditions. J. Hazard. Mat. 2015, 288, 104–112. [Google Scholar] [CrossRef]
- Winters, C.; Guéguen, C.; Noble, A. Equilibrium and kinetic studies of Cu(II) and Ni(II) sorption on living Euglena gracilis. J. Appl. Phycol. 2017, 29, 1391–1398. [Google Scholar] [CrossRef]
- Rodríguez-Zavala, J.S.; García-García, J.D.; Ortiz-Cruz, M.A.; Moreno-Sánchez, R. Molecular mechanisms of resistance to heavy metals in the protist Euglena gracilis. J. Environ. Sci. Health Part A 2007, 42, 1365–1378. [Google Scholar] [CrossRef]
- Mangal, V.; Donaldson, M.E.; Lewis, A.; Saville, B.J.; Guéguen, C. Identifying Euglena gracilis metabolic and transcriptomic adaptations in response to mercury stress. Front. Environ. Sci. 2022, 162, 1–13. [Google Scholar] [CrossRef]
- Naikoo, M.I.; Dar, M.I.; Raghib, F.; Jaleel, H.; Ahmad, B.; Raina, A.; Khan, F.A.; Naushin, F. Role and regulation of plants phenolics in abiotic stress tolerance: An overview. In Plant Signaling Molecules: Role and Regulation under Stressful Environments; Iqbal, M., Khan, R., Reddy, P.S., Ferrante, A., Khan, N.A., Eds.; Elsevier: Amsterdam, The Netherlands, 2019; pp. 157–168. [Google Scholar] [CrossRef]
- Toma, L.; Sanda, G.M.; Niculescu, L.S.; Deleanu, M.; Sima, A.V.; Stancu, C.S. Phenolic compounds exerting lipid-regulatory, anti-inflammatory and epigenetic effects as complementary treatments in cardiovascular diseases. Biomolecules 2020, 10, 641. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Balasundram, N.; Sundram, K.; Samman, S. Phenolic compounds in plants and agri-industrial by-products: Antioxidant activity, occurrence, and potential uses. Food Chem. 2006, 99, 191–203. [Google Scholar] [CrossRef]
- Mangal, V.; Phung, T.; Guéguen, C. An estimation of sulfur concentrations released by three algae (Chlorella vulgaris, Chlamydomonas reinhardtii, Scenedesmus obliquus) in response to variable growth photoperiods. Environ. Sci. Pollut. Res. 2020, 27, 12491–12498. [Google Scholar] [CrossRef]
- Mekinić, I.G.; Skorza, D.; Šimat, V.; Hamed, I.; Cagalj, M.; Perković, Z.P. Phenolic content of brown algae (Pheophyceae) species: Extraction, identification, and quantification. Biomolecules 2019, 9, 244. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, L.; Cheng, D.; Wang, L.; Gao, J.; Zhao, Q.; Wei, W.; Sun, Y. Comparative transcriptomic analysis reveals phenol tolerance mechanism of evolved Chlorella strain. Bioresour. Technol. 2017, 227, 266–272. [Google Scholar] [CrossRef] [PubMed]
- Ruta, L.L.; Farcasanu, I.C. Interaction between polyphenolic antioxidants and saccharomyces cerevisiae cells defective in heavy metal transport across the plasma membrane. Biomolecules 2020, 10, 1512. [Google Scholar] [CrossRef]
- Jung, C.; Maeder, V.; Funk, F.; Frey, F.; Sticher, H.; Frossard, E. Release of phenols form Lupinus albus L. roots exposed to Cu and their possible role in Cu detoxification. Plant Soil 2003, 252, 301–312. [Google Scholar] [CrossRef]
- Sharma, A.; Shahzad, B.; Rehman, A.; Bhardwaj, R.; Landi, M.; Zheng, B. Response of phenylpropanoid pathway and the role of polyphenols in plants under abiotic stress. Molecules 2019, 24, 2452. [Google Scholar] [CrossRef] [Green Version]
- Zhu, J.; Wakisaka, M. Finding of phytase: Understanding growth promotion mechanism of phytic acid to freshwater microalga Euglena gracilis. Bioresour. Technol. 2020, 296, 122343. [Google Scholar] [CrossRef] [PubMed]
- Vello, V.; Chu, W.L.; Lim, P.E.; Majid, N.A.; Phang, S.M. Metabolomic profiles of tropical Chlorella species in response to physiological changes during nitrogen deprivation. J. Appl. Phycol. 2018, 30, 3131–3151. [Google Scholar] [CrossRef]
- Daughney, C.J.; Siciliano, S.D.; Rencz, A.N.; Lean, D.; Fortin, D. Hg(II) adsorption by bacteria: A surface complexation model and its application to shallow acidic lakes and wetlands in Kejimkujik National Park, Nova Scotia, Canada. Environ. Sci. Technol. 2002, 36, 1546–1553. [Google Scholar] [CrossRef]
- Lewis, A.; Guéguen, C. Molecular characterization of water extractable Euglena gracilis cellular material composition using asymmetrical flow field-flow fractionation and high-resolution mass spectrometry. Anal. Bioanal. Chem. 2020, 412, 4143–4153. [Google Scholar] [CrossRef] [PubMed]
- Mangal, V.; Stenzler, B.; Poulain, A.J.; Guéguen, C. Aerobic and anaerobic bacterial mercury uptake is driven by algal organic matter composition and molecular weight. Environ. Sci. Technol. 2019, 53, 157–165. [Google Scholar] [CrossRef] [PubMed]
- Del Mondo, A.; Smerilli, A.; Ambrosino, L.; Albini, A.; Noonan, D.M.; Sansoone, C.; Brunet, C. Insights into phenolic compounds from microalgae: Structural variety and complex beneficial activities form health to nutraceutics. Crit. Rev. Biotechnol. 2021, 41, 155–171. [Google Scholar] [CrossRef] [PubMed]
- Achimón, F.; Dambolena, J.S.; Zygaldo, J.A.; Pizzolitto, R.P. Carbon sources as factors affecting the secondary metabolism of the maize pathogen Fusarium verticillioides. Food Sci. Technol. 2019, 115, 108470. [Google Scholar] [CrossRef]
- Romero-Rodríguez, A.; Robledo-Casados, I.; Sánchez, S. An overview on transcriptional regulators in Streptomyces. Biochem. Biophys. Acta 2015, 1849, 1017–1039. [Google Scholar] [CrossRef]
- Verma, N.; Shukla, S. Impact of various factors responsible for fluctuation in plant secondary metabolites. J. Appl. Res. Med. Aromat Plants 2015, 2, 105–113. [Google Scholar] [CrossRef]
- Frisvad, J.C. Media and growth conditions for induction of secondary metabolite production. In Fungal Secondary Metabolism; Keller, N.P., Turner, G., Eds.; Springer: Berlin/Heidelberg, Germany, 2012; pp. 47–58. [Google Scholar] [CrossRef]
- Julkunen-Tiito, R. Phenolic constituents on the leaves of northern willows: Methods for the analysis of certain phenolics. J. Agric. Food Chem. 1985, 330, 213–217. [Google Scholar] [CrossRef]
- Everette, J.D.; Bryant, Q.M.; Green, A.M.; Abbey, Y.A.; Wangila, G.W.; Walker, R.B. A thorough study of reactivity of various compound classes towards the folin-ciocalteu reagent. J. Agric. Food Chem. 2010, 58, 8139–8144. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parys, S.; Rosenbaum, A.; Kehraus, S.; Reher, G.; Glombitza, K.W.; König, G.M. Evaluation of quantitative methods for the determination of polyphenols in algal extracts. J. Nat. Prod. 2007, 70, 1865–1870. [Google Scholar] [CrossRef]
- Zhong, B.; Robinson, N.A.; Warner, R.D.; Barrow, C.J.; Dunshea, F.R.; Suleria, H.A.R. LC-ESI-QTOF-MS/MS characterization of seaweed phenolics and their antioxidant potential. Mar. Drugs 2020, 18, 331. [Google Scholar] [CrossRef] [PubMed]
- Tohma, H.; Köksal, E.; Kılıç, Ö.; Alan, Y.; Abdullah Yılmaz, M.; Gülçin, İ.; Bursal, E.; Alwasel, S.H. RP-HPLC/MS/MS analysis of the phenolic compounds, antioxidant and antimicrobial activities of Salvia L. Species. Antioxidants 2016, 5, 38. [Google Scholar] [CrossRef] [Green Version]
- Olaveson, M.M.; Pamela, M. Stokes Responses of the acidophilic alga Euglena mutabilis (Euglenophyceae) to carbon enrichment at pH 3. J. Phycol. 1989, 25, 529–539. [Google Scholar] [CrossRef]
- Jha, D.; Jain, V.; Sharma, B.; Kant, A.; Garlapati, V.K. Microalgae-based pharmaceuticals and nutraceuticals: An emerging field with immense market potential. ChemBioEng 2017, 4, 257–272. [Google Scholar] [CrossRef]
- Li, Y.; Tsai, W.; Hsu, Y.; Xie, M.; Chen, J. Comparison of autotrophic and mixotrophic cultivation of green microalgal biodiesel production. Energy Procedia 2014, 52, 371–376. [Google Scholar] [CrossRef] [Green Version]
- Liang, Y.; Sarkany, N.; Cui, Y. Biomass and lipid productivities of Chlorella vulgaris under autotrophic, heterotrophic and mixotrophic growth conditions. Biotechnol. Lett. 2009, 31, 1043–1049. [Google Scholar] [CrossRef] [PubMed]
- Cerventes-Garcia, D.; Troncoso-Rojas, R.; Sánchez-Estrada, A.; González-Mendoza, D.; Grimaldo-Juarez, O. Production of phenolics and flavonoids compounds in Euglena gracilis under copper stress. J. Pure Appl. Microbiol. 2013, 7, 1–8. [Google Scholar]
- Piatczak, E.; Dybowska, M.; Pluciennik, E.; Kosla, K.; Kolniak-Ostek, J.; Kalinowska-Lis, U. Identification and accumulation of phenolic compounds in the leaves and bark of Salic alba (L.) and their biological potential. Biomolecules 2020, 10, 1391. [Google Scholar] [CrossRef]
- Maksimović, Z.; Malenčić, D.; Kovačević, N. Polyphenol contents and antioxidant activity of Maydis stigma extracts. Bioresour. Technol. 2005, 96, 873–877. [Google Scholar] [CrossRef] [PubMed]
- Ngoc, P.C.; Leclercq, L.; Rossi, J.-C.; Desvignes, I.; Hertzog, J.; Fabiano-Tixier, A.-S.; Chemat, F.; Schmitt-Kopplin, P.; Cottet, H. Optimizing water-based extraction of bioactive principles of hawthorn: From experimental laboratory research to homemade preparations. Molecules 2019, 24, 4420. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuhnert, N.; Dairpoosh, F.; Yassin, G.; Golon, A.; Jaiswal, R. What is under the hump? Mass spectrometry based analysis of complex mixtures in processed food—Lessons from the characterization of black tea thearubigins, coffee melanoidines and caramel. Food Funct. 2013, 4, 1130–1147. [Google Scholar] [CrossRef]
- Koch, B.P.; Dittmar, T. From mass to structure: An aromaticity index for high-resolution mass data of natural organic matter. Rapid Commun. Mass Spectrom 2016, 20, 926–932. [Google Scholar] [CrossRef] [Green Version]
- West, T.P. Microbial production of malic acid from biofuel-related coproducts and biomass. Fermentation 2017, 3, 14. [Google Scholar] [CrossRef] [Green Version]
- Abassi, S.; Wang, H.; Kim, H.; Ki, J.S. Molecular cloning and oxidative stress responses of a novel Phi class glutathione S-transferase (GSTF) gene in the freshwater algae closterium ehrenbergii. Environ. Toxicol. 2022, 37, 789–801. [Google Scholar] [CrossRef]
- Tierney, M.S.; Smyth, T.J.; Hayes, M.; Soler-Vila, A.; Croft, A.K.; Brunton, N. Influence of pressurised liquid extraction and solid-liquid extraction methods on the phenolic content and antioxidant activities of Irish macroalgae. Int. J. Food Sci. Technol. 2013, 48, 860–869. [Google Scholar] [CrossRef]
- López, A.; Rico, M.; Rivero, A.; Suárez de Tangil, M. The effects of solvents on the phenolic contents and antioxidant activity of Stypocaulon scoparium algae extracts. Food Chem. 2011, 125, 1104–1109. [Google Scholar] [CrossRef]
- Cerventes-Garcia, D.; Troncoso-Rojas, R.; Sánchez-Estrada, A.; González-Mendoza, D.; Gutierrez-Miceli, F.; Ceceña-Duran, C.; Grimaldo-Juarez, O. Effects of cadmium on total phenolic compounds and flavonoids in Euglena gracilis. Gayana 2016, 80, 1–5. [Google Scholar] [CrossRef]
- Mukwevho, E.; Ferreira, Z.; Ayeleso, A. Potential role of sulfur-containing antioxidant systems in highly oxidative environments. Molecules 2014, 19, 19376–19389. [Google Scholar] [CrossRef] [Green Version]
- Michalak, A. Phenolic Compounds and Their Antioxidant Activity in Plants Growing under Heavy Metal Stress. Polish. J. Environ. Stud. 2006, 15, 523–530. [Google Scholar]
- Navale, A.M.; Paranjape, A.N. Glucose transporters: Physiological and pathological roles. Biophys. Rev. 2016, 8, 5–9. [Google Scholar] [CrossRef] [Green Version]
- Cruzat, V.; Rogero, M.M.; Keane, K.N.; Curi, R.; Newsholme, P. Glutamine: Metabolism and immune function, supplementation and clinical translation. Nutrients 2018, 10, 1564. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Fei, X.; Deng, X. Novel molecular insights into nitrogen starvation-induced triacylglycerols accumulation revealed by differential gene expression analysis in green algae Micractinium pusillum. Biomass Bioenergy 2012, 42, 199–211. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bernard, E.; Guéguen, C. Influence of Carbon Sources on the Phenolic Compound Production by Euglena gracilis Using an Untargeted Metabolomic Approach. Biomolecules 2022, 12, 795. https://doi.org/10.3390/biom12060795
Bernard E, Guéguen C. Influence of Carbon Sources on the Phenolic Compound Production by Euglena gracilis Using an Untargeted Metabolomic Approach. Biomolecules. 2022; 12(6):795. https://doi.org/10.3390/biom12060795
Chicago/Turabian StyleBernard, Eve, and Céline Guéguen. 2022. "Influence of Carbon Sources on the Phenolic Compound Production by Euglena gracilis Using an Untargeted Metabolomic Approach" Biomolecules 12, no. 6: 795. https://doi.org/10.3390/biom12060795
APA StyleBernard, E., & Guéguen, C. (2022). Influence of Carbon Sources on the Phenolic Compound Production by Euglena gracilis Using an Untargeted Metabolomic Approach. Biomolecules, 12(6), 795. https://doi.org/10.3390/biom12060795