Hair Follicle-Related MicroRNA-34a Serum Expression and rs2666433A/G Variant in Patients with Alopecia: A Cross-Sectional Analysis
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethical Statement
2.2. Study Participants
2.3. Blood Sampling and Genomic DNA Isolation
2.4. MIR34A rs2666433A/G Genotyping
2.5. MIR34A Expression Profiling
2.6. Statistical Analysis
3. Results
3.1. Characteristics of the Study Population
3.2. Genotyping of MIR34A Polymorphism
3.3. Impact of rs2666433 Genotype on Disease Risk Transcriptomic Signature of miR-34a
3.4. Transcriptomic Signature of miR-34a
3.5. Association of mir-34a Expression and Variant with Disease Severity
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol. 2018, 9, 402. [Google Scholar] [CrossRef]
- Wang, H.; Chen, Y.H. microRNA Biomarkers in Clinical Study. Biomolecules 2021, 11, 1810. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Wang, X.; Zhang, R. Pathogenesis of Alopecia Areata Based on Bioinformatics Analysis. Indian J. Dermatol. 2019, 64, 1–6. [Google Scholar] [CrossRef]
- Toraih, E.A.; Ameen, H.M.; Hussein, M.H.; Youssef Elabd, A.A.; Mohamed, A.M.; Abdel-Gawad, A.R.; Fawzy, M.S. Association of Autoimmune Regulator Gene Rs2075876 Variant, but Not Gene Expression with Alopecia Areata in Males: A Case-control Study. Immunol. Investig. 2020, 49, 146–165. [Google Scholar] [CrossRef] [PubMed]
- Ismail, N.A.; Toraih, E.A.; Ameen, H.M.; Gomaa, A.H.A.; Marie, R.E.M. Association of Rs231775 Genetic Variant of Cytotoxic T-lymphocyte Associated Protein 4 with Alopecia Areata Disease in Males: A Case-Control Study. Immunol. Investig. 2021, 50, 977–986. [Google Scholar] [CrossRef]
- Yi, R.; O’Carroll, D.; Pasolli, H.A.; Zhang, Z.; Dietrich, F.S.; Tarakhovsky, A.; Fuchs, E. Morphogenesis in skin is governed by discrete sets of differentially expressed microRNAs. Nat. Genet. 2006, 38, 356–362. [Google Scholar] [CrossRef] [PubMed]
- Teta, M.; Choi, Y.S.; Okegbe, T.; Wong, G.; Tam, O.H.; Chong, M.M.; Seykora, J.T.; Nagy, A.; Littman, D.R.; Andl, T.; et al. Inducible deletion of epidermal Dicer and Drosha reveals multiple functions for miRNAs in postnatal skin. Development 2012, 139, 1405–1416. [Google Scholar] [CrossRef]
- Amelio, I.; Lena, A.M.; Bonanno, E.; Melino, G.; Candi, E. miR-24 affects hair follicle morphogenesis targeting Tcf-3. Cell Death Dis. 2013, 4, e922. [Google Scholar] [CrossRef]
- Ahmed, M.I.; Alam, M.; Emelianov, V.U.; Poterlowicz, K.; Patel, A.; Sharov, A.A.; Mardaryev, A.N.; Botchkareva, N.V. MicroRNA-214 controls skin and hair follicle development by modulating the activity of the Wnt pathway. J. Cell Biol. 2014, 207, 549–567. [Google Scholar] [CrossRef]
- Andl, T.; Botchkareva, N.V. MicroRNAs (miRNAs) in the control of HF development and cycling: The next frontiers in hair research. Exp. Dermatol. 2015, 24, 821–826. [Google Scholar] [CrossRef]
- Tavakolpour, S.; Elkaei Behjati, S. The ignored roles of microRNAs in alopecia areata. Dermatol. Ther. 2017, 30, e12488. [Google Scholar] [CrossRef] [PubMed]
- Hochfeld, L.M.; Anhalt, T.; Reinbold, C.S.; Herrera-Rivero, M.; Fricker, N.; Nöthen, M.M.; Heilmann-Heimbach, S. Expression profiling and bioinformatic analyses suggest new target genes and pathways for human hair follicle related microRNAs. BMC Dermatol. 2017, 17, 3. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Cammaerts, S.; Strazisar, M.; De Rijk, P.; Del Favero, J. Genetic variants in microRNA genes: Impact on microRNA expression, function, and disease. Front. Genet. 2015, 6, 186. [Google Scholar] [CrossRef] [PubMed]
- Wei, G.J.; Yuan, M.Q.; Jiang, L.H.; Lu, Y.L.; Liu, C.H.; Luo, H.C.; Huang, H.T.; Qi, Z.Q.; Wei, Y.S. A Genetic Variant of miR-34a Contributes to Susceptibility of Ischemic Stroke Among Chinese Population. Front. Physiol. 2019, 10, 432. [Google Scholar] [CrossRef]
- Kavak, A.; Baykal, C.; Ozarmağan, G.; Akar, U. HLA in alopecia areata. Int. J. Dermatol. 2000, 39, 589–592. [Google Scholar] [CrossRef]
- Olsen, E.A.; Hordinsky, M.K.; Price, V.H.; Roberts, J.L.; Shapiro, J.; Canfield, D.; Duvic, M.; King, L.E.; McMichael, A.J.; Randall, V.A.; et al. Alopecia areata investigational assessment guidelines—Part II. National Alopecia Areata Foundation. J. Am. Acad. Dermatol. 2004, 51, 440–447. [Google Scholar] [CrossRef]
- Seetharam, K.A. Alopecia areata: An update. Indian J. Dermatol. Venereol. Leprol. 2013, 79, 563–575. [Google Scholar] [CrossRef] [PubMed]
- Kutyavin, I.V.; Afonina, I.A.; Mills, A.; Gorn, V.V.; Lukhtanov, E.A.; Belousov, E.S.; Singer, M.J.; Walburger, D.K.; Lokhov, S.G.; Gall, A.A.; et al. 3′-minor groove binder-DNA probes increase sequence specificity at PCR extension temperatures. Nucleic Acids Res. 2000, 28, 655–661. [Google Scholar] [CrossRef]
- Toraih, E.A.; Aly, N.M.; Abdallah, H.Y.; Al-Qahtani, S.A.; Shaalan, A.A.; Hussein, M.H.; Fawzy, M.S. MicroRNA-target cross-talks: Key players in glioblastoma multiforme. Tumour. Biol. 2017, 39, 1010428317726842. [Google Scholar] [CrossRef] [PubMed]
- Toraih, E.A.; Ibrahiem, A.T.; Fawzy, M.S.; Hussein, M.H.; Al-Qahtani, S.A.M.; Shaalan, A.A.M. MicroRNA-34a: A Key Regulator in the Hallmarks of Renal Cell Carcinoma. Oxid. Med. Cell. Longev. 2017, 2017, 3269379. [Google Scholar] [CrossRef]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Fawzy, M.S.; Hussein, M.H.; Abdelaziz, E.Z.; Yamany, H.A.; Ismail, H.; Toraih, E.A. Association of MicroRNA-196a2 Variant with Response to Short-Acting β2-Agonist in COPD: An Egyptian Pilot Study. PLoS ONE 2016, 11, e0152834. [Google Scholar] [CrossRef]
- Zhou, C.; Li, X.; Wang, C.; Zhang, J. Alopecia Areata: An Update on Etiopathogenesis, Diagnosis, and Management. Clin. Rev. Allergy Immunol. 2021, 61, 403–423. [Google Scholar] [CrossRef] [PubMed]
- Betz, R.C.; Petukhova, L.; Ripke, S.; Huang, H.; Menelaou, A.; Redler, S.; Becker, T.; Heilmann, S.; Yamany, T.; Duvic, M.; et al. Genome-wide meta-analysis in alopecia areata resolves HLA associations and reveals two new susceptibility loci. Nat. Commun. 2015, 6, 5966. [Google Scholar] [CrossRef]
- Mohammadi, P.; Nilforoushzadeh, M.A.; Youssef, K.K.; Sharifi-Zarchi, A.; Moradi, S.; Khosravani, P.; Aghdami, R.; Taheri, P.; Hosseini Salekdeh, G.; Baharvand, H.; et al. Defining microRNA signatures of hair follicular stem and progenitor cells in healthy and androgenic alopecia patients. J. Dermatol. Sci. 2021, 101, 49–57. [Google Scholar] [CrossRef] [PubMed]
- Paul, S.; Licona-Vázquez, I.; Serrano-Cano, F.I.; Frías-Reid, N.; Pacheco-Dorantes, C.; Pathak, S.; Chakraborty, S.; Srivastava, A. Current insight into the functions of microRNAs in common human hair loss disorders: A mini review. Hum. Cell 2021, 34, 1040–1050. [Google Scholar] [CrossRef] [PubMed]
- Fawzy, M.S.; Ibrahiem, A.T.; AlSel, B.T.A.; Alghamdi, S.A.; Toraih, E.A. Analysis of microRNA-34a expression profile and rs2666433 variant in colorectal cancer: A pilot study. Sci. Rep. 2020, 10, 16940. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Peng, R.; Li, A.; Zhang, L.; Liu, H.; Peng, H.; Zhang, Z. Sequence variation in microRNA-34a is associated with diabetes mellitus susceptibility in a southwest Chinese Han population. Int. J. Clin. Exp. Pathol. 2018, 11, 1637–1644. [Google Scholar]
- Ismail, N.M.; Toraih, E.A.; Mohammad, M.H.S.; Alshammari, E.M.; Fawzy, M.S. Association of microRNA-34a rs2666433 (A/G) Variant with Systemic Lupus Erythematosus in Female Patients: A Case-Control Study. J. Clin. Med. 2021, 10, 5095. [Google Scholar] [CrossRef]
- Gupta, M.; Mahajan, V.K.; Mehta, K.S.; Chauhan, P.S.; Rawat, R. Peroxisome proliferator-activated receptors (PPARs) and PPAR agonists: The ‘future’ in dermatology therapeutics? Arch. Dermatol. Res. 2015, 307, 767–780. [Google Scholar] [CrossRef] [PubMed]
- Ramot, Y.; Alam, M.; Oláh, A.; Bíró, T.; Ponce, L.; Chéret, J.; Bertolini, M.; Paus, R. Peroxisome Proliferator-Activated Receptor-γ-Mediated Signaling Regulates Mitochondrial Energy Metabolism in Human Hair Follicle Epithelium. J. Investig. Dermatol. 2018, 138, 1656–1659. [Google Scholar] [CrossRef]
- Chéret, J.; Piccini, I.; Hardman-Smart, J.; Ghatak, S.; Alam, M.; Lehmann, J.; Jimenez, F.; Erdmann, H.; Poblet, E.; Botchkareva, N.; et al. Preclinical evidence that the PPARγ modulator, N-Acetyl-GED-0507-34-Levo, may protect human hair follicle epithelial stem cells against lichen planopilaris-associated damage. J. Eur. Acad. Dermatol. Venereol. 2020, 34, e195–e197. [Google Scholar] [CrossRef]
- Ramot, Y.; Bertolini, M.; Boboljova, M.; Uchida, Y.; Paus, R. PPAR-γ signalling as a key mediator of human hair follicle physiology and pathology. Exp. Dermatol. 2020, 29, 312–321. [Google Scholar] [CrossRef] [PubMed]
- Islam, N.; Garza, L.A. Adipose and Hair Function: An aPPARent Connection. J. Investig. Dermatol. 2018, 138, 480–482. [Google Scholar] [CrossRef]
- Clayton, R.W.; Göbel, K.; Niessen, C.M.; Paus, R.; van Steensel, M.A.M.; Lim, X. Homeostasis of the sebaceous gland and mechanisms of acne pathogenesis. Br. J. Dermatol. 2019, 181, 677–690. [Google Scholar] [CrossRef]
- Imanishi, H.; Ansell, D.M.; Chéret, J.; Harries, M.; Bertolini, M.; Sepp, N.; Bíró, T.; Poblet, E.; Jimenez, F.; Hardman, J.; et al. Epithelial-to-Mesenchymal Stem Cell Transition in a Human Organ: Lessons from Lichen Planopilaris. J. Investig. Dermatol. 2018, 138, 511–519. [Google Scholar] [CrossRef]
- Karnik, P.; Tekeste, Z.; McCormick, T.S.; Gilliam, A.C.; Price, V.H.; Cooper, K.D.; Mirmirani, P. Hair follicle stem cell-specific PPARgamma deletion causes scarring alopecia. J. Investig. Dermatol. 2009, 129, 1243–1257. [Google Scholar] [CrossRef] [PubMed]
- Ramot, Y.; Mastrofrancesco, A.; Camera, E.; Desreumaux, P.; Paus, R.; Picardo, M. The role of PPARγ-mediated signalling in skin biology and pathology: New targets and opportunities for clinical dermatology. Exp. Dermatol. 2015, 24, 245–251. [Google Scholar] [CrossRef]
- Gutierrez-Hartmann, A.; Duval, D.L.; Bradford, A.P. ETS transcription factors in endocrine systems. Trends Endocrinol. Metab. 2007, 18, 150–158. [Google Scholar] [CrossRef]
- Lee, T.J.; Kang, H.K.; Berry, J.C.; Joo, H.G.; Park, C.; Miller, M.J.; Choi, K. ER71/ETV2 Promotes Hair Regeneration from Chemotherapeutic Drug-Induced Hair Loss by Enhancing Angiogenesis. Biomol. Ther. 2021, 29, 545–550. [Google Scholar] [CrossRef] [PubMed]
- Choi, Y.S.; Cheng, J.; Segre, J.; Sinha, S. Generation and analysis of Elf5-LacZ mouse: Unique and dynamic expression of Elf5 (ESE-2) in the inner root sheath of cycling hair follicles. Histochem. Cell Biol. 2008, 129, 85–94. [Google Scholar] [CrossRef] [PubMed]
- Goodarzi, H.R.; Abbasi, A.; Saffari, M.; Tabei, M.B.; Noori Daloii, M.R. MicroRNAs take part in pathophysiology and pathogenesis of Male Pattern Baldness. Mol. Biol. Rep. 2010, 37, 2959–2965. [Google Scholar] [CrossRef]
- Aksenenko, M.; Palkina, N.; Komina, A.; Ruksha, T. MiR-92a-1-5p and miR-328-3p Are Up-Regulated in Skin of Female Pattern Hair Loss Patients. Ann. Dermatol. 2019, 31, 256–259. [Google Scholar] [CrossRef]
- Wei, H.; Xu, X.; Yang, S.; Liu, C.; Li, Q.; Jin, P. The potential role of hsa_circ_0001079 in androgenetic alopecia via sponging hsa-miR-136-5p. J. Clin. Lab. Anal. 2022, 36, e24021. [Google Scholar] [CrossRef]
- Wang, E.H.C.; DeStefano, G.M.; Patel, A.V.; Drill, E.; Harel, S.; Cela, C.; Tavazoie, M.; Christiano, A.M. Identification of differentially expressed miRNAs in alopecia areata that target immune-regulatory pathways. Genes Immun. 2017, 18, 100–104. [Google Scholar] [CrossRef]
- Bi, Y.; Liu, G.; Yang, R. MicroRNAs: Novel regulators during the immune response. J. Cell Physiol. 2009, 218, 467–472. [Google Scholar] [CrossRef]
- Mustafa, A.I.; Al-Refaie, A.M.; El-Shimi, O.S.; Fawzy, E.; Sorour, N.E. Diagnostic implications of MicroRNAs; 155, 146 a, and 203 lesional expression in alopecia areata: A preliminary case-controlled study. J. Cosmet. Dermatol. 2021. [Google Scholar] [CrossRef] [PubMed]
- Sheng, Y.; Qi, S.; Hu, R.; Zhao, J.; Rui, W.; Miao, Y.; Ma, J.; Yang, Q. Identification of blood microRNA alterations in patients with severe active alopecia areata. J. Cell. Biochem. 2019, 120, 14421–14430. [Google Scholar] [CrossRef]
- Tafazzoli, A.; Forstner, A.J.; Broadley, D.; Hofmann, A.; Redler, S.; Petukhova, L.; Giehl, K.A.; Kruse, R.; Blaumeiser, B.; Böhm, M.; et al. Genome-Wide MicroRNA Analysis Implicates miR-30b/d in the Etiology of Alopecia Areata. J. Investig. Dermatol. 2018, 138, 549–556. [Google Scholar] [CrossRef]
- Grabarek, B.O.; Dąbala, M.; Kasela, T.; Gralewski, M.; Gładysz, D. Changes in the expression pattern of DUSP1-7 and miRNA regulating their expression in the keratinocytes treated with LPS and adalimumab. Curr. Pharm. Biotechnol. 2021, 23, 873–881. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; Shi, X.; Wang, M.; Chen, H. Study on the Mechanism of miR-34a Affecting the Proliferation, Migration, and Invasion of Human Keloid Fibroblasts by Regulating the Expression of SATB1. J. Healthc. Eng. 2021, 2021, 8741512. [Google Scholar] [CrossRef]
- Li, F.; Li, X.; Qiao, L.; Liu, W.; Xu, C.; Wang, X. MALAT1 regulates miR-34a expression in melanoma cells. Cell Death Dis. 2019, 10, 389. [Google Scholar] [CrossRef]
- Andl, T.; Murchison, E.P.; Liu, F.; Zhang, Y.; Yunta-Gonzalez, M.; Tobias, J.W.; Andl, C.D.; Seykora, J.T.; Hannon, G.J.; Millar, S.E. The miRNA-processing enzyme dicer is essential for the morphogenesis and maintenance of hair follicles. Curr. Biol. 2006, 16, 1041–1049. [Google Scholar] [CrossRef]
- Song, R.; Hu, X.Q.; Zhang, L. Mitochondrial MiRNA in Cardiovascular Function and Disease. Cells 2019, 8, 1475. [Google Scholar] [CrossRef] [PubMed]
- Prie, B.E.; Voiculescu, V.M.; Ionescu-Bozdog, O.B.; Petrutescu, B.; Iosif, L.; Gaman, L.E.; Clatici, V.G.; Stoian, I.; Giurcaneanu, C. Oxidative stress and alopecia areata. J. Med. Life 2015, 8, 43–46. [Google Scholar] [PubMed]
- Lim, Y.C.; Kim, H.; Lim, S.M.; Kim, J.S. Genetic analysis of a novel antioxidant multi-target iron chelator, M30 protecting against chemotherapy-induced alopecia in mice. BMC Cancer 2019, 19, 149. [Google Scholar] [CrossRef]
- Abdelkader, H.A.; Amin, I.; Rashed, L.A.; Samir, M.; Ezzat, M. Histone deacetylase 1 in patients with alopecia areata and acne vulgaris: An epigenetic alteration. Australas J. Dermatol. 2022. [Google Scholar] [CrossRef]
- Di Mario, C.; Petricca, L.; Gigante, M.R.; Costanzi, S.; Vischini, G.; Paglionico, A.; Varriano, V.; Bui, L.; Tanti, G.; Lanzo, L.; et al. POS0110 association of miR-155 AND miR-34a expression in lupus nephritis renal tissue with disease onset and outcomes. Ann. Rheum. Dis. 2021, 80, 266. [Google Scholar] [CrossRef]
- Xie, M.; Wang, J.; Gong, W.; Xu, H.; Pan, X.; Chen, Y.; Ru, S.; Wang, H.; Chen, X.; Zhao, Y.; et al. NF-κB-driven miR-34a impairs Treg/Th17 balance via targeting Foxp3. J. Autoimmun. 2019, 102, 96–113. [Google Scholar] [CrossRef]
- Navarro Quiroz, E.; Navarro Quiroz, R.; Pacheco Lugo, L.; Aroca Martínez, G.; Gómez Escorcia, L.; Gonzalez Torres, H.; Cadena Bonfanti, A.; Marmolejo, M.D.C.; Sanchez, E.; Villarreal Camacho, J.L.; et al. Integrated analysis of microRNA regulation and its interaction with mechanisms of epigenetic regulation in the etiology of systemic lupus erythematosus. PLoS ONE 2019, 14, e0218116. [Google Scholar] [CrossRef]
- Ganzetti, G.; Campanati, A.; Offidani, A. Alopecia Areata: A possible extraintestinal manifestation of Crohn’s disease. J. Crohns Colitis 2012, 6, 962–963. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Krammes, L.; Hart, M.; Rheinheimer, S.; Diener, C.; Menegatti, J.; Grässer, F.; Keller, A.; Meese, E. Induction of the Endoplasmic-Reticulum-Stress Response: MicroRNA-34a Targeting of the IRE1α-Branch. Cells 2020, 9, 1442. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.A.; Song, C.H. Insights Into the Role of Endoplasmic Reticulum Stress in Infectious Diseases. Front. Immunol. 2019, 10, 3147. [Google Scholar] [CrossRef]
- Yamakuchi, M.; Ferlito, M.; Lowenstein, C.J. miR-34a repression of SIRT1 regulates apoptosis. Proc. Natl. Acad. Sci. USA 2008, 105, 13421–13426. [Google Scholar] [CrossRef]
- Zhang, W.; Huang, Q.; Zeng, Z.; Wu, J.; Zhang, Y.; Chen, Z. Sirt1 Inhibits Oxidative Stress in Vascular Endothelial Cells. Oxid. Med. Cell. Longev. 2017, 2017, 7543973. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zhang, X.M.; Zong, D.D.; Ji, X.Y.; Jiang, H.; Zhang, F.Z.; He, S.D. miR-34a-5p up-regulates the IL-1β/COX2/PGE2 inflammation pathway and induces the release of CGRP via inhibition of SIRT1 in rat trigeminal ganglion neurons. FEBS Open Bio 2021, 11, 300–311. [Google Scholar] [CrossRef]
- Hoffmann, R. The potential role of cytokines and T cells in alopecia areata. J. Investig. Dermatol. Symp. Proc. 1999, 4, 235–238. [Google Scholar] [CrossRef]
- Becatti, M.; Fiorillo, C.; Barygina, V.; Cecchi, C.; Lotti, T.; Prignano, F.; Silvestro, A.; Nassi, P.; Taddei, N. SIRT1 regulates MAPK pathways in vitiligo skin: Insight into the molecular pathways of cell survival. J. Cell. Mol. Med. 2014, 18, 514–529. [Google Scholar] [CrossRef]
- Cao, C.; Lu, S.; Kivlin, R.; Wallin, B.; Card, E.; Bagdasarian, A.; Tamakloe, T.; Wang, W.J.; Song, X.; Chu, W.M.; et al. SIRT1 confers protection against UVB- and H2O2-induced cell death via modulation of p53 and JNK in cultured skin keratinocytes. J. Cell. Mol. Med. 2009, 13, 3632–3643. [Google Scholar] [CrossRef]
- Zhang, D.; Qiu, X.; Li, J.; Zheng, S.; Li, L.; Zhao, H. TGF-β secreted by tumor-associated macrophages promotes proliferation and invasion of colorectal cancer via miR-34a-VEGF axis. Cell Cycle 2018, 17, 2766–2778. [Google Scholar] [CrossRef] [PubMed]
- Goldman, C.K.; Tsai, J.C.; Soroceanu, L.; Gillespie, G.Y. Loss of vascular endothelial growth factor in human alopecia hair follicles. J. Investig. Dermatol. 1995, 104, 18S–20S. [Google Scholar] [CrossRef]
- Simonetti, O.; Lucarini, G.; Bernardini, M.L.; Simoncini, C.; Biagini, G.; Offidani, A. Expression of vascular endothelial growth factor, apoptosis inhibitors (survivin and p16) and CCL27 in alopecia areata before and after diphencyprone treatment: An immunohistochemical study. Br. J. Dermatol. 2004, 150, 940–948. [Google Scholar] [CrossRef]
- Deng, W.; Hu, T.; Han, L.; Liu, B.; Tang, X.; Chen, H.; Chen, X.; Wan, M. miRNA microarray profiling in patients with androgenic alopecia and the effects of miR-133b on hair growth. Exp. Mol. Pathol. 2021, 118, 104589. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Sun, Y.; Guo, C.; Liu, T.; Fei, X.; Chang, C. Androgen receptor regulates ASS1P3/miR-34a-5p/ASS1 signaling to promote renal cell carcinoma cell growth. Cell Death Dis. 2019, 10, 339. [Google Scholar] [CrossRef] [PubMed]
- Peng, F.; Huang, X.; Li, P.; Tang, J. miR-34a inhibits proliferation of prostate cancer LNCaP cells by regulating androgen receptor gene. Chin. J. Clin. Pharmacol. Ther. 2021, 26, 10–17. [Google Scholar]

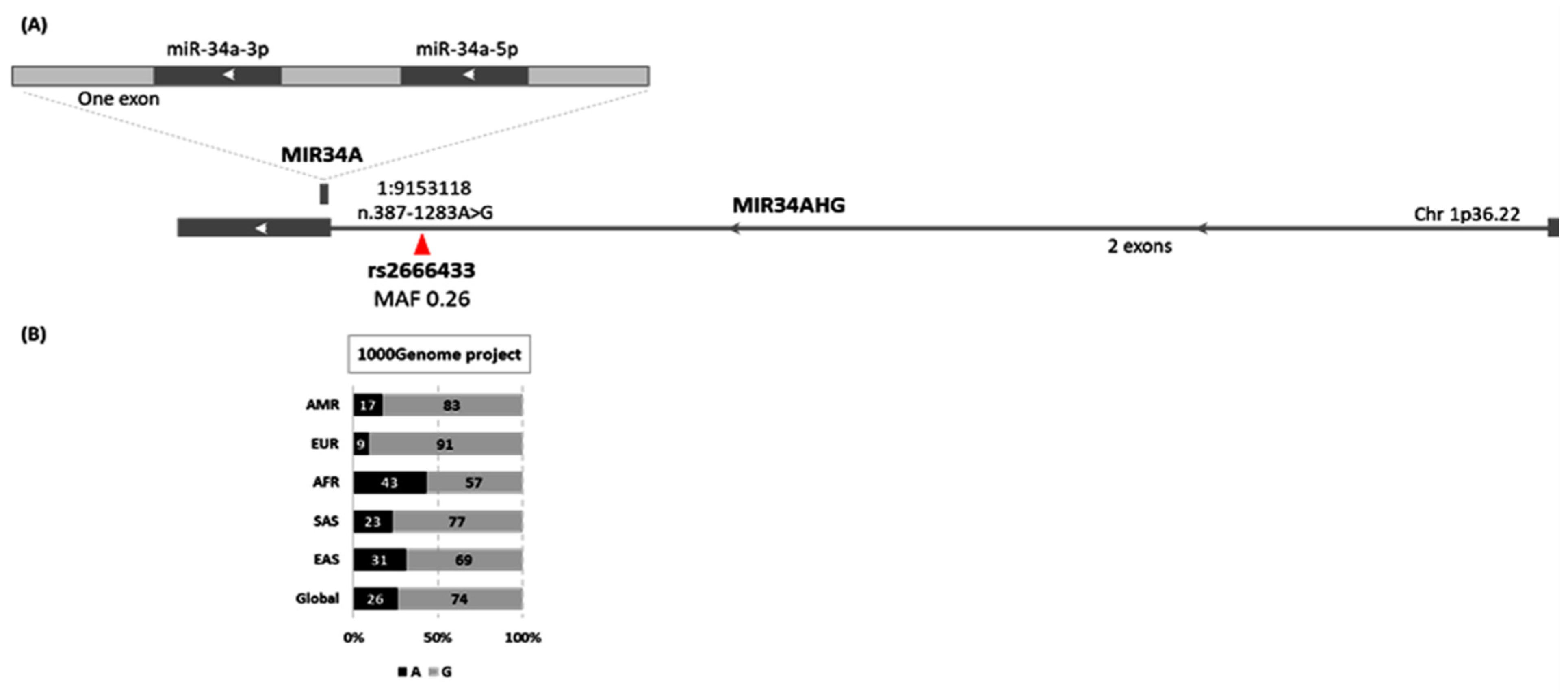
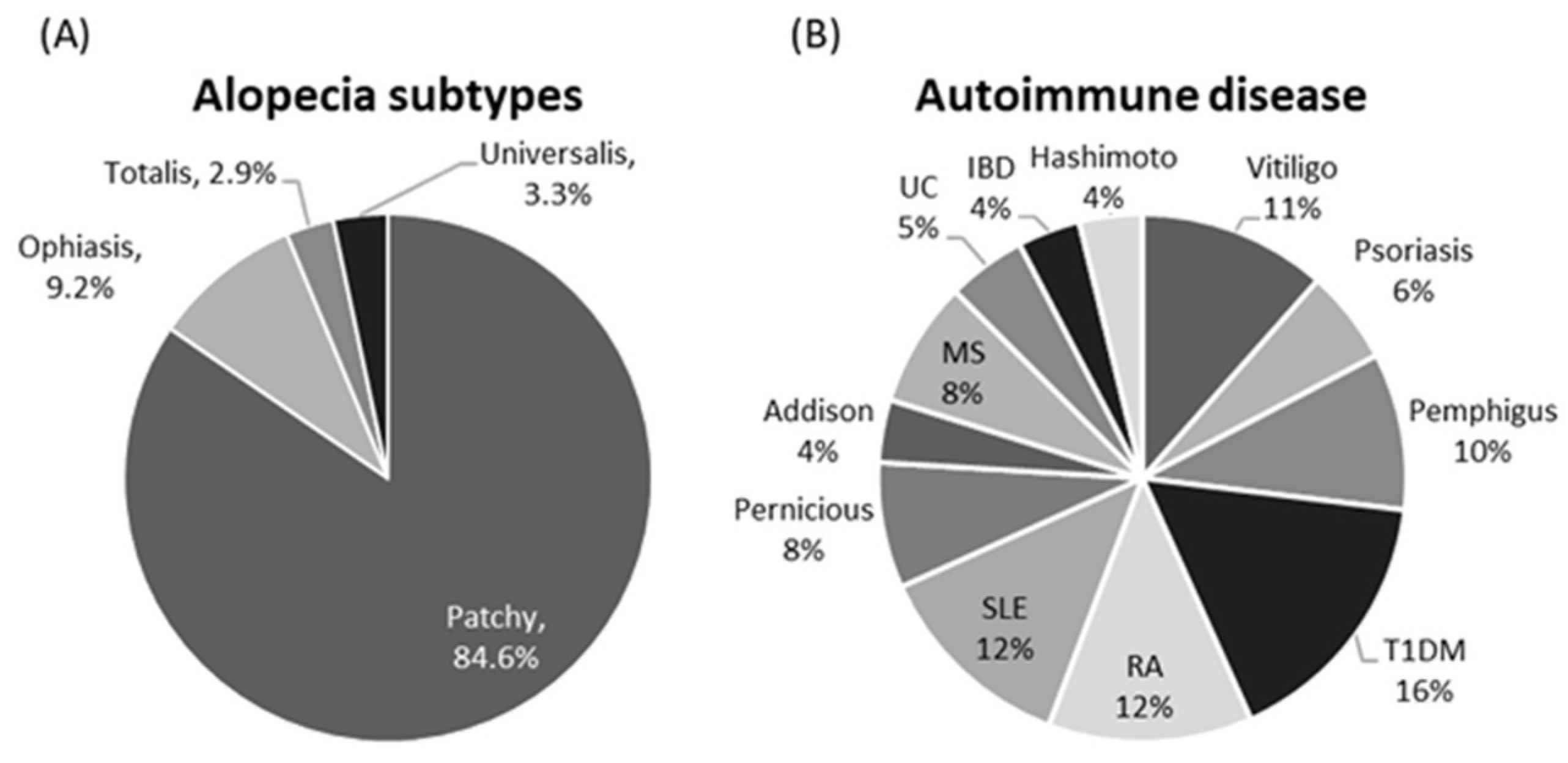
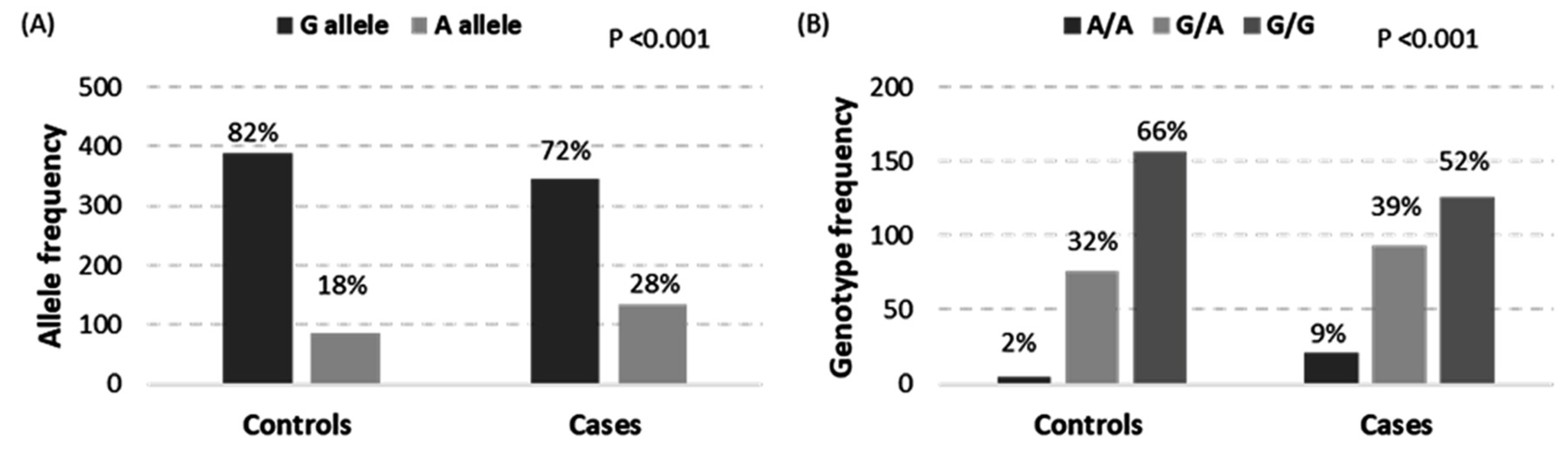
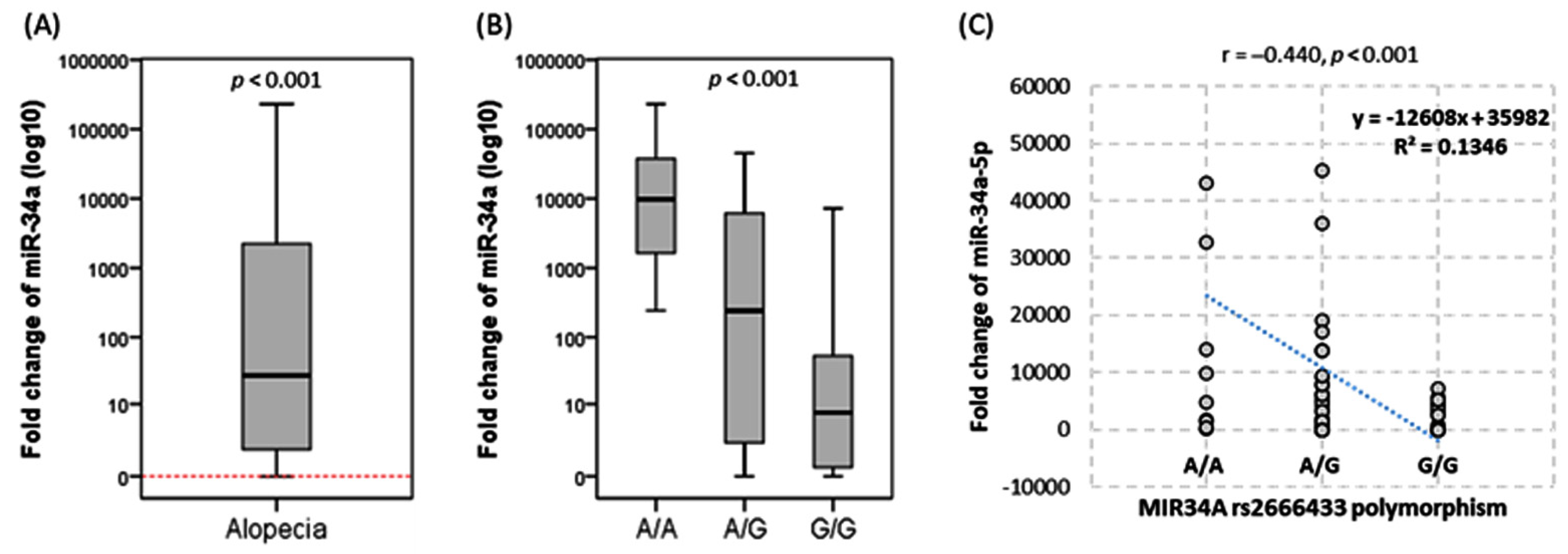
| Variables | Categories | Controls | AA | p-Value | OR (95% CI) |
|---|---|---|---|---|---|
| Age, years | Mean ± SD | 29.3 ± 5.4 | 30.7 ± 7.1 | 0.09 | |
| ≤30 years | 142 (59.2) | 120 (50) | 0.05 | ||
| >30 years | 98 (40.8) | 120 (50) | 1.44 (0.99–2.07) | ||
| Sex | Male | 194 (80.8) | 208 (86.7) | 0.10 | |
| Female | 46 (19.2) | 32 (13.3) | 0.64 (0.39–1.06) | ||
| BMI, Kg/m2 | Mean ± SD | 25.4 ± 2.6 | 25.8 ± 2.8 | 0.08 | |
| Obesity | Negative | 230 (95.8) | 222 (92.5) | 0.17 | |
| Positive | 10 (4.2) | 18 (7.5) | 1.85 (0.84–4.12) | ||
| Residence | Port-said | 18 (7.5) | 13 (5.4) | 0.22 | |
| Suez | 21 (8.8) | 15 (6.3) | |||
| Ismailia | 59 (24.6) | 77 (32.1) | |||
| Cairo | 142 (59.2) | 135 (56.3) | |||
| Occupation | Student | 96 (40) | 106 (44.2) | 0.17 | |
| Unemployed | 97 (40.4) | 102 (42.5) | |||
| Employed | 47 (19.6) | 32 (13.3) | |||
| Family history | Alopecia | 0 (0) | 106 (44.2) | NA | |
| Autoimmune dis | 0 (0) | 103 (42.9) | NA |
| Characteristics | Patchy and Aphiasis | Totalis and Universalis | p-Value | OR (95% CI) | |
|---|---|---|---|---|---|
| Number | 225 | 15 | |||
| Demographic characteristics | |||||
| Age, year | ≤30 years | 113 (50.2) | 7 (46.7) | 0.37 | 1.50 (0.67–3.33) |
| >30 years | 112 (49.8) | 8 (53.3) | |||
| Sex | Male | 195 (86.7) | 13 (86.7) | 1.00 | 1.00 (0.98–1.10) |
| Female | 30 (13.3) | 2 (13.3) | |||
| Obesity | Negative | 207 (92) | 15 (100) | 0.61 | 0.85 (0.63–1.16) |
| Positive | 18 (8) | 0 (0) | |||
| Residence | Port-said | 12 (5.3) | 1 (6.7) | 0.68 | |
| Suez | 13 (5.8) | 2 (13.3) | |||
| Ismailia | 73 (32.4) | 4 (26.7) | |||
| Cairo | 127 (56.4) | 8 (53.3) | |||
| Occupation | Student | 97 (43.1) | 9 (60) | 0.82 | |
| Unemployed | 96 (42.7) | 6 (40) | |||
| Employed | 32 (14.2) | 0 (0) | |||
| Family history of alopecia | Negative | 126 (56) | 8 (53.3) | 0.53 | 0.83 (0.58–1.19) |
| Positive | 99 (44) | 7 (46.7) | |||
| Family history of autoimmune disease | Negative | 129 (57.3) | 8 (53.3) | 0.79 | 0.83 (0.53–1.19) |
| Positive | 96 (42.7) | 7 (46.7) | |||
| Prior episode of alopecia | Negative | 73 (32.4) | 8 (53.3) | 0.15 | 0.42 (0.87–1.02) |
| Positive | 152 (67.6) | 7 (46.7) | |||
| Duration of disease, month | ≤1 years | 190 (84.4) | 15 (100) | 0.40 | 0.92 (0.89–0.96) |
| >1 years | 35 (15.6) | 0 (0) | |||
| Age at onset | ≤20 years | 205 (91.1) | 12 (80) | 0.163 | 2.56 (0.66–9.84) |
| >20 years | 20 (8.9) | 3 (20) | |||
| Disease characteristics | |||||
| Nail changes | Negative | 145 (64.4) | 12 (80) | 0.27 | 0.83 (0.58–1.19) |
| Positive | 80 (35.6) | 3 (20) | |||
| Itching | Negative | 169 (75.1) | 15 (100) | 0.40 | 0.80 (0.51–1.24) |
| Positive | 56 (24.9) | 0 (0) | |||
| Scalp infection | Negative | 164 (72.9) | 10 (66.7) | 0.56 | 1.34 (0.44–4.09) |
| Positive | 61 (27.1) | 5 (33.3) | |||
| Concomitant comorbidity | None | 154 (68.4) | 12 (80) | 0.64 | |
| Single | 47 (20.9) | 2 (13.3) | |||
| Multiple | 24 (10.7) | 1 (6.7) | |||
| Hypertension | Negative | 205 (91.1) | 14 (93.3) | 0.76 | 0.73 (0.09–5.86) |
| Positive | 20 (8.9) | 1 (6.7) | |||
| Atopy | Negative | 171 (76) | 12 (80) | 0.50 | 0.79 (0.21–2.91) |
| Positive | 54 (24) | 3 (20) | |||
| Emotional stress | Negative | 59 (26.2) | 1 (6.7) | 0.12 | 4.9 (0.64–38.6) |
| Positive | 166 (73.8) | 14 (93.3) | |||
| SALT score, % | Median (quartiles) | 8 (6–11) | 100 | <0.001 | |
| Prognostic index | Mean ± SD | 1.69 ± 1.17 | 2.26 ± 0.88 | 0.027 | |
| DLQI score | Mean ± SD | 13.1 ± 10.0 | 12.4 ± 9.8 | 0.52 | |
| Responded to treatment | Negative | 63 (28) | 1 (6.7) | 0.07 | 5.44 (0.70–4.2) |
| Positive | 162 (72) | 14 (93.3) | |||
| Model | Genotype | Controls | Cases | Crude OR (95% CI) | p-Value | Adjusted OR (95% CI) | p-Value |
|---|---|---|---|---|---|---|---|
| Codominant | G/G | 156 (65.8%) | 126 (52.5%) | 1.00 | 4 × 10−4 | 1.00 | 0.0041 |
| A/G | 76 (32.1%) | 93 (38.8%) | 1.52 (1.03–2.22) | 1.83 (1.14–2.93) | |||
| A/A | 5 (2.1%) | 21 (8.8%) | 5.20 (1.91–14.18) | 4.19 (1.33–13.19) | |||
| Dominant | G/G | 156 (65.8%) | 126 (52.5%) | 1.00 | 0.003 | 1.00 | 0.0028 |
| A/G-A/A | 81 (34.2%) | 114 (47.5%) | 1.74 (1.20–2.52) | 2.00 (1.27–3.15) | |||
| Recessive | G/G-A/G | 232 (97.9%) | 219 (91.2%) | 1.00 | 9 × 10−4 | 1.00 | 0.03 |
| A/A | 5 (2.1%) | 21 (8.8%) | 4.45 (1.65–12.00) | 3.36 (1.08–10.48) | |||
| Overdominant | G/G-A/A | 161 (67.9%) | 147 (61.2%) | 1.00 | 0.13 | 1.00 | 0.033 |
| A/G | 76 (32.1%) | 93 (38.8%) | 1.34 (0.92–1.95) | 1.65 (1.04–2.63) | |||
| Log-additive | - | - | - | 1.77 (1.30–2.43) | 2 × 10−4 | 1.91 (1.30–2.82) | 0.001 |
| Model | Genotype | Mild | Moderate/Severe | Crude OR (95% CI) | p-Value | Adjusted OR (95% CI) | p-Value |
|---|---|---|---|---|---|---|---|
| Codominant | G/G | 72 (62.6%) | 54 (43.2%) | 1.00 | 0.009 | 1.00 | 0.017 |
| A/G | 34 (29.6%) | 59 (47.2%) | 2.31 (1.33–4.01) | 2.33 (1.29–4.22) | |||
| A/A | 9 (7.8%) | 12 (9.6%) | 1.78 (0.70–4.52) | 1.70 (0.64–4.50) | |||
| Dominant | G/G | 72 (62.6%) | 54 (43.2%) | 1.00 | 0.002 | 1.00 | 0.005 |
| A/G-A/A | 43 (37.4%) | 71 (56.8%) | 2.20 (1.31–3.69) | 2.18 (1.25–3.80) | |||
| Recessive | G/G-A/G | 106 (92.2%) | 113 (90.4%) | 1.00 | 0.63 | 1.00 | 0.69 |
| A/A | 9 (7.8%) | 12 (9.6%) | 1.25 (0.51–3.09) | 1.21 (0.47–3.12) | |||
| Overdominant | G/G-A/A | 81 (70.4%) | 66 (52.8%) | 1.00 | 0.004 | 1.00 | 0.008 |
| A/G | 34 (29.6%) | 59 (47.2%) | 2.13 (1.25–3.63) | 2.16 (1.21–3.84) | |||
| Log-additive | - | - | - | 1.68 (1.12–2.52) | 0.011 | 1.64 (1.07–2.51) | 0.021 |
| Characteristics | No. of Cases | Fold Change | p-Value | Genotype | p-Value | |||
|---|---|---|---|---|---|---|---|---|
| Median (Quartiles) | AA | AG | GG | |||||
| Age | ≤30 years | 120 (50) | 35.2 (1.8–4238.7) | 0.26 | 10 (47.6) | 50 (53.8) | 60 (47.6) | 0.65 |
| >30 years | 120 (50) | 14.1 (0.4–983.4) | 11 (52.4) | 43 (46.2) | 66 (52.4) | |||
| Sex | Male | 208 (86.7) | 20.1 (1.5–1698.5) | 0.33 | 17 (81) | 83 (89.2) | 108 (85.7) | 0.54 |
| Female | 32 (13.3) | 877.4 (0.9–5442) | 4 (19) | 10 (10.8) | 18 (14.3) | |||
| Obesity | Negative | 222 (92.5) | 17.2 (1.3–2687.7) | 0.81 | 20 (95.2) | 90 (96.8) | 112 (88.9) | 0.08 |
| Positive | 18 (7.5) | 161.2 (2.1–249.1) | 1 (4.8) | 3 (3.2) | 14 (11.1) | |||
| Family history of alopecia | Negative | 134 (55.8) | 30.1 (1.6–3501.9) | 0.33 | 10 (47.6) | 60 (64.5) | 64 (50.8) | 0.09 |
| Positive | 106 (44.2) | 13.3 (0.4–1067.1) | 11 (52.4) | 33 (35.5) | 62 (49.2) | |||
| Family history autoimmune dis | Negative | 137 (57.1) | 29.6 (2–3400.3) | 0.34 | 10 (47.6) | 60 (64.5) | 67 (53.2) | 0.16 |
| Positive | 103 (42.9) | 25.2 (0.4–457.2) | 11 (52.4) | 33 (35.5) | 59 (46.8) | |||
| Age at onset | ≤20 years | 217 (90.4) | 29.6 (1.5–2601.5) | 0.83 | 20 (95.2) | 82 (88.2) | 115 (91.3) | 0.54 |
| >20 years | 23 (9.6) | 17.2 (0.9–2540.4) | 1 (4.8) | 11 (11.8) | 11 (8.7) | |||
| Duration of disease | ≤12 months | 205 (85.4) | 29.6 (1.5–2601.5) | 0.22 | 19 (90.5) | 78 (83.9) | 108 (85.7) | 0.73 |
| >12 months | 35 (14.6) | 17.2 (0.9–2540.4) | 2 (9.5) | 15 (16.1) | 18 (14.3) | |||
| Prior episode of alopecia | Negative | 81 (33.8) | 10.4 (1.2–1480.7) | 0.86 | 7 (33.3) | 33 (35.5) | 41 (32.5) | 0.90 |
| Positive | 159 (66.3) | 34.9 (1.6–2687.7) | 14 (66.7) | 60 (64.5) | 85 (67.5) | |||
| Nail changes | Negative | 157 (65.4) | 11.5 (0.7–1703.3) | 0.25 | 13 (61.9) | 67 (72) | 77 (61.1) | 0.22 |
| Positive | 83 (34.6) | 51.5 (2–3677.2) | 8 (38.1) | 26 (28) | 49 (38.9) | |||
| Itching | Negative | 184 (76.7) | 29.6 (1.3–2882.4) | 0.93 | 16 (76.2) | 77 (82.8) | 91 (72.2) | 0.09 |
| Positive | 56 (23.3) | 25.2 (1.9–504.2) | 5 (23.8) | 16 (17.2) | 35 (27.8) | |||
| Scalp infection | Negative | 174 (72.5) | 14.9 (0.7–2601.5) | 0.57 | 13 (61.9) | 65 (69.9) | 96 (76.2) | 0.30 |
| Positive | 66 (27.5) | 29.6 (2.1–2302.6) | 8 (38.1) | 28 (30.1) | 30 (23.8) | |||
| Atopy | Negative | 183 (76.3) | 14.6 (0.4–2451.1) | 0.16 | 16 (76.2) | 72 (77.4) | 95 (75.4) | 0.94 |
| Positive | 57 (23.8) | 51.5 (3.3–2601.5) | 5 (23.8) | 21 (22.6) | 31 (24.6) | |||
| Hypertension | Negative | 219 (91.3) | 14.9 (0.9–1640.1) | 0.05 | 21 (100) | 82 (88.2) | 116 (92.1) | 0.20 |
| Positive | 21 (8.8) | 249.1 (7.7–7219.4) | 0 (0) | 11 (11.8) | 10 (7.9) | |||
| Emotional stress | Negative | 60 (25) | 14.1 (0.6–1783.6) | 0.74 | 6 (28.6) | 17 (18.3) | 37 (29.4) | 0.16 |
| Positive | 180 (75) | 29.6 (1.6–2806.6) | 15 (71.4) | 76 (81.7) | 89 (70.6) | |||
| Concomitant autoimmune disease | Negative | 166 (69.2) | 3.7 (0.3–1384.3) | 0.001 | 14 (66.7) | 63 (67.7) | 89 (70.6) | 0.87 |
| Positive | 74 (30.8) | 64.7 (6.6–4804.9) | 7 (33.3) | 30 (32.3) | 37 (29.4) | |||
| T1DM | Negative | 223 (92.9) | 30.6 (1.3–1917.5) | 0.92 | 20 (95.2) | 89 (95.7) | 114 (90.5) | 0.30 |
| Positive | 17 (7.1) | 10.4 (2.1–3897.3) | 1 (4.8) | 4 (4.3) | 12 (9.5) | |||
| SLE | Negative | 227 (94.6) | 14.6 (1.1–1480.7) | 0.007 | 21 (100) | 84 (90.3) | 122 (96.8) | 0.05 |
| Positive | 13 (5.4) | 4191.1 (179.9–13704) | 0 (0) | 9 (9.7) | 4 (3.2) | |||
| RA | Negative | 227 (94.6) | 29.6 (1.2–2925.5) | 0.48 | 21 (100) | 90 (96.8) | 116 (92.1) | 0.16 |
| Positive | 13 (5.4) | 3.4 (1.9–48.3) | 0 (0) | 3 (3.2) | 10 (7.9) | |||
| Vitiligo | Negative | 228 (95) | 14.6 (1.1–1739) | 0.049 | 20 (95.2) | 85 (91.4) | 123 (97.6) | 0.11 |
| Positive | 12 (5) | 887.7 (47.8–8190) | 1 (4.8) | 8 (8.6) | 3 (2.4) | |||
| Responded to treatment | Negative | 64 (26.7) | 13.3 (0.4–887.3) | 0.15 | 3 (14.3) | 23 (24.7) | 38 (30.2) | 0.27 |
| Positive | 176 (73.3) | 34.9 (1.7–3197.1) | 18 (85.7) | 70 (75.3) | 88 (69.8) | |||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Maher, S.A.; Ismail, N.A.; Toraih, E.A.; Habib, A.H.; Gouda, N.S.; Gomaa, A.H.A.; Fawzy, M.S.; Helal, G.M. Hair Follicle-Related MicroRNA-34a Serum Expression and rs2666433A/G Variant in Patients with Alopecia: A Cross-Sectional Analysis. Biomolecules 2022, 12, 602. https://doi.org/10.3390/biom12050602
Maher SA, Ismail NA, Toraih EA, Habib AH, Gouda NS, Gomaa AHA, Fawzy MS, Helal GM. Hair Follicle-Related MicroRNA-34a Serum Expression and rs2666433A/G Variant in Patients with Alopecia: A Cross-Sectional Analysis. Biomolecules. 2022; 12(5):602. https://doi.org/10.3390/biom12050602
Chicago/Turabian StyleMaher, Shymaa Ahmed, Nader Ali Ismail, Eman A. Toraih, Alaa H. Habib, Nawal S. Gouda, Amal H. A. Gomaa, Manal S. Fawzy, and Ghada M. Helal. 2022. "Hair Follicle-Related MicroRNA-34a Serum Expression and rs2666433A/G Variant in Patients with Alopecia: A Cross-Sectional Analysis" Biomolecules 12, no. 5: 602. https://doi.org/10.3390/biom12050602
APA StyleMaher, S. A., Ismail, N. A., Toraih, E. A., Habib, A. H., Gouda, N. S., Gomaa, A. H. A., Fawzy, M. S., & Helal, G. M. (2022). Hair Follicle-Related MicroRNA-34a Serum Expression and rs2666433A/G Variant in Patients with Alopecia: A Cross-Sectional Analysis. Biomolecules, 12(5), 602. https://doi.org/10.3390/biom12050602









