Plg-RKT Expression in Human Breast Cancer Tissues
Abstract
1. Introduction
2. Materials and Methods
2.1. Tissue Microarrays
2.2. Antibodies
2.3. Immunohistochemistry and Calculation of Immunoscores
2.4. Cell Lines
2.5. Western Blotting
2.6. Flow Cytometric Analysis
2.7. Statistical Analysis
3. Results
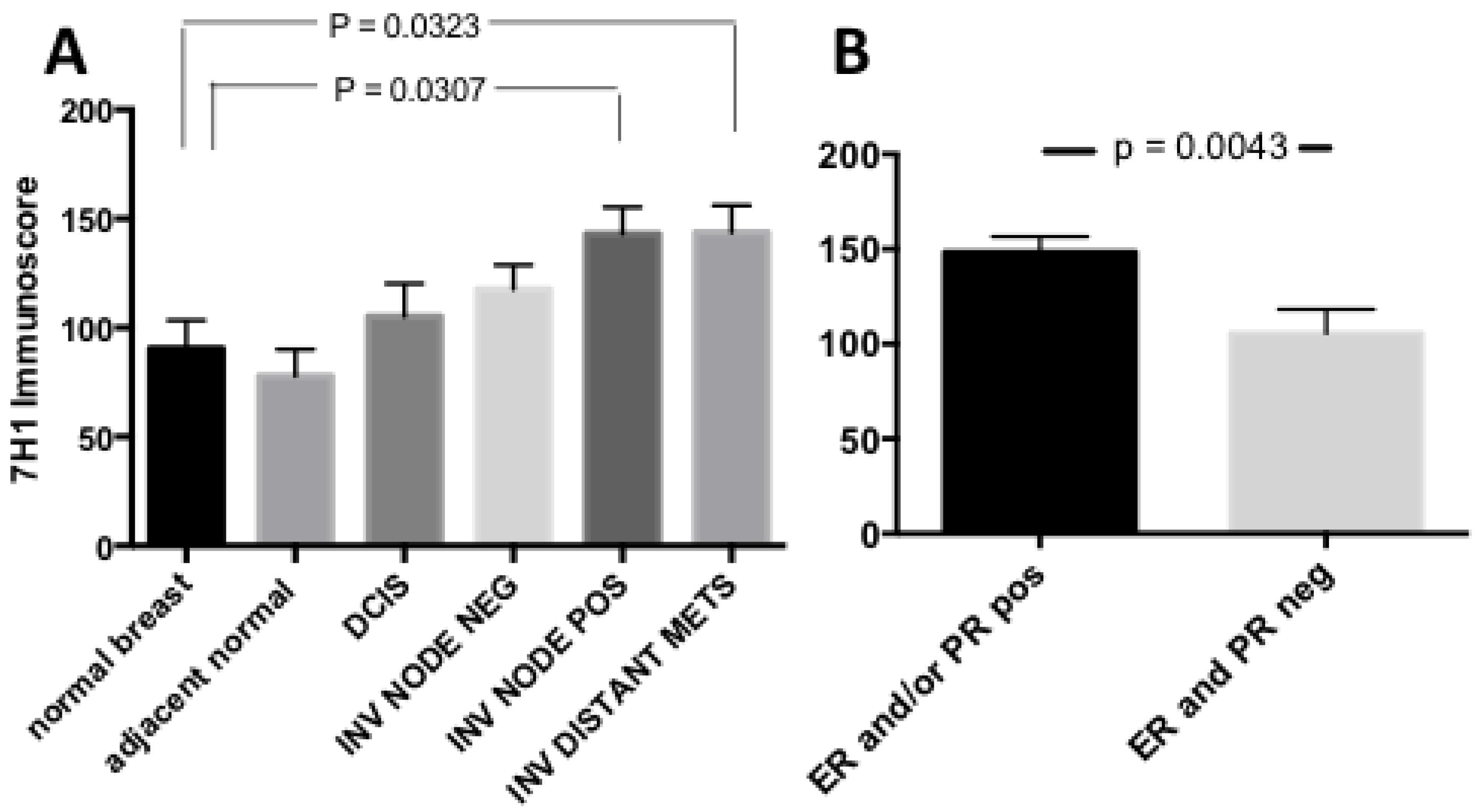
3.1. Plg-RKT Expression in Human Breast Cancer Tissue Microarrays
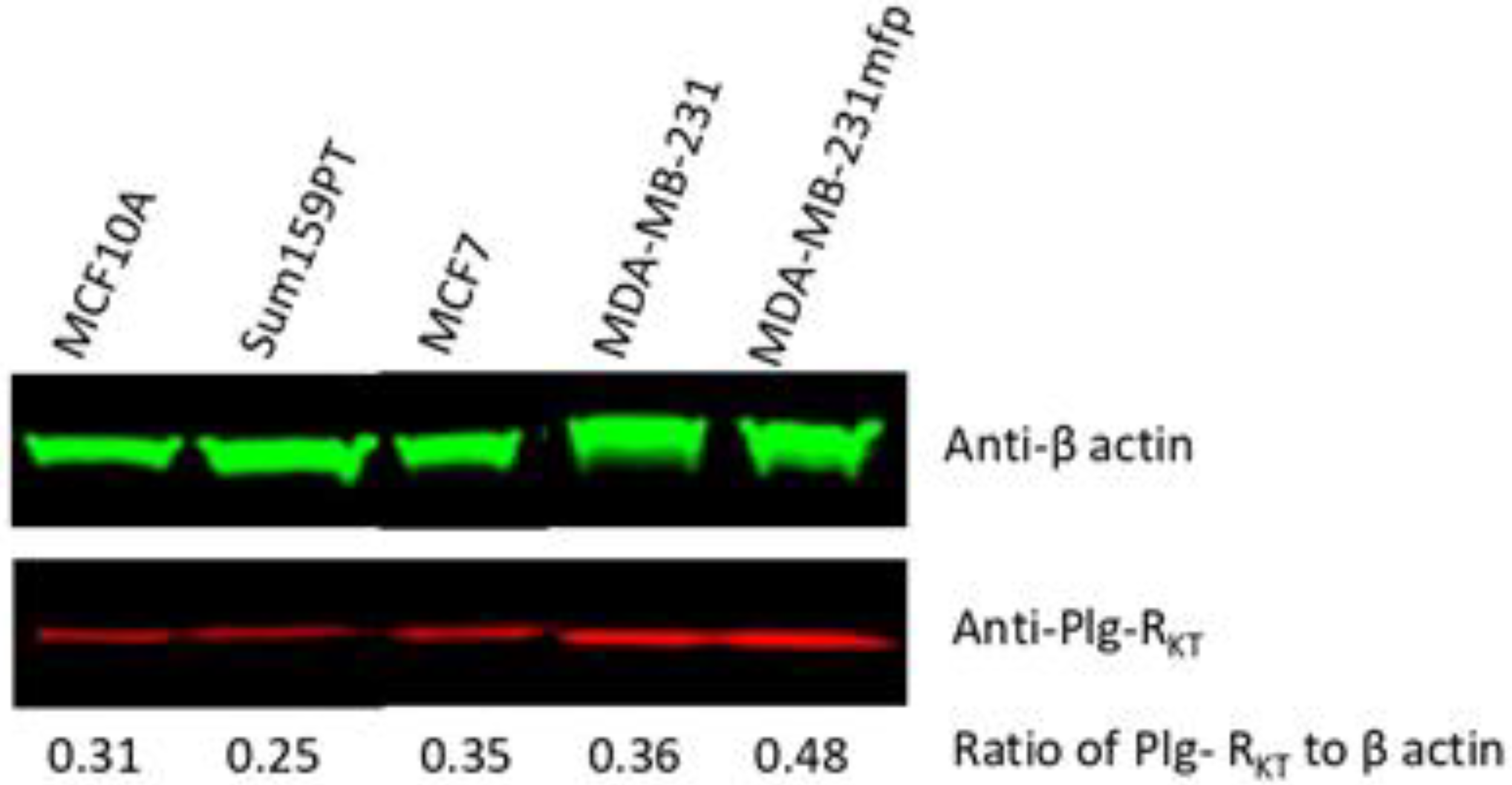
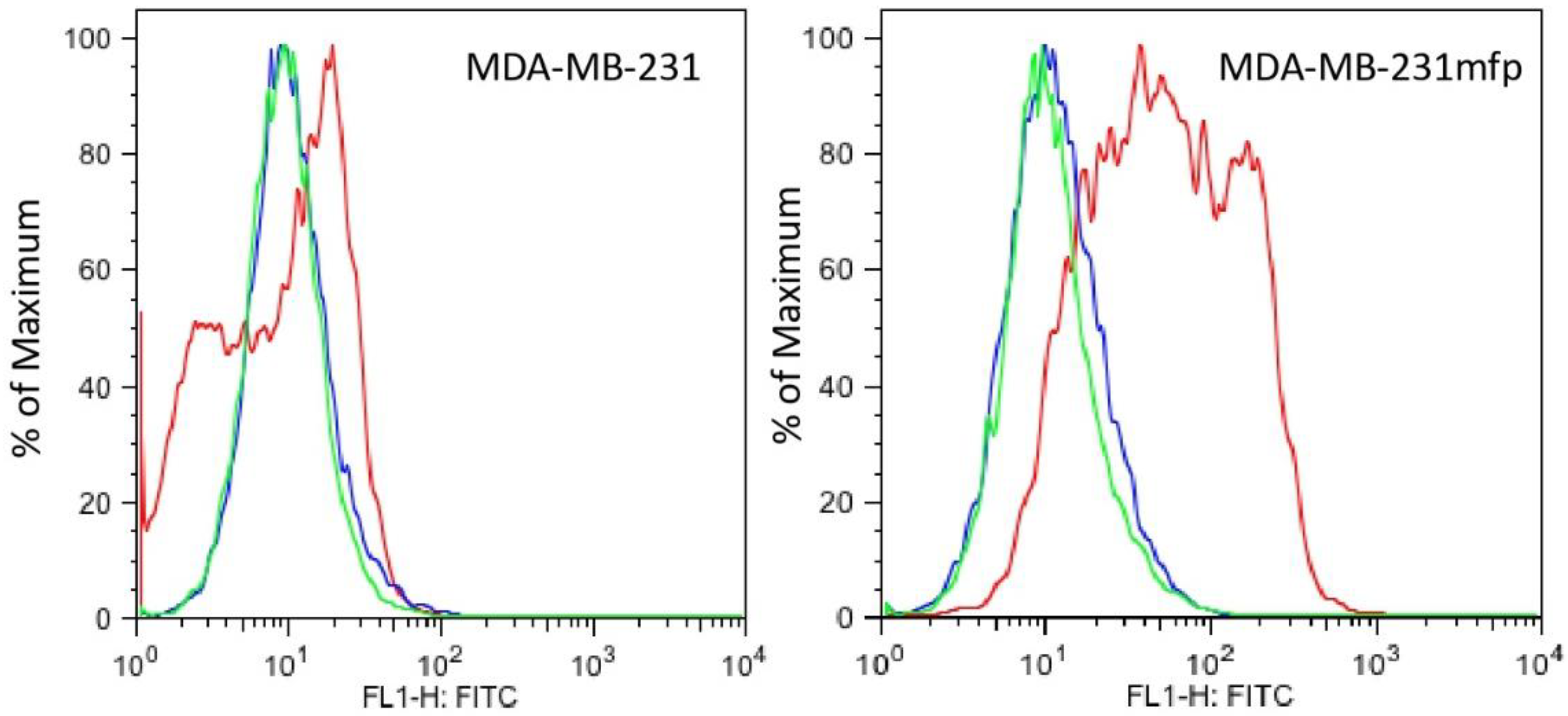
3.2. Plg-RKT Expression in Human Breast Cancer Cell Lines
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Fay, W.P.; Garg, N.; Sunkar, M. Vascular functions of the plasminogen activation system. Arterioscler. Thromb. Vasc. Biol. 2007, 27, 1231–1237. [Google Scholar] [CrossRef] [PubMed]
- Kwaan, H.C.; McMahon, B. The role of plasminogen-plasmin system in cancer. Cancer Treat. Res. 2009, 148, 43–66. [Google Scholar] [CrossRef] [PubMed]
- Andreasen, P.A.; Egelund, R.; Petersen, H.H. The plasminogen activation system in tumor growth, invasion, and metastasis. Cell. Mol. Life Sci. 2000, 57, 25–40. [Google Scholar] [CrossRef] [PubMed]
- Dano, K.; Behrendt, N.; Hoyer-Hansen, G.; Johnsen, M.; Lund, L.R.; Ploug, M.; Romer, J. Plasminogen activation and cancer. Thromb. Haemost. 2005, 93, 676–681. [Google Scholar] [CrossRef] [PubMed]
- Han, B.; Nakamura, M.; Mori, I.; Nakamura, Y.; Kakudo, K. Urokinase-type plasminogen activator system and breast cancer (Review). Oncol. Rep. 2005, 14, 105–112. [Google Scholar]
- Harbeck, N.; Kates, R.E.; Schmitt, M.; Gauger, K.; Kiechle, M.; Janicke, F.; Thomassen, C.; Look, M.P.; Foekens, J.A. Urokinase-type plasminogen activator and its inhibitor type 1 predict disease outcome and therapy response in primary breast cancer. Clin. Breast Cancer 2004, 5, 348–352. [Google Scholar] [CrossRef]
- Duffy, M.J.; McGowan, P.M.; Harbeck, N.; Thomssen, C.; Schmitt, M. uPA and PAI-1 as biomarkers in breast cancer: Validated for clinical use in level-of-evidence-1 studies. Breast Cancer Res. 2014, 16, 428. [Google Scholar] [CrossRef]
- Gouri, A.; Dekaken, A.; El Bairi, K.; Aissaoui, A.; Laabed, N.; Chefrour, M.; Ciccolini, J.; Milano, G.; Benharkat, S. Plasminogen Activator System and Breast Cancer: Potential Role in Therapy Decision Making and Precision Medicine. Biomark. Insights 2016, 11, 105–111. [Google Scholar] [CrossRef]
- Li Santi, A.; Napolitano, F.; Montuori, N.; Ragno, P. The Urokinase Receptor: A Multifunctional Receptor in Cancer Cell Biology. Therapeutic Implications. Int. J. Mol. Sci. 2021, 22, 4111. [Google Scholar] [CrossRef]
- Miles, L.A.; Hawley, S.B.; Baik, N.; Andronicos, N.M.; Castellino, F.J.; Parmer, R.J. Plasminogen receptors: The sine qua non of cell surface plasminogen activation. Front. Biosci. 2005, 10, 1754–1762. [Google Scholar]
- Miles, L.A.; Parmer, R.J. Plasminogen receptors: The first quarter century. Semin. Thromb. Hemost. 2013, 39, 329–337. [Google Scholar] [CrossRef] [PubMed]
- Félez, J.; Miles, L.A.; Fábregas, P.; Jardi, M.; Plow, E.F.; Lijnen, R.H. Characterization of cellular binding sites and interactive regions within reactants required for enhancement of plasminogen activation by tPA on the surface of leukocytic cells. Thromb. Haemost. 1996, 76, 577–584. [Google Scholar] [CrossRef] [PubMed]
- Godier, A.; Hunt, B.J. Plasminogen receptors and their role in the pathogenesis of inflammatory, autoimmune and malignant disease. J. Thromb. Haemost. 2013, 11, 26–34. [Google Scholar] [CrossRef]
- Didiasova, M.; Wujak, L.; Wygrecka, M.; Zakrzewicz, D. From plasminogen to plasmin: Role of plasminogen receptors in human cancer. Int. J. Mol. Sci. 2014, 15, 21229–21252. [Google Scholar] [CrossRef] [PubMed]
- Rein-Smith, C.M.; Church, F.C. Emerging pathophysiological roles for fibrinolysis. Curr. Opin. Hematol. 2014, 21, 438–444. [Google Scholar] [CrossRef] [PubMed]
- Kumari, S.; Malla, R. New Insight on the Role of Plasminogen Receptor in Cancer Progression. Cancer Growth Metastasis 2015, 8, 35–42. [Google Scholar] [CrossRef]
- Gonias, S.L.; Zampieri, C. Plasminogen Receptors in Human Malignancies: Effects on Prognosis and Feasibility as Targets for Drug Development. Curr. Drug Targets 2020, 21, 647–656. [Google Scholar] [CrossRef]
- Bharadwaj, A.G.; Holloway, R.W.; Miller, V.A.; Waisman, D.M. Plasmin and Plasminogen System in the Tumor Microenvironment: Implications for Cancer Diagnosis, Prognosis, and Therapy. Cancers 2021, 13, 1838. [Google Scholar] [CrossRef]
- Andronicos, N.M.; Chen, E.I.; Baik, N.; Bai, H.; Parmer, C.M.; Kiosses, W.B.; Kamps, M.P.; Yates, J.R., III; Parmer, R.J.; Miles, L.A. Proteomics-based discovery of a novel, structurally unique, and developmentally regulated plasminogen receptor, Plg-RKT, a major regulator of cell surface plasminogen activation. Blood 2010, 115, 1319–1330. [Google Scholar] [CrossRef]
- Miles, L.A.; Andronicos, N.M.; Chen, E.I.; Baik, N.; Bai, H.; Parmer, C.M.; Lighvani, S.; Nangia, S.; Kiosses, W.B.; Kamps, M.P.; et al. Identification of the novel plasminogen receptor, Plg-RKT. In Proteomics/Book 1: Human Diseases and Protein Functions; Man, T.K., Flores, R.J., Eds.; Intech: Rijeka, Croatia, 2012; pp. 219–238. ISBN 978-953-307-832-8. [Google Scholar]
- Bai, H.; Baik, N.; Kiosses, W.B.; Krajewski, S.; Miles, L.A.; Parmer, R.J. The novel plasminogen receptor, plasminogen receptor(KT) (Plg-R(KT)), regulates catecholamine release. J. Biol. Chem 2011, 286, 33125–33133. [Google Scholar] [CrossRef]
- Lighvani, S.; Baik, N.; Diggs, J.E.; Khaldoyanidi, S.; Parmer, R.J.; Miles, L.A. Regulation of macrophage migration by a novel plasminogen receptor Plg-RKT. Blood 2011, 118, 5622–5630. [Google Scholar] [CrossRef] [PubMed]
- Miles, L.A.; Baik, N.; Lighvani, S.; Khaldoyanidi, S.; Varki, N.M.; Bai, H.; Mueller, B.M.; Parmer, R.J. Deficiency of plasminogen receptor, Plg-RKT, causes defects in plasminogen binding and inflammatory macrophage recruitment in vivo. J. Thromb. Haemost. 2017, 15, 155–162. [Google Scholar] [CrossRef] [PubMed]
- Thaler, B.; Baik, N.; Hohensinner, P.J.; Baumgartner, J.; Panzenbock, A.; Stojkovic, S.; Demyanets, S.; Huk, I.; Rega-Kaun, G.; Kaun, C.; et al. Differential expression of Plg-RKT and its effects on migration of proinflammatory monocyte and macrophage subsets. Blood 2019, 134, 561–567. [Google Scholar] [CrossRef] [PubMed]
- Vago, J.P.; Sugimoto, M.A.; Lima, K.M.; Negreiros-Lima, G.L.; Baik, N.; Teixeira, M.M.; Perretti, M.; Parmer, R.J.; Miles, L.A.; Sousa, L.P. Plasminogen and the Plasminogen receptor, Plg-RKT, regulate macrophage phenotypic and functional changes. Front. Immunol. 2019, 10, 1458. [Google Scholar] [CrossRef]
- Krajewski, S.; Krajewska, M.; Ellerby, L.M.; Welsh, K.; Xie, Z.; Deveraux, Q.L.; Salvesen, G.S.; Bredesen, D.E.; Rosenthal, R.E.; Fiskum, G.; et al. Release of caspase-9 from mitochondria during neuronal apoptosis and cerebral ischemia. Proc. Natl. Acad. Sci. USA 1999, 96, 5752–5757. [Google Scholar] [CrossRef]
- Krajewska, M.; Zapata, J.M.; Meinhold-Heerlein, I.; Hedayat, H.; Monks, A.; Bettendorf, H.; Shabaik, A.; Bubendorf, L.; Kallioniemi, O.P.; Kim, H.; et al. Expression of Bcl-2 family member Bid in normal and malignant tissues. Neoplasia 2002, 4, 129–140. [Google Scholar] [CrossRef] [PubMed]
- Jessani, N.; Humphrey, M.; McDonald, W.H.; Niessen, S.; Masuda, K.; Gangadharan, B.; Yates, J.R., III; Mueller, B.M.; Cravatt, B.F. Carcinoma and stromal enzyme activity profiles associated with breast tumor growth in vivo. Proc. Natl. Acad. Sci. USA 2004, 101, 13756–13761. [Google Scholar] [CrossRef]
- Jessani, N.; Niessen, S.; Mueller, B.M.; Cravatt, B.F. Breast cancer cell lines grown in vivo: What goes in isn’t always the same as what comes out. Cell Cycle 2005, 4, 253–255. [Google Scholar] [CrossRef][Green Version]
- Suarez-Arroyo, I.J.; Feliz-Mosquea, Y.R.; Perez-Laspiur, J.; Arju, R.; Giashuddin, S.; Maldonado-Martinez, G.; Cubano, L.A.; Schneider, R.J.; Martinez-Montemayor, M.M. The proteome signature of the inflammatory breast cancer plasma membrane identifies novel molecular markers of disease. Am. J. Cancer Res. 2016, 6, 1720–1740. [Google Scholar]
- Collen, D. On the regulation and control of fibrinolysis. Thromb. Haemost. 1980, 43, 77–89. [Google Scholar] [CrossRef]
- Amens, J.N.; Bahçecioglu, G.; Zorlutuna, P. Immune System Effects on Breast Cancer. Cell. Mol. Bioeng. 2021, 14, 279–292. [Google Scholar] [CrossRef] [PubMed]
- Miles, L.A.; Baik, N.; Bai, H.; Makarenkova, H.P.; Kiosses, W.B.; Krajewski, S.; Castellino, F.J.; Valenzuela, A.; Varki, N.M.; Mueller, B.M.; et al. The Plasminogen Receptor, Plg-RKT, is Essential for Mammary Lobuloalveolar Development and Lactation. J. Thromb. Haemost. 2018, 16, 919–932. [Google Scholar] [CrossRef] [PubMed]
- Breast cancer and breastfeeding: Collaborative reanalysis of individual data from 47 epidemiological studies in 30 countries, including 50302 women with breast cancer and 96973 women without the disease. Lancet 2002, 360, 187–195. [CrossRef]
- Gonzalez-Jimenez, E.; Garcia, P.A.; Aguilar, M.J.; Padilla, C.A.; Alvarez, J. Breastfeeding and the prevention of breast cancer: A retrospective review of clinical histories. J. Clin. Nurs. 2013, 23, 2397–2403. [Google Scholar] [CrossRef]
- Bartick, M.C.; Stuebe, A.M.; Schwarz, E.B.; Luongo, C.; Reinhold, A.G.; Foster, E.M. Cost analysis of maternal disease associated with suboptimal breastfeeding. Obstet. Gynecol 2013, 122, 111–119. [Google Scholar] [CrossRef] [PubMed]
- Qiu, R.; Zhong, Y.; Hu, M.; Wu, B. Breastfeeding and Reduced Risk of Breast Cancer: A Systematic Review and Meta-Analysis. Comput. Math. Methods Med. 2022, 2022, 8500910. [Google Scholar] [CrossRef] [PubMed]
- Xing, P.; Li, J.; Jin, F. A case-control study of reproductive factors associated with subtypes of breast cancer in Northeast China. Med. Oncol. 2010, 27, 926–931. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Miles, L.A.; Krajewski, S.; Baik, N.; Parmer, R.J.; Mueller, B.M. Plg-RKT Expression in Human Breast Cancer Tissues. Biomolecules 2022, 12, 503. https://doi.org/10.3390/biom12040503
Miles LA, Krajewski S, Baik N, Parmer RJ, Mueller BM. Plg-RKT Expression in Human Breast Cancer Tissues. Biomolecules. 2022; 12(4):503. https://doi.org/10.3390/biom12040503
Chicago/Turabian StyleMiles, Lindsey A., Stan Krajewski, Nagyung Baik, Robert J. Parmer, and Barbara M. Mueller. 2022. "Plg-RKT Expression in Human Breast Cancer Tissues" Biomolecules 12, no. 4: 503. https://doi.org/10.3390/biom12040503
APA StyleMiles, L. A., Krajewski, S., Baik, N., Parmer, R. J., & Mueller, B. M. (2022). Plg-RKT Expression in Human Breast Cancer Tissues. Biomolecules, 12(4), 503. https://doi.org/10.3390/biom12040503





