Structural Comparative Modeling of Multi-Domain F508del CFTR
Abstract
:1. Introduction
2. Materials and Methods
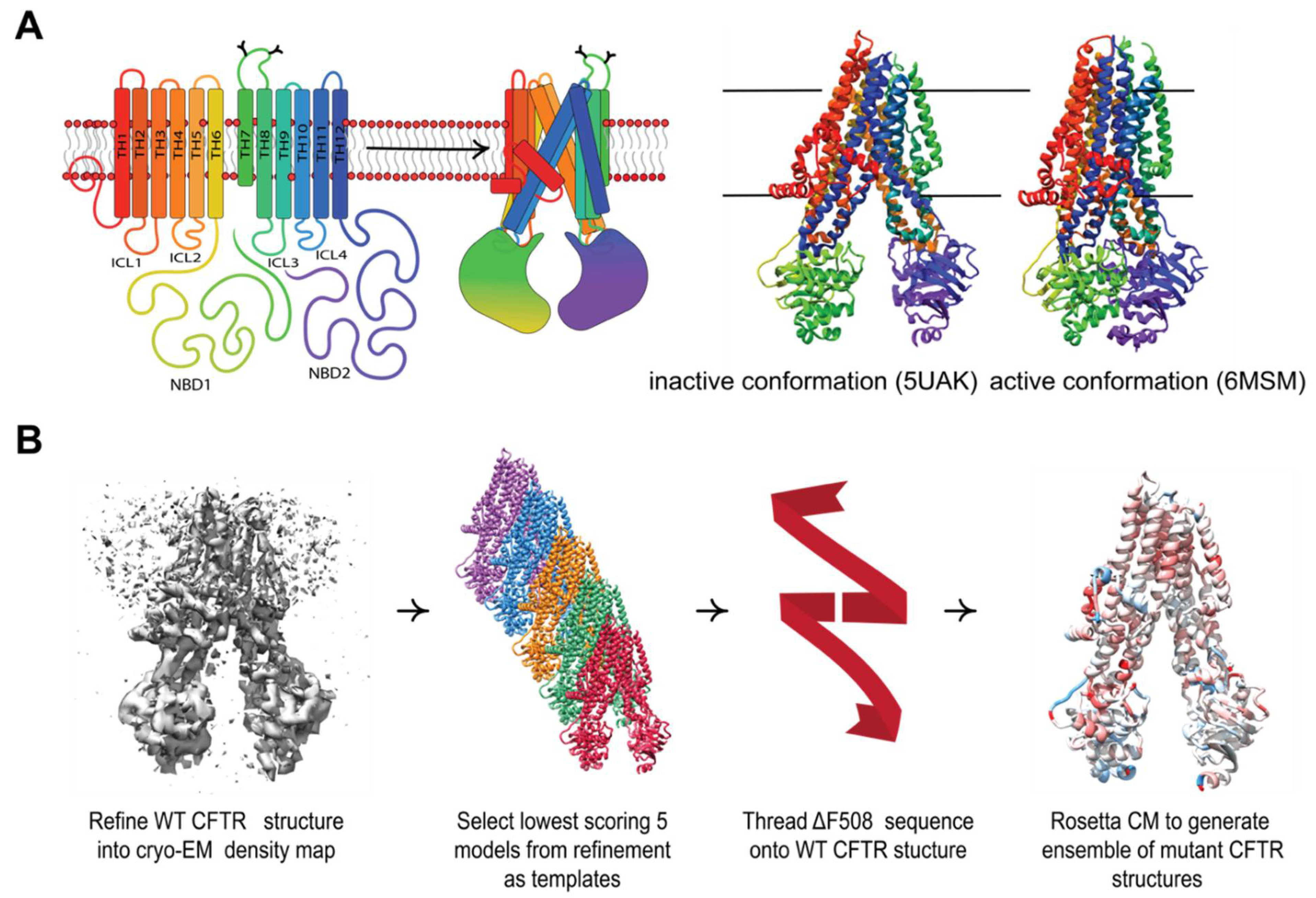
2.1. Protein Structural Data Preparation
2.2. Cryo-EM Refinement
2.3. Optimization of Cryo-EM Refinement Parameters
2.4. In Silico Mutagenesis
2.5. Rosetta Comparative Modeling
2.6. Calculation of Protein Stability Metrics
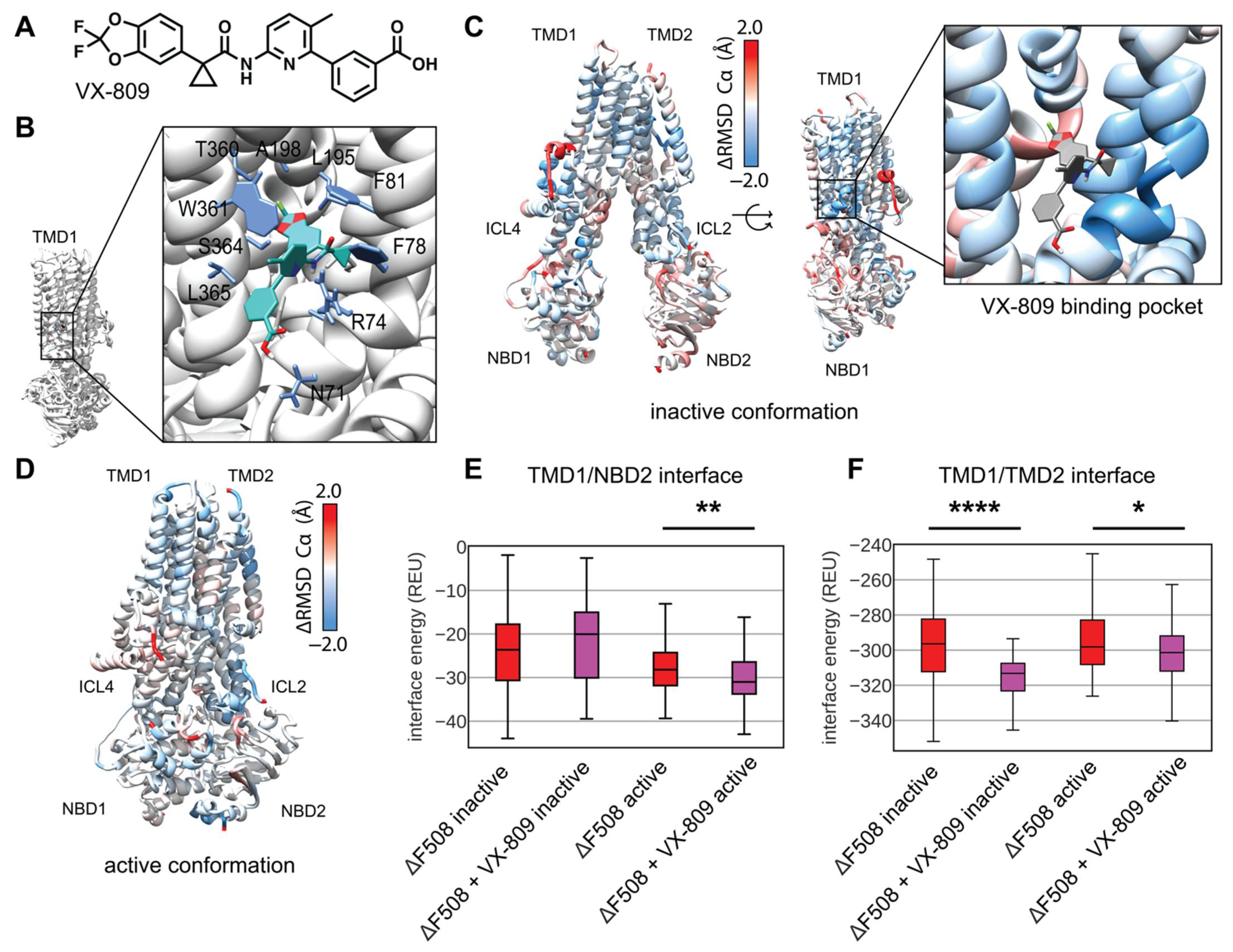
2.7. Docking and Parameterizing VX-809
3. Results
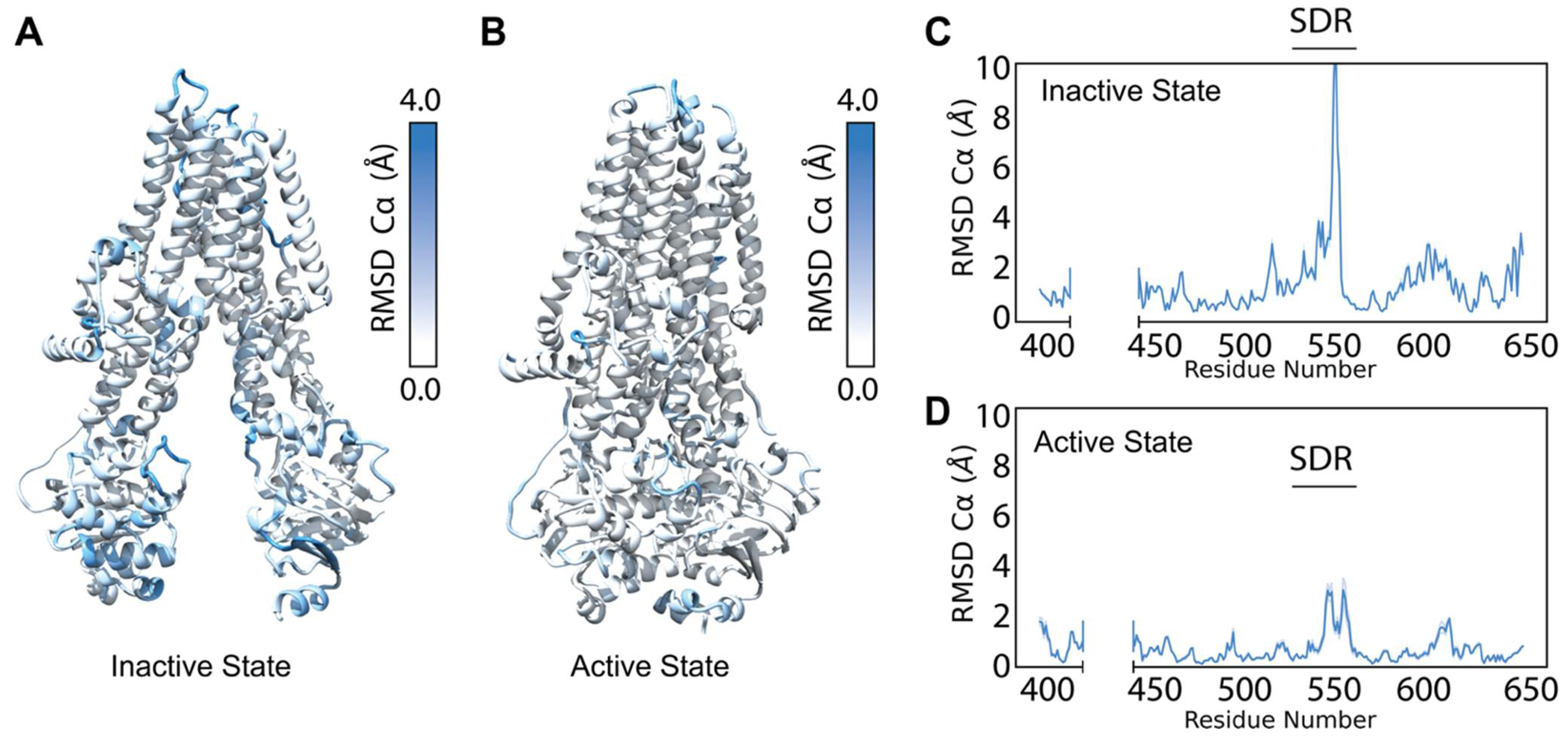
3.1. Refining CFTR Models into Available Cryo-EM Density Data
3.2. Testing F508del Modeling with CFTR NBD1 Second Site Suppressor Mutations
3.3. F508del Destabilizes Inactive and Active State of Human CFTR
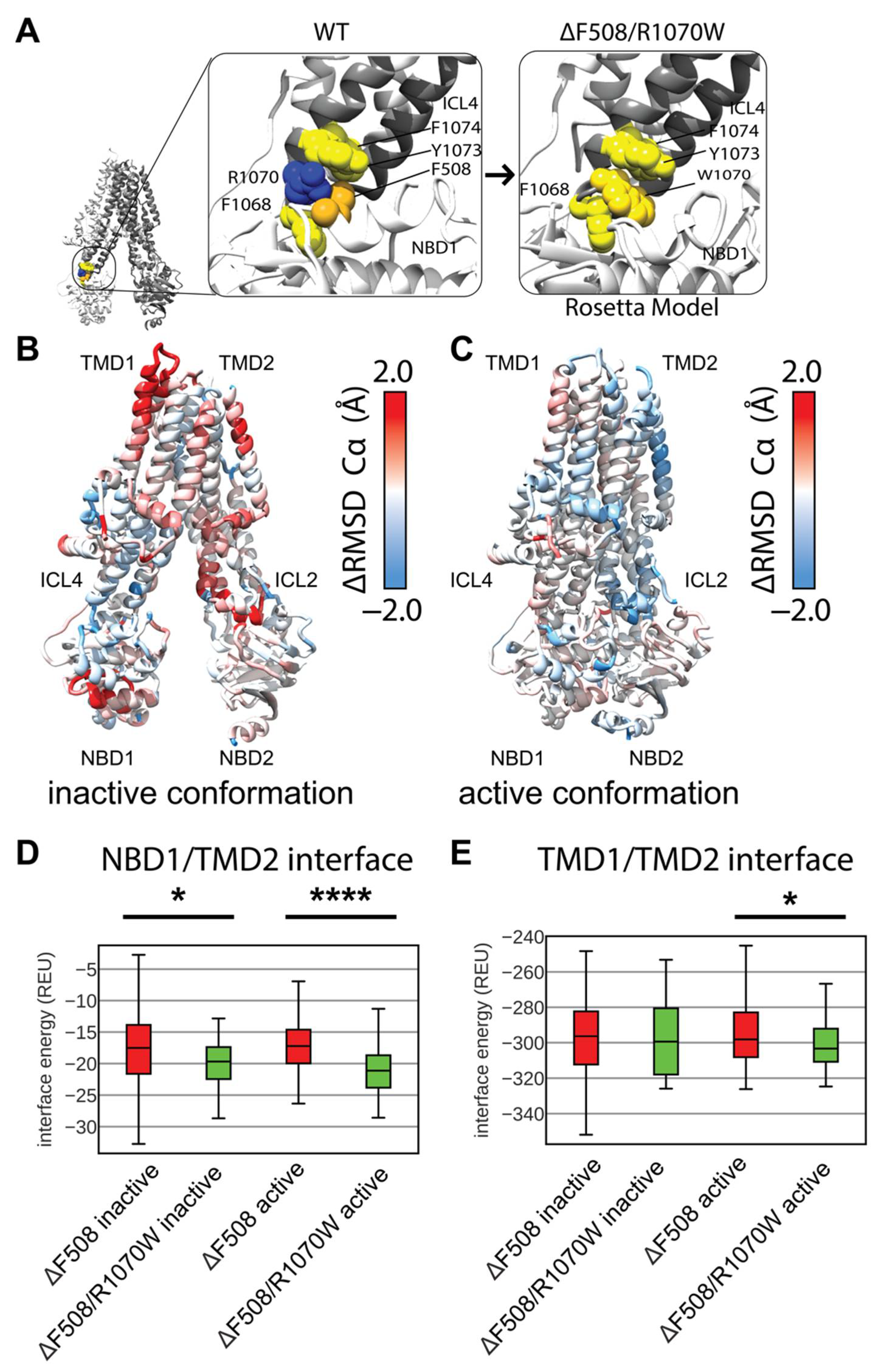
3.4. Modeling F508del/R1070W in Multi-Domain CFTR Lowers Interactions Energy at the NBD1/TMD2 Interface
3.5. Modeling Multi-Domain F508del CFTR Bound to VX-809
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cutting, G.R. Cystic fibrosis genetics: From molecular understanding to clinical application. Nat. Rev. Genet. 2014, 16, 45–56. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, F.; Zhang, Z.; Csanády, L.; Gadsby, D.C.; Chen, J. Molecular Structure of the Human CFTR Ion Channel. Cell 2017, 169, 85–95.e8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Riordan, J.R.; Rommens, J.M.; Kerem, B.; Alon, N.; Grzelczak, Z.; Zielenski, J.; Zielenski, J.; Lok, S.; Plavsic, N.; Chou, J.; et al. Identification of the Cystic Fibrosis Gene: Cloning and Characterization of Complementary DNA. Science 1989, 245, 1066–1073. [Google Scholar] [CrossRef] [PubMed]
- Welsh, M.; Smith, A.E. Molecular mechanisms of CFTR chloride channel dysfunction in cystic fibrosis. Cell 1993, 73, 1251–1254. [Google Scholar] [CrossRef]
- Koch, C.; Høiby, N. Pathogenesis of cystic fibrosis. Lancet 1993, 341, 1065–1069. Available online: https://www.sciencedirect.com/science/article/pii/014067369392422P (accessed on 7 December 2020). [CrossRef]
- Zhang, Z.; Liu, F.; Chen, J. Molecular structure of the ATP-bound, phosphorylated human CFTR. Proc. Natl. Acad. Sci. USA 2018, 115, 12757–12762. [Google Scholar] [CrossRef] [Green Version]
- Van Goor, F.; Hadida, S.; Grootenhuis, P.D.J.; Burton, B.; Stack, J.H.; Straley, K.S.; Decker, C.J.; Miller, M.; McCartney, J.; Olson, E.R.; et al. Correction of the F508del-CFTR protein processing defect in vitro by the investigational drug VX-809. Proc. Natl. Acad. Sci. USA 2011, 108, 18843–18848. [Google Scholar] [CrossRef] [Green Version]
- Van Goor, F. Rescue of CF airway epithelial cell functino in vitro by a CFTR potentiator, VX-770. Proc. Natl. Acad. Sci. USA 2009, 20, 16–17. [Google Scholar]
- Pinto, M.C.; AL Silva, I.; Figueira, M.F.; Amaral, M.D.; Lopes-Pacheco, M. Pharmacological Modulation of Ion Channels for the Treatment of Cystic Fibrosis. J. Exp. Pharmacol. 2021, 13, 693–723. [Google Scholar] [CrossRef]
- Guerra, L.; Favia, M.; Di Gioia, S.; Laselva, O.; Bisogno, A.; Casavola, V.; Colombo, C.; Conese, M. The preclinical discovery and development of the combination of ivacaftor + tezacaftor used to treat cystic fibrosis. Expert Opin. Drug Discov. 2020, 15, 873–891. [Google Scholar] [CrossRef]
- Jain, R.; Kazmerski, T.M.; Aitken, M.L.; West, N.; Wilson, A.; Bozkanat, K.M.; Montemayor, K.; von Berg, K.; Sjoberg, J.; Poranski, M.; et al. Challenges Faced by Women with Cystic Fibrosis. Clin. Chest Med. 2021, 42, 517–530. [Google Scholar] [CrossRef] [PubMed]
- Rotolo, S.M.; Duehlmeyer, S.; Slack, S.M.; Jacobs, H.R.; Heckman, B. Testicular pain following initiation of elexacaftor/tezacaftor/ivacaftor in males with cystic fibrosis. J. Cyst. Fibros. 2020, 19, e39–e41. [Google Scholar] [CrossRef] [PubMed]
- Dagenais, R.V.E.; Su, V.C.H.; Quon, B.S. Real-World Safety of CFTR Modulators in the Treatment of Cystic Fibrosis: A Systematic Review. J. Clin. Med. 2020, 10, 23. [Google Scholar] [CrossRef] [PubMed]
- Laselva, O.; Qureshi, Z.; Zeng, Z.-W.; Petrotchenko, E.V.; Ramjeesingh, M.; Hamilton, C.M.; Huan, L.-J.; Borchers, C.H.; Pomès, R.; Young, R.; et al. Identification of binding sites for ivacaftor on the cystic fibrosis transmembrane conductance regulator. iScience 2021, 24, 102542. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Zhang, Z.; Levit, A.; Levring, J.; Touhara, K.K.; Shoichet, B.K.; Chen, J. Structural identification of a hotspot on CFTR for potentiation. Science 2019, 364, 1184–1188. [Google Scholar] [CrossRef]
- Fiedorczuk, K.; Chen, J. Mechanism of CFTR correction by type I folding correctors. Cell 2022, 185, 158–168.e11. [Google Scholar] [CrossRef]
- Baatallah, N.; Elbahnsi, A.; Mornon, J.-P.; Chevalier, B.; Pranke, I.; Servel, N.; Zelli, R.; Décout, J.-L.; Edelman, A.; Sermet-Gaudelus, I.; et al. Pharmacological chaperones improve intra-domain stability and inter-domain assembly via distinct binding sites to rescue misfolded CFTR. Cell. Mol. Life Sci. 2021, 78, 7813–7829. [Google Scholar] [CrossRef]
- Okiyoneda, T.; Veit, G.; Dekkers, J.F.; Bagdany, M.; Soya, N.; Xu, H.; Roldan, A.; Verkman, A.S.; Kurth, M.J.; Simon, A.; et al. Mechanism-based corrector combination restores ΔF508-CFTR folding and function. Nat. Chem. Biol. 2013, 9, 444–454. [Google Scholar] [CrossRef] [Green Version]
- Veit, G.; Roldan, A.; Hancock, M.A.; Da Fonte, D.F.; Xu, H.; Hussein, M.; Frenkiel, S.; Matouk, E.; Velkov, T.; Lukacs, G.L. Allosteric folding correction of F508del and rare CFTR mutants by elexacaftor-tezacaftor-ivacaftor (Trikafta) combination. JCI Insight 2020, 5, e139983. [Google Scholar] [CrossRef]
- Rabeh, W.; Bossard, F.; Xu, H.; Okiyoneda, T.; Bagdany, M.; Mulvihill, C.M.; Du, K.; di Bernardo, S.; Liu, Y.; Konermann, L.; et al. Correction of Both NBD1 Energetics and Domain Interface Is Required to Restore ΔF508 CFTR Folding and Function. Cell 2012, 148, 150–163. [Google Scholar] [CrossRef] [Green Version]
- Mendoza, J.L.; Schmidt, A.; Li, Q.; Nuvaga, E.; Barrett, T.; Bridges, R.J.; Feranchak, A.P.; Brautigam, C.A.; Thomas, P.J. Requirements for Efficient Correction of ΔF508 CFTR Revealed by Analyses of Evolved Sequences. Cell 2012, 148, 164–174. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Laselva, O.; Molinski, S.; Casavola, V.; Bear, C.E. Correctors of the Major Cystic Fibrosis Mutant Interact through Membrane-Spanning Domains. Mol. Pharmacol. 2018, 93, 612–618. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hudson, R.P.; Dawson, J.E.; Chong, P.A.; Yang, Z.; Millen, L.; Thomas, P.J.; Brouillette, C.G.; Forman-Kay, J.D. Direct binding of the Corrector VX-809 to Human CFTR NBD1: Evidence of an Allosteric coupling between the Binding site and the NBD1:CL4 Interface. Mol. Pharmacol. 2017, 92, 124–135. [Google Scholar] [CrossRef] [Green Version]
- Bahia, M.S.; Khazanov, N.; Zhou, Q.; Yang, Z.; Wang, C.; Hong, J.S.; Rab, A.; Sorscher, E.J.; Brouillette, C.G.; Hunt, J.F.; et al. Stability Prediction for Mutations in the Cytosolic Domains of Cystic Fibrosis Transmembrane Conductance Regulator. J. Chem. Inf. Model. 2021, 61, 1762–1777. [Google Scholar] [CrossRef]
- Estácio, S.G.; Martiniano, H.F.M.C.; Faísca, P.F.N. Thermal unfolding simulations of NBD1 domain variants reveal structural motifs associated with the impaired folding of F508del-CFTR. Mol. BioSyst. 2016, 12, 2834–2848. [Google Scholar] [CrossRef]
- Zhenin, M.; Noy, E.; Senderowitz, H. REMD Simulations Reveal the Dynamic Profile and Mechanism of Action of Deleterious, Rescuing, and Stabilizing Perturbations to NBD1 from CFTR. J. Chem. Inf. Model. 2015, 55, 2349–2364. [Google Scholar] [CrossRef]
- Abreu, B.; Lopes, E.F.; Oliveira, A.S.F.; Soares, C.M. F508del disturbs the dynamics of the nucleotide binding domains of CFTR before and after ATP hydrolysis. Proteins Struct. Funct. Bioinform. 2019, 88, 113–126. [Google Scholar] [CrossRef]
- Odera, M.; Furuta, T.; Sohma, Y.; Sakurai, M. Molecular dynamics simulation study on the structural instability of the most common cystic fibrosis-associated mutant ΔF508-CFTR. Biophys. Phys. 2018, 15, 33–44. [Google Scholar] [CrossRef] [Green Version]
- Wang, R.Y.R.; Song, Y.; Barad, B.A.; Cheng, Y.; Fraser, J.S.; DiMaio, F. Automated structure refinement of macromolecular assemblies from cryo-EM maps using Rosetta. eLife 2016, 5, 1–22. [Google Scholar] [CrossRef]
- Song, Y.; DiMaio, F.; Wang, R.Y.-R.; Kim, D.; Miles, C.; Brunette, T.; Thompson, J.; Baker, D. High-Resolution Comparative Modeling with RosettaCM. Structure 2013, 21, 1735–1742. [Google Scholar] [CrossRef] [Green Version]
- Protasevich, I.; Yang, Z.; Wang, C.; Atwell, S.; Zhao, X.; Emtage, S.; Wetmore, D.; Hunt, J.F.; Brouillette, C.G. Thermal unfolding studies show the disease causing F508del mutation in CFTR thermodynamically destabilizes nucleotide-binding domain 1. Protein Sci. 2010, 19, 1917–1931. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Farinha, C.M.; Canato, S. From the endoplasmic reticulum to the plasma membrane: Mechanisms of CFTR folding and trafficking. Cell. Mol. Life Sci. 2017, 74, 39–55. [Google Scholar] [CrossRef] [PubMed]
- Du, K.; Sharma, M.; Lukacs, G. The ΔF508 cystic fibrosis mutation impairs domain-domain interactions and arrests post-translational folding of CFTR. Nat. Struct. Mol. Biol. 2004, 12, 17–25. [Google Scholar] [CrossRef] [PubMed]
- Hwang, T.-C.; Yeh, J.-T.; Zhang, J.; Yu, Y.-C.; Yeh, H.-I.; Destefano, S. Structural mechanisms of CFTR function and dysfunction. J. Gen. Physiol. 2018, 150, 539–570. [Google Scholar] [CrossRef] [Green Version]
- Yarov-Yarovoy, V.; Schonbrun, J.; Baker, D. Multipass membrane protein structure prediction using Rosetta. Proteins Struct. Funct. Bioinform. 2005, 62, 1010–1025. [Google Scholar] [CrossRef] [Green Version]
- Barth, P.; Schonbrun, J.; Baker, D. Toward high-resolution prediction and design of transmembrane helical protein structures. Proc. Natl. Acad. Sci. USA 2007, 104, 15682–15687. [Google Scholar] [CrossRef] [Green Version]
- Viklund, H.; Elofsson, A. OCTOPUS: Improving topology prediction by two-track ANN-based preference scores and an extended topological grammar. Bioinformatics 2008, 24, 1662–1668. [Google Scholar] [CrossRef] [Green Version]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [Green Version]
- Mendenhall, J.; Brown, B.P.; Kothiwale, S.; Meiler, J. BCL::Conf: Improved Open-Source Knowledge-Based Conformation Sampling Using the Crystallography Open Database. J. Chem. Inf. Model. 2020, 61, 189–201. [Google Scholar] [CrossRef]
- Lemmon, G.; Meiler, J. Rosetta Ligand Docking with Flexible XML Protocols BT–Computational Drug Discovery and Design; Baron, R., Ed.; Springer: New York, NY, USA, 2012; pp. 143–155. [Google Scholar] [CrossRef] [Green Version]
- Bender, B.J.; Marlow, B.; Meiler, J. Improving homology modeling from low-sequence identity templates in Rosetta: A case study in GPCRs. PLoS Comput. Biol. 2020, 16, e1007597. [Google Scholar] [CrossRef]
- Farkas, B.; Tordai, H.; Padányi, R.; Tordai, A.; Gera, J.; Paragi, G.; Hegedűs, T. Discovering the chloride pathway in the CFTR channel. Cell. Mol. Life Sci. 2019, 77, 765–778. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aleksandrov, A.A.; Kota, P.; Cui, L.; Jensen, T.; Alekseev, A.E.; Reyes, S.; He, L.; Gentzsch, M.; Aleksandrov, L.A.; Dokholyan, N.V.; et al. Allosteric Modulation Balances Thermodynamic Stability and Restores Function of ΔF508 CFTR. J. Mol. Biol. 2012, 419, 41–60. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Denning, G.; Anderson, M.; Amara, J.F.; Marshall, J.; Smith, A.E.; Welsh, M. Processing of mutant cystic fibrosis transmembrane conductance regulator is temperature-sensitive. Nature 1992, 358, 761–764. [Google Scholar] [CrossRef] [PubMed]
- Lewis, H.A.; Zhao, X.; Wang, C.; Sauder, J.M.; Rooney, I.; Noland, B.W.; Lorimer, D.; Kearins, M.C.; Conners, K.; Condon, B.; et al. Impact of the ΔF508 Mutation in First Nucleotide-binding Domain of Human Cystic Fibrosis Transmembrane Conductance Regulator on Domain Folding and Structure. J. Biol. Chem. 2005, 280, 1346–1353. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lewis, H.; Wang, C.; Zhao, X.; Hamuro, Y.; Conners, K.; Kearins, M.; Lu, F.; Sauder, J.; Molnar, K.; Coales, S.; et al. Structure and Dynamics of NBD1 from CFTR Characterized Using Crystallography and Hydrogen/Deuterium Exchange Mass Spectrometry. J. Mol. Biol. 2010, 396, 406–430. [Google Scholar] [CrossRef] [PubMed]
- Khushoo, A.; Yang, Z.; Johnson, A.E.; Skach, W.R. Ligand-Driven Vectorial Folding of Ribosome-Bound Human CFTR NBD1. Mol. Cell 2011, 41, 682–692. [Google Scholar] [CrossRef] [Green Version]
- Thibodeau, P.; Richardson, J.M., III; Wang, W.; Millen, L.; Watson, J.; Mendoza, J.; Du, K.; Fischman, S.; Senderowitz, H.; Lukacs, G.; et al. The Cystic Fibrosis-causing Mutation ΔF508 Affects Multiple Steps in Cystic Fibrosis Transmembrane Conductance Regulator Biogenesis. J. Biol. Chem. 2010, 285, 35825–35835. [Google Scholar] [CrossRef] [Green Version]
- He, L.; Kota, P.; Aleksandrov, A.A.; Cui, L.; Jensen, T.; Dokholyan, N.V.; Riordan, J.R. Correctors of ΔF508 CFTR restore global conformational maturation without thermally stabilizing the mutant protein. FASEB J. 2013, 27, 536–545. [Google Scholar] [CrossRef] [Green Version]
- Lopes-Pacheco, M.; Silva, I.A.; Turner, M.J.; Carlile, G.W.; Sondo, E.; Thomas, D.Y.; Pedemonte, N.; Hanrahan, J.W.; Amaral, M.D. Characterization of the mechanism of action of RDR01752, a novel corrector of F508del-CFTR. Biochem. Pharmacol. 2020, 180, 114133. [Google Scholar] [CrossRef]
- Loo, T.W.; Bartlett, M.C.; Clarke, D.M. Corrector VX-809 stabilizes the first transmembrane domain of CFTR. Biochem. Pharmacol. 2013, 86, 612–619. [Google Scholar] [CrossRef]
- Eckford, P.D.; Ramjeesingh, M.; Molinski, S.; Pasyk, S.; Dekkers, J.F.; Li, C.; Ahmadi, S.; Ip, W.; Chung, T.E.; Du, K.; et al. VX-809 and Related Corrector Compounds Exhibit Secondary Activity Stabilizing Active F508del-CFTR after Its Partial Rescue to the Cell Surface. Chem. Biol. 2014, 21, 666–678. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Combs, S.A.; DeLuca, S.L.; DeLuca, S.H.; Lemmon, G.; Nannemann, D.; Nguyen, E.; Willis, J.R.; Sheehan, J.H.; Meiler, J. Small-molecule ligand docking into comparative models with Rosetta. Nat. Protoc. 2013, 8, 1277–1298. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- DeLuca, S.; Khar, K.; Meiler, J. Fully Flexible Docking of Medium Sized Ligand Libraries with RosettaLigand. PLoS ONE 2015, 10, e0132508. [Google Scholar] [CrossRef]
- Brown, B.P.; Mendenhall, J.; Geanes, A.R.; Meiler, J. General Purpose Structure-Based Drug Discovery Neural Network Score Functions with Human-Interpretable Pharmacophore Maps. J. Chem. Inf. Model. 2021, 61, 603–620. [Google Scholar] [CrossRef]
- Aleksandrov, A.A.; Kota, P.; Aleksandrov, L.A.; He, L.; Jensen, T.; Cui, L.; Gentzsch, M.; Dokholyan, N.V.; Riordan, J.R. Regulatory Insertion Removal Restores Maturation, Stability and Function of ΔF508 CFTR. J. Mol. Biol. 2010, 401, 194–210. [Google Scholar] [CrossRef] [Green Version]
- Scholl, D.; Sigoillot, M.; Overtus, M.; Martinez, R.C.; Martens, C.; Wang, Y.; Pardon, E.; Laeremans, T.; Garcia-Pino, A.; Steyaert, J.; et al. A topological switch in CFTR modulates channel activity and sensitivity to unfolding. Nat. Chem. Biol. 2021, 17, 989–997. [Google Scholar] [CrossRef]
- Veit, G.; Xu, H.; Dreano, E.; Avramescu, R.G.; Bagdany, M.; Beitel, L.K.; Roldan, A.; Hancock, M.; Lay, C.; Li, W.; et al. Structure-guided combination therapy to potently improve the function of mutant CFTRs. Nat. Med. 2018, 24, 1732–1742. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
McDonald, E.F.; Woods, H.; Smith, S.T.; Kim, M.; Schoeder, C.T.; Plate, L.; Meiler, J. Structural Comparative Modeling of Multi-Domain F508del CFTR. Biomolecules 2022, 12, 471. https://doi.org/10.3390/biom12030471
McDonald EF, Woods H, Smith ST, Kim M, Schoeder CT, Plate L, Meiler J. Structural Comparative Modeling of Multi-Domain F508del CFTR. Biomolecules. 2022; 12(3):471. https://doi.org/10.3390/biom12030471
Chicago/Turabian StyleMcDonald, Eli Fritz, Hope Woods, Shannon T. Smith, Minsoo Kim, Clara T. Schoeder, Lars Plate, and Jens Meiler. 2022. "Structural Comparative Modeling of Multi-Domain F508del CFTR" Biomolecules 12, no. 3: 471. https://doi.org/10.3390/biom12030471
APA StyleMcDonald, E. F., Woods, H., Smith, S. T., Kim, M., Schoeder, C. T., Plate, L., & Meiler, J. (2022). Structural Comparative Modeling of Multi-Domain F508del CFTR. Biomolecules, 12(3), 471. https://doi.org/10.3390/biom12030471






