Endogenous Human Proteins Interfering with Amyloid Formation
Abstract
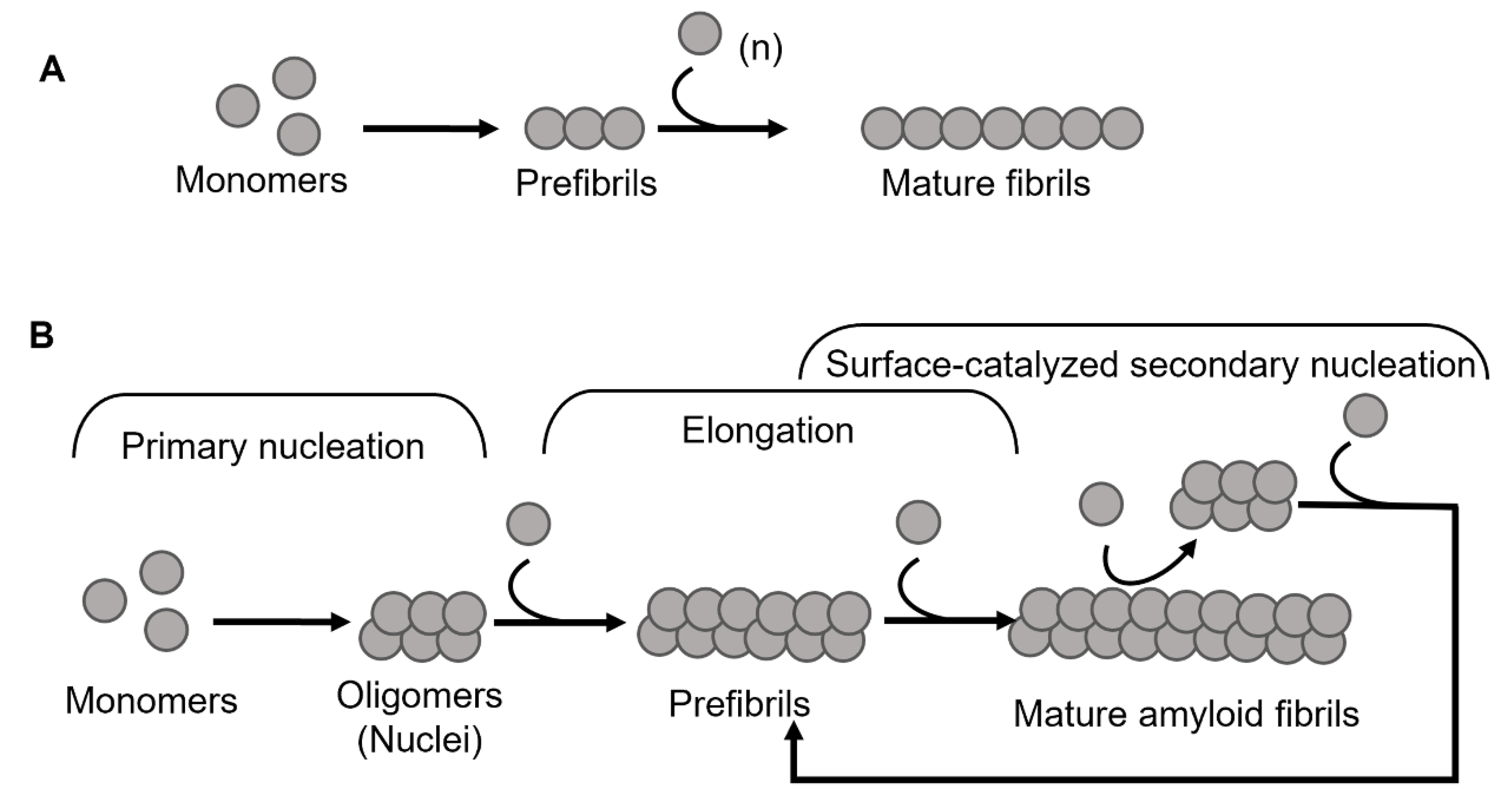
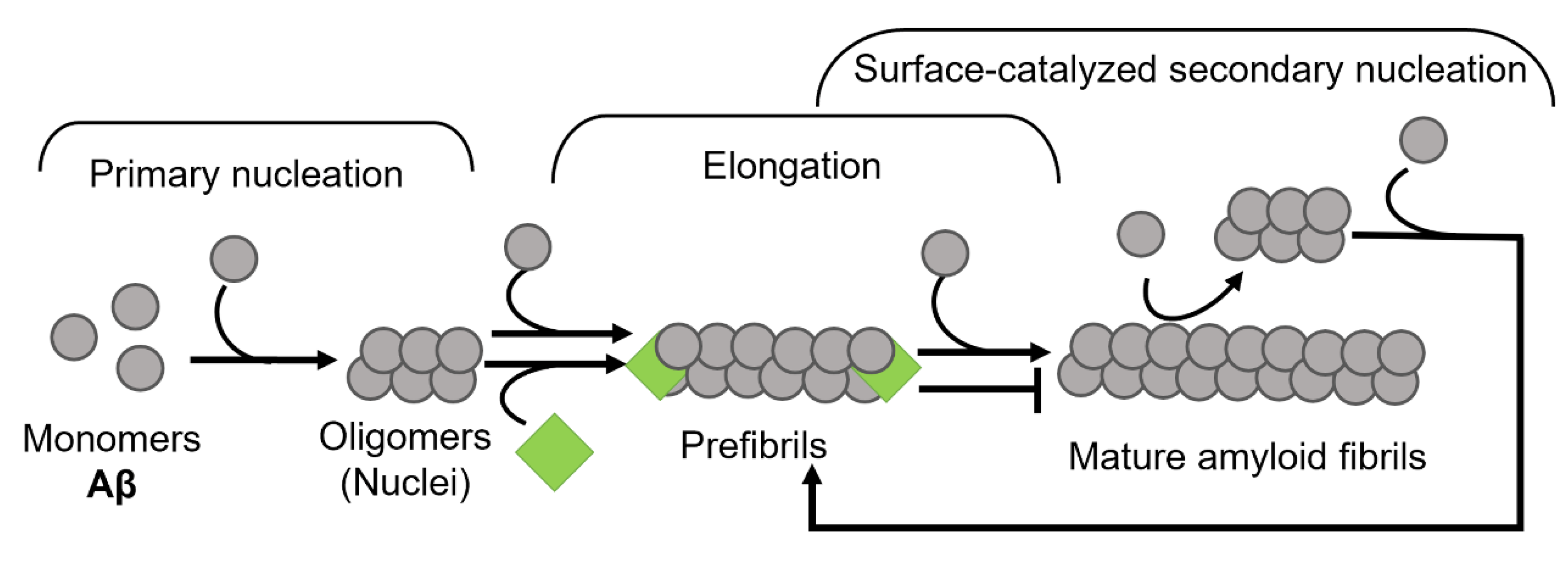
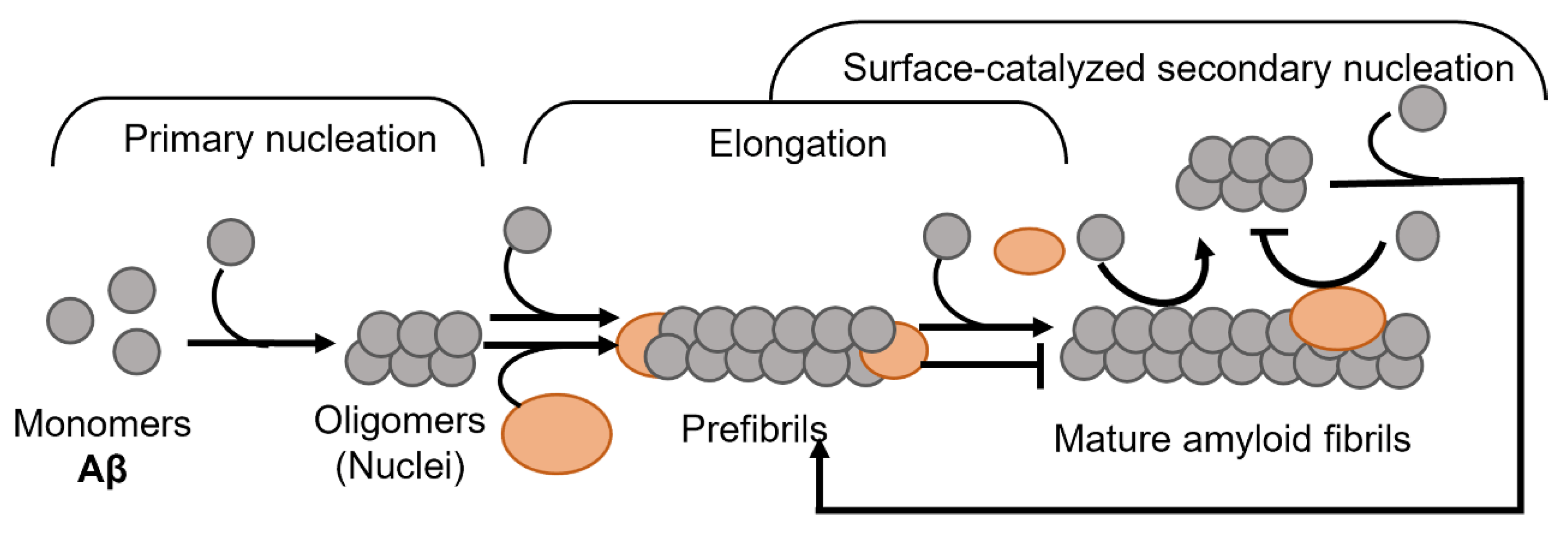
:1. Introduction
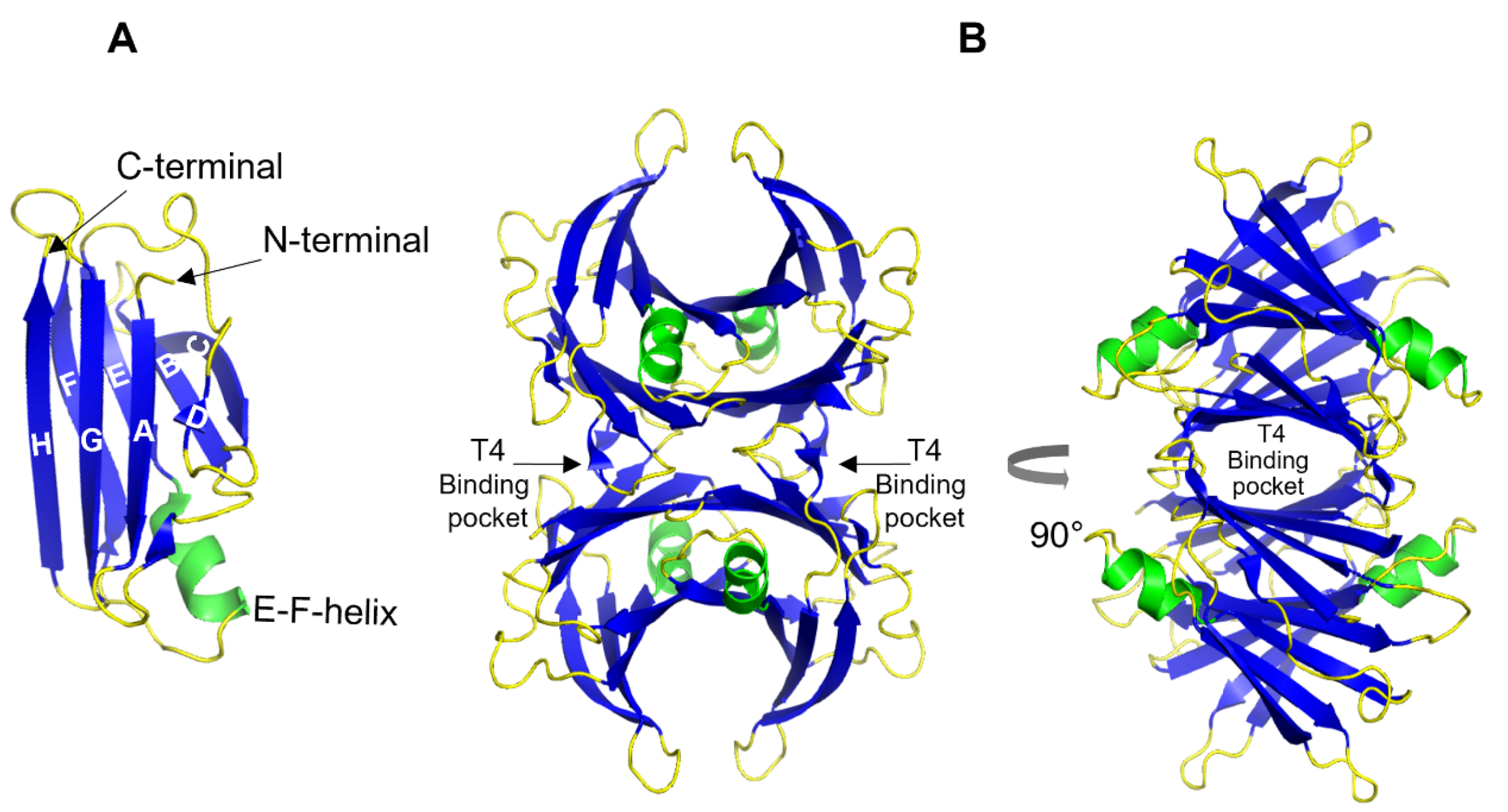
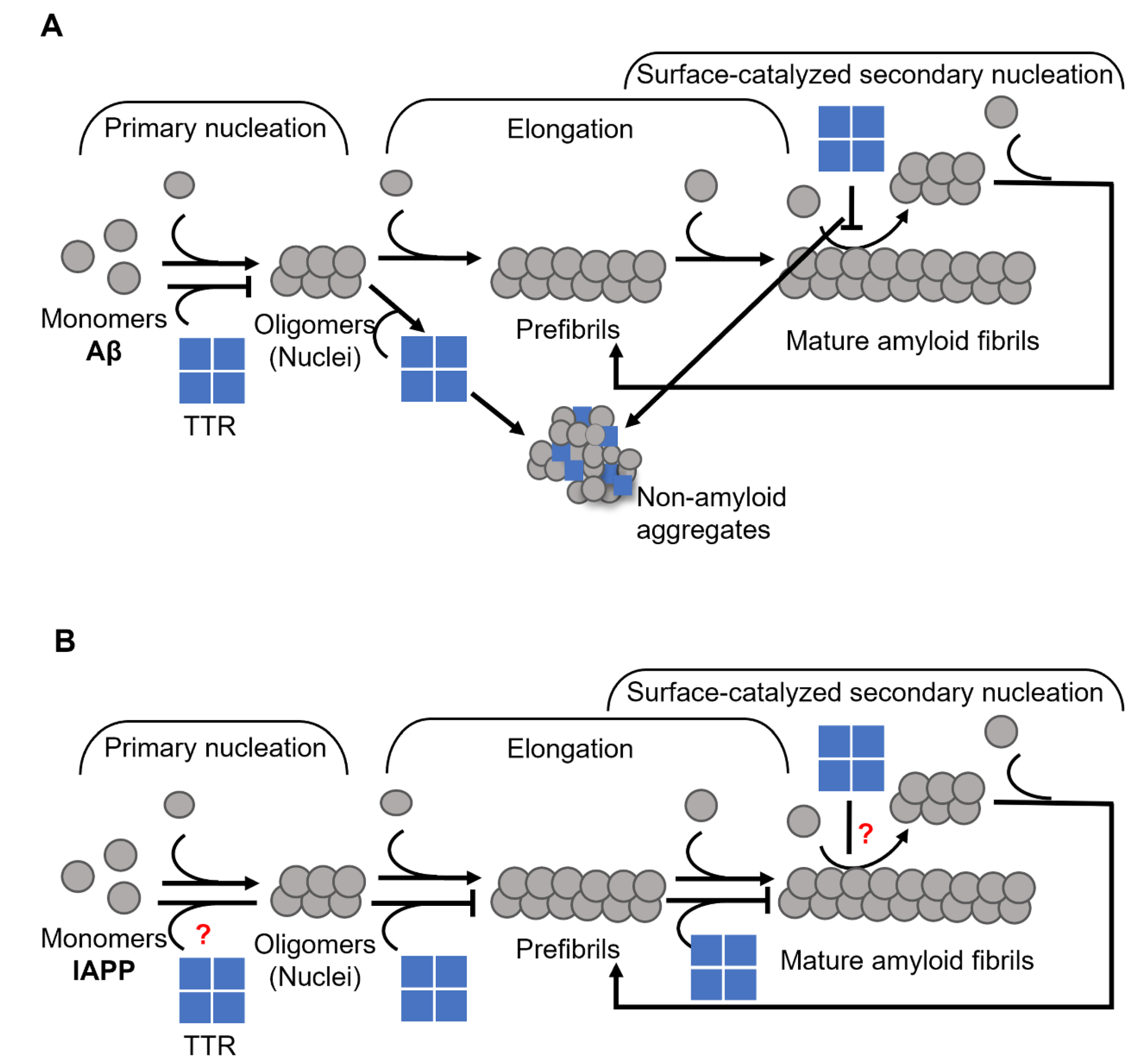
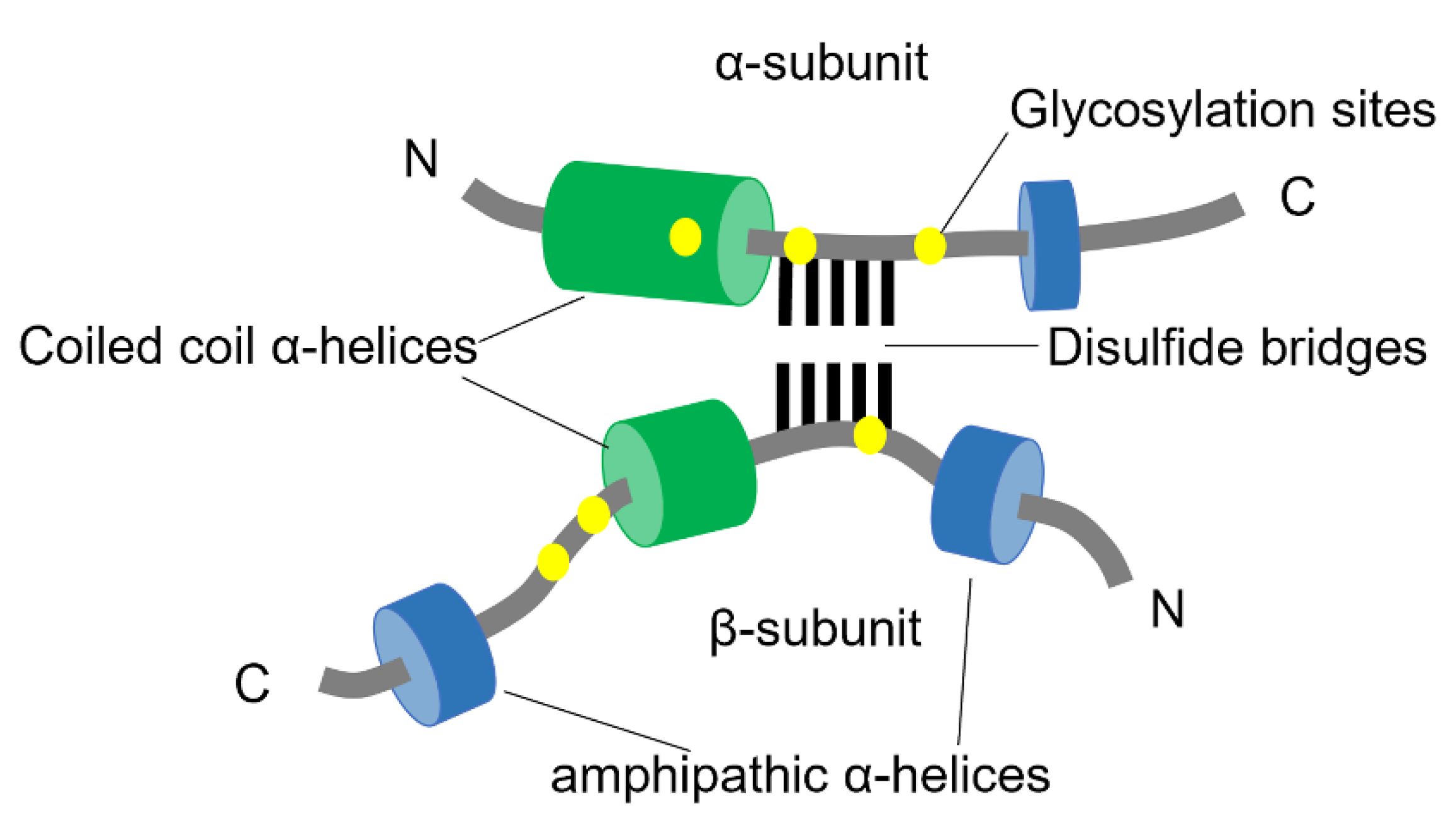
2. Transthyretin
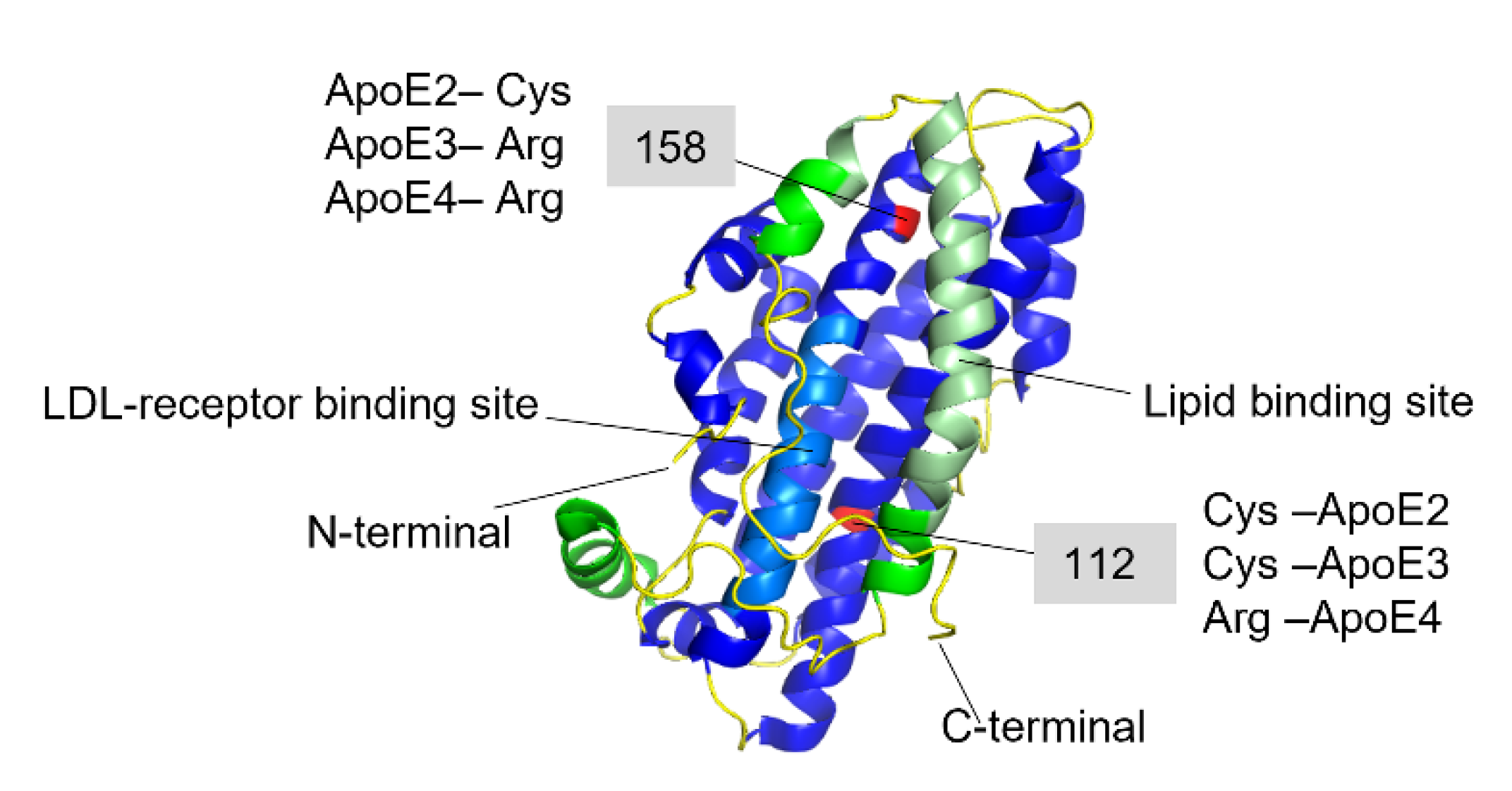
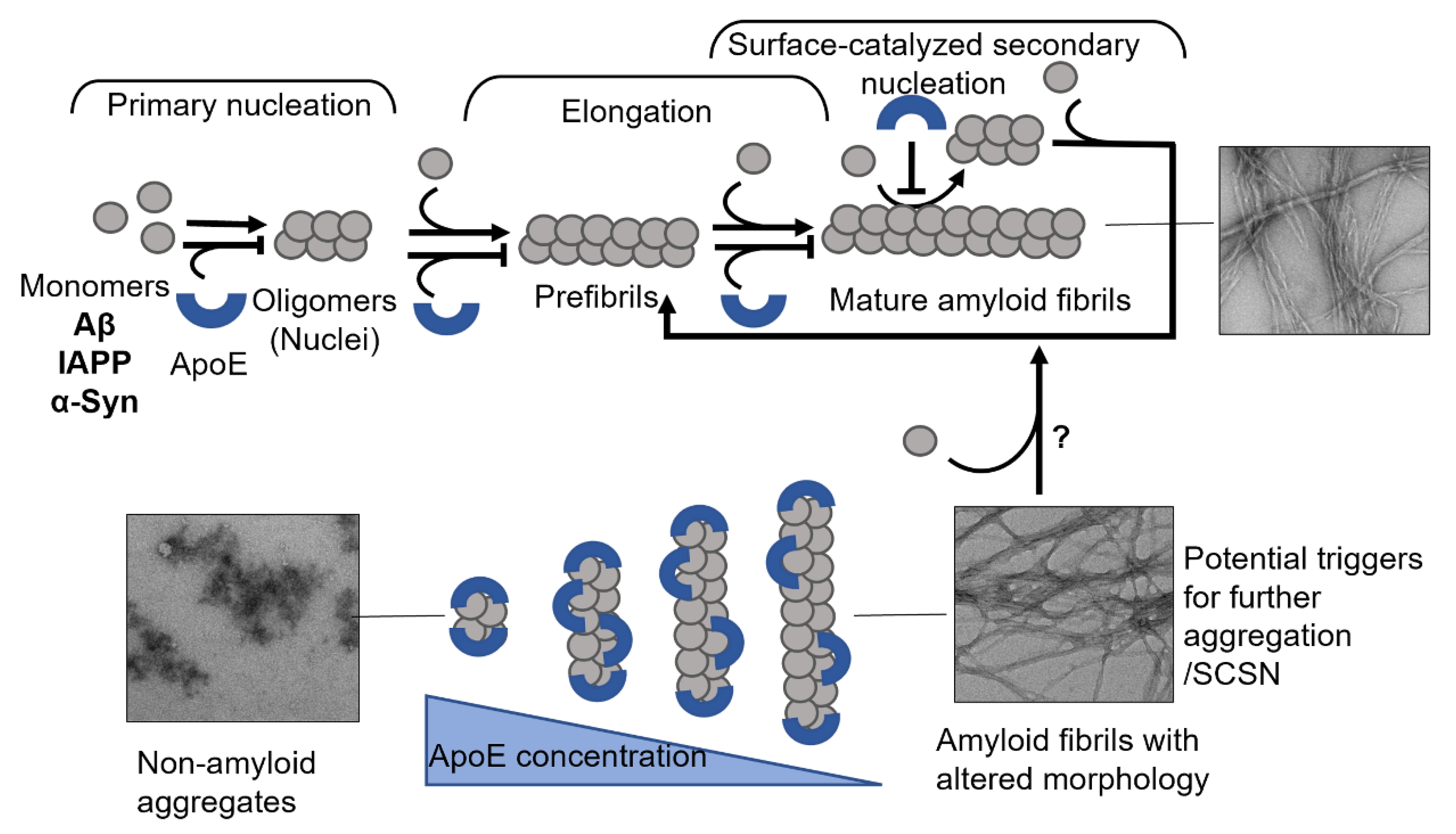
3. Apolipoprotein E
4. Clusterin
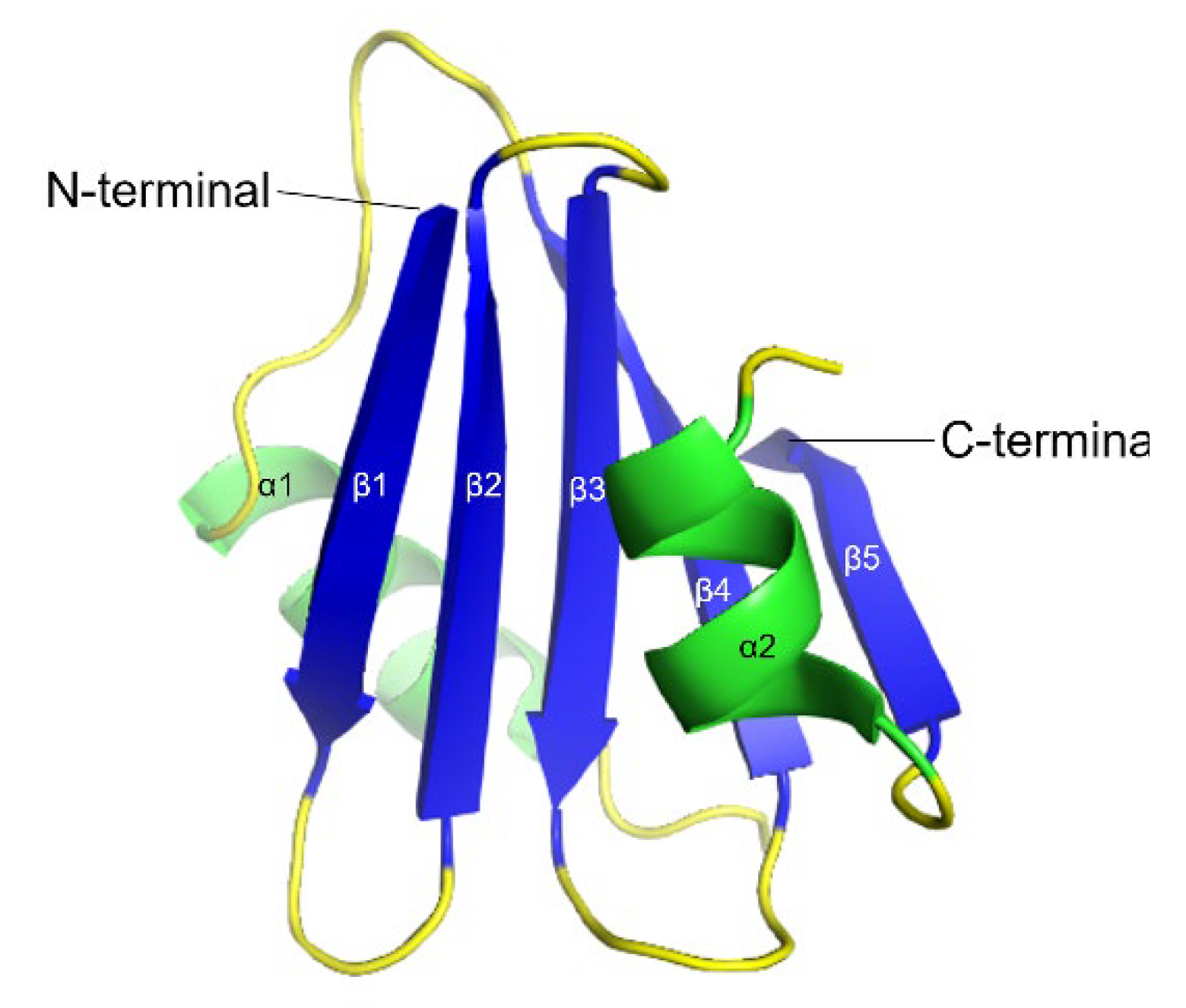
5. Brichos
6. Concluding Remarks
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Chiti, F.; Dobson, C.M. Protein Misfolding, Amyloid Formation, and Human Disease: A Summary of Progress Over the Last Decade. Annu. Rev. Biochem. 2017, 86, 27–68. [Google Scholar] [CrossRef] [PubMed]
- Bély, M.; Makovitzky, J. Sensitivity and specificity of Congo red staining according to Romhányi. Comparison with Puchtler’s or Bennhold’s methods. Acta Histochem. 2006, 108, 175–180. [Google Scholar] [CrossRef]
- Frieden, C. Protein aggregation processes: In search of the mechanism. Protein Sci. 2007, 16, 2334–2344. [Google Scholar] [CrossRef]
- Powers, E.; Powers, D.L. The Kinetics of Nucleated Polymerizations at High Concentrations: Amyloid Fibril Formation Near and Above the “Supercritical Concentration”. Biophys. J. 2006, 91, 122–132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hurshman, A.R.; White, J.T.; Powers, E.T.; Kelly, J.W. Transthyretin Aggregation under Partially Denaturing Conditions Is a Downhill Polymerization. Biochemistry 2004, 43, 7365–7381. [Google Scholar] [CrossRef] [PubMed]
- Cohen, S.I.A.; Linse, S.; Luheshi, L.M.; Hellstrand, E.; White, D.A.; Rajah, L.; Otzen, D.E.; Vendruscolo, M.; Dobson, C.M.; Knowles, T.P.J. Proliferation of amyloid β42 aggregates occurs through a secondary nucleation mechanism. Proc. Natl. Acad. Sci. USA 2013, 110, 9758–9763. [Google Scholar] [CrossRef] [Green Version]
- Gaspar, R.; Meisl, G.; Buell, A.K.; Young, L.; Kaminski, C.F.; Knowles, T.P.J.; Sparr, E.; Linse, S. Secondary nucleation of monomers on fibril surface dominates α-synuclein aggregation and provides autocatalytic amyloid amplification. Q. Rev. Biophys. 2017, 50, e6. [Google Scholar] [CrossRef] [Green Version]
- Camargo, D.C.R.; Chia, S.; Menzies, J.; Mannini, B.; Meisl, G.; Lundqvist, M.; Pohl, C.; Bernfur, K.; Lattanzi, V.; Habchi, J.; et al. Surface-Catalyzed Secondary Nucleation Dominates the Generation of Toxic IAPP Aggregates. Front. Mol. Biosci. 2021, 8, 757425. [Google Scholar] [CrossRef]
- Xue, W.-F.; Homans, S.W.; Radford, S.E. Systematic analysis of nucleation-dependent polymerization reveals new insights into the mechanism of amyloid self-assembly. Proc. Natl. Acad. Sci. USA 2008, 105, 8926–8931. [Google Scholar] [CrossRef] [Green Version]
- Sousa, M.M.; Fernandes, R.; Palha, J.A.; Taboada, A.; Vieira, P.; Saraiva, M.J. Evidence for Early Cytotoxic Aggregates in Transgenic Mice for Human Transthyretin Leu55Pro. Am. J. Pathol. 2002, 161, 1935–1948. [Google Scholar] [CrossRef] [Green Version]
- Jin, S.; Kedia, N.; Illes-Toth, E.; Haralampiev, I.; Prisner, S.; Herrmann, A.; Wanker, E.; Bieschke, J. Amyloid-β(1–42) Aggregation Initiates Its Cellular Uptake and Cytotoxicity. J. Biol. Chem. 2016, 291, 19590–19606. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kayed, R.; Lasagna-Reeves, C.A. Molecular Mechanisms of Amyloid Oligomers Toxicity. J. Alzheimer’s Dis. 2013, 33 (Suppl. 1), S67–S78. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baglioni, S.; Casamenti, F.; Bucciantini, M.; Luheshi, L.M.; Taddei, N.; Chiti, F.; Dobson, C.M.; Stefani, M. Prefibrillar Amyloid Aggregates Could Be Generic Toxins in Higher Organisms. J. Neurosci. 2006, 26, 8160–8167. [Google Scholar] [CrossRef] [PubMed]
- Haass, C.; Selkoe, D.J. Soluble protein oligomers in neurodegeneration: Lessons from the Alzheimer’s amyloid β-peptide. Nat. Rev. Mol. Cell Biol. 2007, 8, 101–112. [Google Scholar] [CrossRef] [PubMed]
- Roberts, B.; Lind, M.; Wagen, A.Z.; Rembach, A.; Frugier, T.; Li, Q.-X.; Ryan, T.M.; McLean, C.A.; Doecke, J.D.; Rowe, C.C.; et al. Biochemically-defined pools of amyloid-β in sporadic Alzheimer’s disease: Correlation with amyloid PET. Brain 2017, 140, 1486–1498. [Google Scholar] [CrossRef] [Green Version]
- Bergkvist, L.; Richards, D.R.; Bernardo-Gancedo, A.; Kumita, J.R.; Nilsson, P.R.; Brorsson, A.-C. Serum amyloid P component promotes formation of distinct aggregated lysozyme morphologies and reduces toxicity in Drosophila flies expressing F57I lysozyme. PLoS ONE 2020, 15, e0227227. [Google Scholar] [CrossRef]
- Härd, T.; Lendel, C. Inhibition of Amyloid Formation. J. Mol. Biol. 2012, 421, 441–465. [Google Scholar] [CrossRef]
- Härd, T. Amyloid Fibrils: Formation, Polymorphism, and Inhibition. J. Phys. Chem. Lett. 2014, 5, 607–614. [Google Scholar] [CrossRef]
- Habchi, J.; Chia, S.; Limbocker, R.; Mannini, B.; Ahn, M.; Perni, M.; Hansson, O.; Arosio, P.; Kumita, J.R.; Challa, P.K.; et al. Systematic development of small molecules to inhibit specific microscopic steps of Aβ42 aggregation in Alzheimer’s disease. Proc. Natl. Acad. Sci. USA 2017, 114, E200–E208. [Google Scholar] [CrossRef] [Green Version]
- Heller, G.T.; Aprile, F.A.; Michaels, T.C.T.; Limbocker, R.; Perni, M.; Ruggeri, F.S.; Mannini, B.; Löhr, T.; Bonomi, M.; Camilloni, C.; et al. Small-molecule sequestration of amyloid-β as a drug discovery strategy for Alzheimer’s disease. Sci. Adv. 2020, 6, eabb5924. [Google Scholar] [CrossRef]
- Adolfsson, D.E.; Tyagi, M.; Singh, P.; Deuschmann, A.; Ådén, J.; Gharibyan, A.L.; Jayaweera, S.W.; Lindgren, A.E.G.; Olofsson, A.; Almqvist, F. Intramolecular Povarov Reactions for the Synthesis of Chromenopyridine Fused 2-Pyridone Polyheterocycles Binding to α-Synuclein and Amyloid-β Fibrils. J. Org. Chem. 2020, 85, 14174–14189. [Google Scholar] [CrossRef] [PubMed]
- Velander, P.; Wu, L.; Henderson, F.; Zhang, S.; Bevan, D.; Xu, B. Natural product-based amyloid inhibitors. Biochem. Pharmacol. 2017, 139, 40–55. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, L.; Yang, C.; Zheng, J.; Chen, Y.; Xiao, Y.; Huang, K. Non-polyphenolic natural inhibitors of amyloid aggregation. Eur. J. Med. Chem. 2020, 192, 112197. [Google Scholar] [CrossRef]
- Gilman, S.; Koller, M.; Black, R.S.; Jenkins, L.; Griffith, S.G.; Fox, N.C.; Eisner, L.; Kirby, L.; Rovira, M.B.; Forette, F.; et al. Clinical effects of Aβ immunization (AN1792) in patients with AD in an interrupted trial. Neurology 2005, 64, 1553–1562. [Google Scholar] [CrossRef] [PubMed]
- Bouter, Y.; Noguerola, J.S.L.; Tucholla, P.; Crespi, G.A.N.; Parker, M.; Wiltfang, J.; Miles, L.A.; Bayer, T.A. Abeta targets of the biosimilar antibodies of Bapineuzumab, Crenezumab, Solanezumab in comparison to an antibody against N-truncated Abeta in sporadic Alzheimer disease cases and mouse models. Acta Neuropathol. 2015, 130, 713–729. [Google Scholar] [CrossRef] [PubMed]
- Bohrmann, B.; Baumann, K.; Benz, J.; Gerber, F.; Huber, W.; Knoflach, F.; Messer, J.; Oroszlan, K.; Rauchenberger, R.; Richter, W.F.; et al. Gantenerumab: A Novel Human Anti-Aβ Antibody Demonstrates Sustained Cerebral Amyloid-β Binding and Elicits Cell-Mediated Removal of Human Amyloid-β. J. Alzheimer’s Dis. 2012, 28, 49–69. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Z.; Gao, G.; Duan, C.; Yang, H. Progress of immunotherapy of anti-α-synuclein in Parkinson’s disease. Biomed. Pharmacother. 2019, 115, 108843. [Google Scholar] [CrossRef]
- Vaikath, N.N.; Hmila, I.; Gupta, V.; Erskine, D.; Ingelsson, M.; El-Agnaf, O.M.A. Antibodies against alpha-synuclein: Tools and therapies. J. Neurochem. 2019, 150, 612–625. [Google Scholar] [CrossRef] [Green Version]
- Panza, F.; Lozupone, M.; Seripa, D.; Imbimbo, B. Amyloid-β immunotherapy for alzheimer disease: Is it now a long shot? Ann. Neurol. 2019, 85, 303–315. [Google Scholar] [CrossRef]
- Goldgaber, D.; Schwarzman, A.I.; Bhasin, R.; Gregori, L.; Schmechel, D.; Saunders, A.M.; Roses, A.D.; Strittmatter, W.J. Sequestration of Amyloid β-Peptidea. Ann. N. Y. Acad. Sci. 1993, 695, 139–143. [Google Scholar] [CrossRef]
- Strittmatter, W.J.; Saunders, A.M.; Schmechel, D.; Pericak-Vance, M.; Enghild, J.J.; Salvesen, G.S.; Roses, A.D. Apolipoprotein E: High-avidity binding to beta-amyloid and increased frequency of type 4 allele in late-onset familial Alzheimer disease. Proc. Natl. Acad. Sci. USA 1993, 90, 1977–1981. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schwarzman, A.L.; Gregori, L.; Vitek, M.P.; Lyubski, S.; Strittmatter, W.J.; Enghilde, J.J.; Bhasin, R.; Silverman, J.; Weisgraber, K.H.; Coyle, P.K.; et al. Transthyretin sequesters amyloid beta protein and prevents amyloid formation. Proc. Natl. Acad. Sci. USA 1994, 91, 8368–8372. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mangrolia, P.; Yang, D.T.; Murphy, R.M. Transthyretin variants with improved inhibition of β-amyloid aggregation. Protein Eng. Des. Sel. 2016, 29, 209–218. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, X.; Zhang, X.; Ladiwala, A.R.A.; Du, D.; Yadav, J.K.; Tessier, P.; Wright, P.; Kelly, J.W.; Buxbaum, J.N. Mechanisms of Transthyretin Inhibition of β-Amyloid Aggregation In Vitro. J. Neurosci. 2013, 33, 19423–19433. [Google Scholar] [CrossRef] [PubMed]
- Cascella, R.; Conti, S.; Mannini, B.; Li, X.; Buxbaum, J.N.; Tiribilli, B.; Chiti, F.; Cecchi, C. Transthyretin suppresses the toxicity of oligomers formed by misfolded proteins in vitro. Biochim. Biophys. Acta 2013, 1832, 2302–2314. [Google Scholar] [CrossRef] [Green Version]
- Wood, S.J.; MacKenzie, L.; Maleeff, B.; Hurle, M.R.; Wetzel, R. Selective inhibition of Abeta fibril formation. J. Biol. Chem. 1996, 271, 4086–4092. [Google Scholar] [CrossRef] [Green Version]
- Nilsson, L.; Pamren, A.; Islam, T.; Brannstrom, K.; Golchin, S.A.; Pettersson, N.; Iakovleva, I.; Sandblad, L.; Gharibyan, A.L.; Olofsson, A. Transthyretin Interferes with Abeta Amyloid Formation by Redirecting Oligomeric Nuclei into Non-Amyloid Aggregates. J. Mol. Biol. 2018, 430, 2722–2733. [Google Scholar] [CrossRef]
- Ma, J.; Yee, A.; Brewer, H.B., Jr.; Das, S.; Potter, H. Amyloid-associated proteins alpha 1-antichymotrypsin and apolipoprotein E promote assembly of Alzheimer beta-protein into filaments. Nature 1994, 372, 92–94. [Google Scholar] [CrossRef]
- Wisniewski, T.; Frangione, B. Apolipoprotein E: A pathological chaperone protein in patients with cerebral and systemic amyloid. Neurosci. Lett. 1992, 135, 235–238. [Google Scholar] [CrossRef]
- Hashimoto, T.; Serrano-Pozo, A.; Hori, Y.; Adams, K.W.; Takeda, S.; Banerji, A.O.; Mitani, A.; Joyner, D.; Thyssen, D.H.; Bacskai, B.J.; et al. Apolipoprotein E, Especially Apolipoprotein E4, Increases the Oligomerization of Amyloid β Peptide. J. Neurosci. 2012, 32, 15181–15192. [Google Scholar] [CrossRef] [Green Version]
- Ghosh, S.; Sil, T.B.; Dolai, S.; Garai, K. High-affinity multivalent interactions between apolipoprotein E and the oligomers of amyloid-beta. FEBS J. 2019, 286, 4737–4753. [Google Scholar] [CrossRef]
- Garai, K.; Verghese, P.B.; Baban, B.; Holtzman, D.M.; Frieden, C. The Binding of Apolipoprotein E to Oligomers and Fibrils of Amyloid-β Alters the Kinetics of Amyloid Aggregation. Biochemistry 2014, 53, 6323–6331. [Google Scholar] [CrossRef] [PubMed]
- Beeg, M.; Stravalaci, M.; Romeo, M.; Carrá, A.D.; Cagnotto, A.; Rossi, A.; Diomede, L.; Salmona, M.; Gobbi, M. Clusterin Binds to Aβ1–42 Oligomers with High Affinity and Interferes with Peptide Aggregation by Inhibiting Primary and Secondary Nucleation. J. Biol. Chem. 2016, 291, 6958–6966. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hammad, S.M.; Ranganathan, S.; Loukinova, E.; Twal, W.O.; Argraves, W.S. Interaction of apolipoprotein J-amyloid beta-peptide complex with low density lipoprotein receptor-related protein-2/megalin. A mechanism to prevent pathological accumulation of amyloid beta-peptide. J. Biol. Chem. 1997, 272, 18644–18649. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Biverstål, H.; Dolfe, L.; Hermansson, E.; Leppert, A.; Reifenrath, M.; Winblad, B.; Presto, J.; Johansson, J. Dissociation of a BRICHOS trimer into monomers leads to increased inhibitory effect on Aβ42 fibril formation. Biochim. Biophys. Acta 2015, 1854, 835–843. [Google Scholar] [CrossRef] [PubMed]
- Willander, H.; Presto, J.; Askarieh, G.; Biverstal, H.; Frohm, B.; Knight, S.; Johansson, J.; Linse, S. BRICHOS Domains Efficiently Delay Fibrillation of Amyloid β-Peptide. J. Biol. Chem. 2012, 287, 31608–31617. [Google Scholar] [CrossRef] [Green Version]
- Peng, S.; Fitzen, M.; Jörnvall, H.; Johansson, J. The extracellular domain of Bri2 (ITM2B) binds the ABri peptide (1–23) and amyloid β-peptide (Aβ1–40): Implications for Bri2 effects on processing of amyloid precursor protein and Aβ aggregation. Biochem. Biophys. Res. Commun. 2010, 393, 356–361. [Google Scholar] [CrossRef]
- Del Campo, M.; Hoozemans, J.; Dekkers, L.-L.; Rozemuller, A.J.; Korth, C.; Müller-Schiffmann, A.; Scheltens, P.; Blankenstein, M.; Jimenez, C.R.; Veerhuis, R.; et al. BRI2-BRICHOS is increased in human amyloid plaques in early stages of Alzheimer’s disease. Neurobiol. Aging 2014, 35, 1596–1604. [Google Scholar] [CrossRef]
- Aleshire, S.L.; Bradley, C.A.; Richardson, L.D.; Parl, F.F. Localization of human prealbumin in choroid plexus epithelium. J. Histochem. Cytochem. Off. J. Histochem. Soc. 1983, 31, 608–612. [Google Scholar] [CrossRef]
- Dickson, P.W.; Schreiber, G. High levels of messenger RNA for transthyretin (prealbumin) in human choroid plexus. Neurosci. Lett. 1986, 66, 311–315. [Google Scholar] [CrossRef]
- Westermark, G.T.; Westermark, P. Transthyretin and Amyloid in the Islets of Langerhans in Type-2 Diabetes. Exp. Diabetes Res. 2008, 2008, 429274. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dwork, A.J.; Cavallaro, T.; Martone, R.L.; Goodman, D.S.; Schon, E.A.; Herbert, J. Distribution of transthyretin in the rat eye. Investig. Ophthalmol. Vis. Sci. 1990, 31, 489–496. [Google Scholar]
- Murakami, T.; Ohsawa, Y.; Zhenghua, L.; Yamamura, K.-I.; Sunada, Y. The transthyretin gene is expressed in Schwann cells of peripheral nerves. Brain Res. 2010, 1348, 222–225. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Masliah, E.; Reixach, N.; Buxbaum, J.N. Neuronal Production of Transthyretin in Human and Murine Alzheimer’s Disease: Is It Protective? J. Neurosci. 2011, 31, 12483–12490. [Google Scholar] [CrossRef] [Green Version]
- Blake, C.C.F.; Geisow, M.J.; Oatley, S.J.; Rérat, B.; Rérat, C. Structure of prealbumin: Secondary, tertiary and quaternary interactions determined by Fourier refinement at 1.8 Å. J. Mol. Biol. 1978, 121, 339–356. [Google Scholar] [CrossRef]
- Blake, C.C.F.; Swan, I.D.A.; Rerat, C.; Berthou, J.; Laurent, A.; Rerat, B. An X-ray study of the subunit structure of prealbumin. J. Mol. Biol. 1971, 61, 217–224. [Google Scholar] [CrossRef]
- Yee, A.W.; Aldeghi, M.; Blakeley, M.P.; Ostermann, A.; Mas, P.J.; Moulin, M.; De Sanctis, D.; Bowler, M.W.; Mueller-Dieckmann, C.; Mitchell, E.P.; et al. A molecular mechanism for transthyretin amyloidogenesis. Nat. Commun. 2019, 10, 925. [Google Scholar] [CrossRef] [Green Version]
- Westermark, P.; Sletten, K.; Johansson, B.; Cornwell, G.G. Fibril in senile systemic amyloidosis is derived from normal transthyretin. Proc. Natl. Acad. Sci. USA 1990, 87, 2843–2845. [Google Scholar] [CrossRef] [Green Version]
- Ando, Y.; Nakamura, M.; Araki, S. Transthyretin-Related Familial Amyloidotic Polyneuropathy. Arch. Neurol. 2005, 62, 1057–1062. [Google Scholar] [CrossRef] [Green Version]
- Skinner, M. Familial amyloidotic cardiomyopathy. J. Lab. Clin. Med. 1991, 117, 171–172. [Google Scholar]
- Quintas, A.; Saraiva, M.J.; Brito, R.M.M. The Tetrameric Protein Transthyretin Dissociates to a Non-native Monomer in Solution: A novel model for amyloidogenesis. J. Biol. Chem. 1999, 274, 32943–32949. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Colon, W.; Kelly, J.W. Partial denaturation of transthyretin is sufficient for amyloid fibril formation in vitro. Biochemistry 1992, 31, 8654–8660. [Google Scholar] [CrossRef] [PubMed]
- Lai, Z.; Colón, W.; Kelly, J.W. The Acid-Mediated Denaturation Pathway of Transthyretin Yields a Conformational Intermediate That Can Self-Assemble into Amyloid. Biochemistry 1996, 35, 6470–6482. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.; Cho, H.S.; Lashuel, H.A.; Kelly, J.W.; Wemmer, D.E. A glimpse of a possible amyloidogenic intermediate of transthyretin. Nat. Struct. Biol. 2000, 7, 754–757. [Google Scholar] [PubMed] [Green Version]
- Jiang, X.; Smith, C.S.; Petrassi, H.M.; Hammarström, P.; White, J.T.; Sacchettini, J.C.; Kelly, J.W. An Engineered Transthyretin Monomer that Is Nonamyloidogenic, Unless It Is Partially Denatured. Biochemistry 2001, 40, 11442–11452. [Google Scholar] [CrossRef]
- Buxbaum, J.N.; Johansson, J. Transthyretin and BRICHOS: The Paradox of Amyloidogenic Proteins with Anti-Amyloidogenic Activity for Aβ in the Central Nervous System. Front. Neurosci. 2017, 11, 119. [Google Scholar] [CrossRef]
- Costa, R.; Gonçalves, A.; Saraiva, M.; Cardoso, I. Transthyretin binding to A-Beta peptide—Impact on A-Beta fibrillogenesis and toxicity. FEBS Lett. 2008, 582, 936–942. [Google Scholar] [CrossRef] [Green Version]
- Du, J.; Cho, P.Y.; Yang, D.T.; Murphy, R.M. Identification of beta-amyloid-binding sites on transthyretin. Protein Eng. Des. Sel. 2012, 25, 337–345. [Google Scholar] [CrossRef] [Green Version]
- Du, J.; Murphy, R.M. Characterization of the Interaction of β-Amyloid with Transthyretin Monomers and Tetramers. Biochemistry 2010, 49, 8276–8289. [Google Scholar] [CrossRef] [Green Version]
- Ghadami, S.A.; Chia, S.; Ruggeri, F.S.; Meisl, G.; Bemporad, F.; Habchi, J.; Cascella, R.; Dobson, C.M.; Vendruscolo, M.; Knowles, T.P.J.; et al. Transthyretin Inhibits Primary and Secondary Nucleations of Amyloid-β Peptide Aggregation and Reduces the Toxicity of Its Oligomers. Biomacromolecules 2020, 21, 1112–1125. [Google Scholar] [CrossRef]
- Li, X.; Buxbaum, J.N. Transthyretin and the brain re-visited: Is neuronal synthesis of transthyretin protective in Alzheimer’s disease? Mol. Neurodegener. 2011, 6, 79. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gião, T.; Saavedra, J.; Cotrina, E.; Quintana, J.; Llop, J.; Arsequell, G.; Cardoso, I. Undiscovered Roles for Transthyretin: From a Transporter Protein to a New Therapeutic Target for Alzheimer’s Disease. Int. J. Mol. Sci. 2020, 21, 2075. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gloeckner, S.F.; Meyne, F.; Wagner, F.; Heinemann, U.; Krasnianski, A.; Meissner, B.; Zerr, I. Quantitative analysis of transthyretin, tau and amyloid-beta in patients with dementia. J. Alzheimer’s Dis. 2008, 14, 17–25. [Google Scholar] [CrossRef] [PubMed]
- Merched, A.; Serot, J.M.; Visvikis, S.; Aguillon, D.; Faure, G.; Siest, G. Apolipoprotein E, transthyretin and actin in the CSF of Alzheimer’s patients: Relation with the senile plaques and cytoskeleton biochemistry. FEBS Lett. 1998, 425, 225–228. [Google Scholar] [CrossRef] [Green Version]
- Palha, J.; Moreira, P.; Wisniewski, T.; Frangione, B.; Saraiva, M.J. Transthyretin gene in Alzheimer’s disease patients. Neurosci. Lett. 1996, 204, 212–214. [Google Scholar] [CrossRef]
- Buxbaum, J.N.; Ye, Z.; Reixach, N.; Friske, L.; Levy, C.; Das, P.; Golde, T.; Masliah, E.; Roberts, A.R.; Bartfai, T. Transthyretin protects Alzheimer’s mice from the behavioral and biochemical effects of Aβ toxicity. Proc. Natl. Acad. Sci. USA 2008, 105, 2681–2686. [Google Scholar] [CrossRef] [Green Version]
- Stein, T.; Anders, N.J.; DeCarli, C.; Chan, S.L.; Mattson, M.P.; Johnson, J.A. Neutralization of Transthyretin Reverses the Neuroprotective Effects of Secreted Amyloid Precursor Protein (APP) in APPSw Mice Resulting in Tau Phosphorylation and Loss of Hippocampal Neurons: Support for the Amyloid Hypothesis. J. Neurosci. 2004, 24, 7707–7717. [Google Scholar] [CrossRef] [Green Version]
- Choi, S.H.; Leight, S.N.; Lee, V.M.-Y.; Li, T.; Wong, P.C.; Johnson, J.A.; Saraiva, M.J.; Sisodia, S.S. Accelerated Aβ Deposition in APPswe/PS1ΔE9 Mice with Hemizygous Deletions of TTR (Transthyretin). J. Neurosci. 2007, 27, 7006–7010. [Google Scholar] [CrossRef] [Green Version]
- Kerridge, C.; Belyaev, N.D.; Nalivaeva, N.N.; Turner, A.J. The Aβ-clearance protein transthyretin, like neprilysin, is epigenetically regulated by the amyloid precursor protein intracellular domain. J. Neurochem. 2014, 130, 419–431. [Google Scholar] [CrossRef]
- Mazur-Kolecka, B.; Frackowiak, J.; Wiśniewski, H.M. Apolipoproteins E3 and E4 induce, and transthyretin prevents accumulation of the Alzheimer’s β-amyloid peptide in cultured vascular smooth muscle cells. Brain Res. 1995, 698, 217–222. [Google Scholar] [CrossRef]
- Giunta, S.; Valli, M.; Galeazzi, R.; Fattoretti, P.; Corder, E.H.; Galeazzi, L. Transthyretin inhibition of amyloid beta aggregation and toxicity. Clin. Biochem. 2005, 38, 1112–1119. [Google Scholar] [CrossRef] [PubMed]
- Cao, Q.; Anderson, D.H.; Liang, W.Y.; Chou, J.A.; Saelices, L. The inhibition of cellular toxicity of amyloid-β by dissociated transthyretin. J. Biol. Chem. 2020, 295, 14015–14024. [Google Scholar] [CrossRef] [PubMed]
- Garai, K.; Posey, A.E.; Li, X.; Buxbaum, J.N.; Pappu, R.V. Inhibition of amyloid beta fibril formation by monomeric human transthyretin. Protein Sci. 2018, 27, 1252–1261. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, D.T.; Joshi, G.; Cho, P.Y.; Johnson, J.A.; Murphy, R.M. Transthyretin as both a Sensor and a Scavenger of β-Amyloid Oligomers. Biochemistry 2013, 52, 2849–2861. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schwarzman, A.L.; Tsiper, M.; Wente, H.; Wang, A.; Vitek, M.P.; Vasiliev, V.; Goldgaber, D. Amyloidogenic and anti-amyloidogenic properties of recombinant transthyretin variants. Amyloid 2004, 11, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Alemi, M.; Silva, S.C.; Santana, I.; Cardoso, I. Transthyretin stability is critical in assisting beta amyloid clearance- Relevance of transthyretin stabilization in Alzheimer’s disease. CNS Neurosci. Ther. 2017, 23, 605–619. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rejc, L.; Gómez-Vallejo, V.; Rios, X.; Cossío, U.; Baz, Z.; Mujica, E.; Gião, T.; Cotrina, E.Y.; Jiménez-Barbero, J.; Quintana, J.; et al. Oral Treatment with Iododiflunisal Delays Hippocampal Amyloid-β Formation in a Transgenic Mouse Model of Alzheimer’s Disease: A Longitudinal in vivo Molecular Imaging Study1. J. Alzheimer’s Dis. 2020, 77, 99–112. [Google Scholar] [CrossRef]
- Ribeiro, C.A.; Oliveira, S.M.; Guido, L.F.; Magalhães, A.; Valencia, G.; Arsequell, G.; Saraiva, M.J.; Cardoso, I. Transthyretin Stabilization by Iododiflunisal Promotes Amyloid-β Peptide Clearance, Decreases its Deposition, and Ameliorates Cognitive Deficits in an Alzheimer’s Disease Mouse Model. J. Alzheimer’s Dis. 2014, 39, 357–370. [Google Scholar] [CrossRef]
- Ribeiro, C.A.; Santana, I.; Oliveira, C.; Baldeiras, I.; Moreira, J.; Saraiva, M.J.; Cardoso, I. Transthyretin Decrease in Plasma of MCI and AD Patients: Investigation of Mechanisms for Disease Modulation. Curr. Alzheimer Res. 2012, 9, 881–889. [Google Scholar] [CrossRef]
- Saponaro, F.; Kim, J.H.; Chiellini, G. Transthyretin Stabilization: An Emerging Strategy for the Treatment of Alzheimer’s Disease? Int. J. Mol. Sci. 2020, 21, 8672. [Google Scholar] [CrossRef]
- Wasana Jayaweera, S.; Surano, S.; Pettersson, N.; Oskarsson, E.; Lettius, L.; Gharibyan, A.L.; Anan, I.; Olofsson, A. Mechanisms of Transthyretin Inhibition of IAPP AmyloidFormation. Biomolecules 2021, 11, 411. [Google Scholar] [CrossRef] [PubMed]
- Pate, K.M.; Kim, B.J.; Shusta, E.V.; Murphy, R.M. Transthyretin Mimetics as Anti-β-Amyloid Agents: A Comparison of Peptide and Protein Approaches. ChemMedChem 2018, 13, 968–979. [Google Scholar] [CrossRef] [PubMed]
- Westermark, P.; Wernstedt, C.; Wilander, E.; Hayden, D.W.; O’Brien, T.; Johnson, K.H. Amyloid fibrils in human insulinoma and islets of Langerhans of the diabetic cat are derived from a neuropeptide-like protein also present in normal islet cells. Proc. Natl. Acad. Sci. USA 1987, 84, 3881–3885. [Google Scholar] [CrossRef] [Green Version]
- Westermark, P.; Andersson, A.; Westermark, G.T. Islet Amyloid Polypeptide, Islet Amyloid, and Diabetes Mellitus. Physiol. Rev. 2011, 91, 795–826. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Westermark, P.; Grimelius, L. The pancreatic islet cells in insular amyloidosis in human diabetic and non-diabetic adults. Acta Pathol. Microbiol. Scand. Sect. A Pathol. 1973, 81, 291–300. [Google Scholar] [CrossRef] [PubMed]
- Jacobsson, B.; Collins, V.P.; Grimelius, L.; Pettersson, T.; Sandstedt, B.; Carlström, A. Transthyretin immunoreactivity in human and porcine liver, choroid plexus, and pancreatic islets. J. Histochem. Cytochem. 1989, 37, 31–37. [Google Scholar] [CrossRef]
- Jain, N.; Ådén, J.; Nagamatsu, K.; Evans, M.L.; Li, X.; McMichael, B.; Ivanova, M.I.; Almqvist, F.; Buxbaum, J.N.; Chapman, M.R. Inhibition of curli assembly and Escherichia coli biofilm formation by the human systemic amyloid precursor transthyretin. Proc. Natl. Acad. Sci. USA 2017, 114, 12184–12189. [Google Scholar] [CrossRef] [Green Version]
- Adams, D.; Polydefkis, M.; González-Duarte, A.; Wixner, J.; Kristen, A.V.; Schmidt, H.H.; Berk, J.L.; López, I.A.L.; Dispenzieri, A.; Quan, D.; et al. Long-term safety and efficacy of patisiran for hereditary transthyretin-mediated amyloidosis with polyneuropathy: 12-month results of an open-label extension study. Lancet Neurol. 2021, 20, 49–59. [Google Scholar] [CrossRef]
- Huang, Y.; Mahley, R.W. Apolipoprotein E: Structure and function in lipid metabolism, neurobiology, and Alzheimer’s diseases. Neurobiol. Dis. 2014, 72 Pt A, 3–12. [Google Scholar] [CrossRef] [Green Version]
- Fernandez, C.G.; Hamby, M.E.; McReynolds, M.L.; Ray, W.J. The Role of APOE4 in Disrupting the Homeostatic Functions of Astrocytes and Microglia in Aging and Alzheimer’s Disease. Front. Aging Neurosci. 2019, 11, 14. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.; Strickland, M.R.; Soranno, A.; Holtzman, D.M. Apolipoprotein E: Structural Insights and Links to Alzheimer Disease Pathogenesis. Neuron 2021, 109, 205–221. [Google Scholar] [CrossRef] [PubMed]
- Weisgraber, K.H. Apolipoprotein E: Structure-Function Relationships. Adv. Protein Chem. 1994, 45, 249–302. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Weisgraber, K.H.; Mucke, L.; Mahley, R.W. Apolipoprotein E: Diversity of Cellular Origins, Structural and Biophysical Properties, and Effects in Alzheimer’s Disease. J. Mol. Neurosci. 2004, 23, 189–204. [Google Scholar] [CrossRef]
- Arendt, T.; Schindler, C.; Brückner, M.K.; Eschrich, K.; Bigl, V.; Zedlick, D.; Marcova, L. Plastic Neuronal Remodeling Is Impaired in Patients with Alzheimer’s Disease Carrying Apolipoprotein ε4 Allele. J. Neurosci. 1997, 17, 516–529. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, J.-T.; Tan, L.; Hardy, J. Apolipoprotein E in Alzheimer’s Disease: An Update. Annu. Rev. Neurosci. 2014, 37, 79–100. [Google Scholar] [CrossRef]
- Mahley, R.W. Apolipoprotein E: From cardiovascular disease to neurodegenerative disorders. J. Mol. Med. 2016, 94, 739–746. [Google Scholar] [CrossRef] [Green Version]
- Corder, E.H.; Saunders, A.M.; Strittmatter, W.J.; Schmechel, D.E.; Gaskell, P.C.; Small, G.W.; Roses, A.D.; Haines, J.L.; Pericak-Vance, M.A. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer’s disease in late onset families. Science 1993, 261, 921–923. [Google Scholar] [CrossRef]
- Jones, P.B.; Adams, K.W.; Rozkalne, A.; Spires-Jones, T.L.; Hshieh, T.T.; Hashimoto, T.; von Armin, C.A.; Mielke, M.; Bacskai, B.J.; Hyman, B.T. Apolipoprotein E: Isoform specific differences in tertiary structure and interaction with amyloid-beta in human Alzheimer brain. PLoS ONE 2011, 6, e14586. [Google Scholar] [CrossRef]
- Bales, K.R.; Liu, F.; Wu, S.; Lin, S.; Koger, D.; DeLong, C.; Hansen, J.C.; Sullivan, P.M.; Paul, S.M. Human APOE isoform-dependent effects on brain beta-amyloid levels in PDAPP transgenic mice. J. Neurosci. 2009, 29, 6771–6779. [Google Scholar] [CrossRef]
- Castellano, J.M.; Kim, J.; Stewart, F.R.; Jiang, H.; DeMattos, R.B.; Patterson, B.W.; Fagan, A.M.; Morris, J.C.; Mawuenyega, K.G.; Cruchaga, C.; et al. Human apoE isoforms differentially regulate brain amyloid-beta peptide clearance. Sci. Transl. Med. 2011, 3, 89ra57. [Google Scholar] [CrossRef] [Green Version]
- Lee, C.Y.; Tse, W.; Smith, J.D.; Landreth, G.E. Apolipoprotein E promotes beta-amyloid trafficking and degradation by modulating microglial cholesterol levels. J. Biol. Chem. 2012, 287, 2032–2044. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bruinsma, I.B.; Wilhelmus, M.M.; Kox, M.; Veerhuis, R.; de Waal, R.M.; Verbeek, M.M. Apolipoprotein E protects cultured pericytes and astrocytes from D-Abeta(1-40)-mediated cell death. Brain Res. 2010, 1315, 169–180. [Google Scholar] [CrossRef] [PubMed]
- Ly, S.; Altman, R.; Petrlova, J.; Lin, Y.; Hilt, S.; Huser, T.; Laurence, T.A.; Voss, J.C. Binding of apolipoprotein E inhibits the oligomer growth of amyloid-beta peptide in solution as determined by fluorescence cross-correlation spectroscopy. J. Biol. Chem. 2013, 288, 11628–11635. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liao, F.; Yoon, H.; Kim, J. Apolipoprotein E metabolism and functions in brain and its role in Alzheimer’s disease. Curr. Opin. Lipidol. 2017, 28, 60–67. [Google Scholar] [CrossRef] [Green Version]
- Ma, Q.; Zhao, Z.; Sagare, A.P.; Wu, Y.; Wang, M.; Owens, N.C.; Verghese, P.B.; Herz, J.; Holtzman, D.M.; Zlokovic, B.V. Blood-brain barrier-associated pericytes internalize and clear aggregated amyloid-beta42 by LRP1-dependent apolipoprotein E isoform-specific mechanism. Mol. Neurodegener. 2018, 13, 57. [Google Scholar] [CrossRef] [Green Version]
- Potter, H.; Ma, J.; Das, S.; Kayyali, U. The involvement of amyloid associated proteins in the formation of beta-protein filaments in Alzheimer’s disease. Prog. Clin. Biol. Res. 1994, 390, 57–71. [Google Scholar]
- Holtzman, D.M.; Fagan, A.M.; Mackey, B.; Tenkova, T.; Sartorius, L.; Paul, S.M.; Bales, K.; Ashe, K.H.; Irizarry, M.C.; Hyman, B.T. Apolipoprotein E facilitates neuritic and cerebrovascular plaque formation in an Alzheimer’s disease model. Ann. Neurol. 2000, 47, 739–747. [Google Scholar] [CrossRef]
- Rall, S.C., Jr.; Weisgraber, K.H.; Mahley, R.W. Human apolipoprotein E. The complete amino acid sequence. J. Biol. Chem. 1982, 257, 4171–4178. [Google Scholar] [CrossRef]
- Weisgraber, K.H.; Rall, S.C., Jr.; Mahley, R.W. Human E apoprotein heterogeneity. Cysteine-arginine interchanges in the amino acid sequence of the apo-E isoforms. J. Biol. Chem. 1981, 256, 9077–9083. [Google Scholar] [CrossRef]
- Raulin, A.-C.; Kraft, L.; Al-Hilaly, Y.; Xue, W.-F.; McGeehan, J.; Atack, J.R.; Serpell, L. The Molecular Basis for Apolipoprotein E4 as the Major Risk Factor for Late-Onset Alzheimer’s Disease. J. Mol. Biol. 2019, 431, 2248–2265. [Google Scholar] [CrossRef]
- Frieden, C.; Garai, K. Concerning the structure of apoE. Protein Sci. 2013, 22, 1820–1825. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saito, H.; Dhanasekaran, P.; Baldwin, F.; Weisgraber, K.H.; Phillips, M.C.; Lund-Katz, S. Effects of Polymorphism on the Lipid Interaction of Human Apolipoprotein E. J. Biol. Chem. 2003, 278, 40723–40729. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, J.; Li, Q.; Wang, J. Topology of human apolipoprotein E3 uniquely regulates its diverse biological functions. Proc. Natl. Acad. Sci. USA 2011, 108, 14813–14818. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Henry, N.; Krammer, E.-M.; Stengel, F.; Adams, Q.; Van Liefferinge, F.; Hubin, E.; Chaves, R.; Efremov, R.; Aebersold, R.; Vandenbussche, G.; et al. Lipidated apolipoprotein E4 structure and its receptor binding mechanism determined by a combined cross-linking coupled to mass spectrometry and molecular dynamics approach. PLoS Comput. Biol. 2018, 14, e1006165. [Google Scholar] [CrossRef] [PubMed]
- Weisgraber, K.H. Apolipoprotein E distribution among human plasma lipoproteins: Role of the cysteine-arginine interchange at residue 112. J. Lipid Res. 1990, 31, 1503–1511. [Google Scholar] [CrossRef]
- Dong, L.-M.; Parkin, S.; Trakhanov, S.D.; Rupp, B.; Simmons, T.; Arnold, K.S.; Newhouse, Y.M.; Innerarity, T.L.; Weisgraber, K.H. Novel mechanism for defective receptor binding of apolipoprotein E2 in type III hyperlipoproteinemia. Nat. Struct. Mol. Biol. 1996, 3, 718–722. [Google Scholar] [CrossRef]
- Dong, L.-M.; Weisgraber, K.H. Human Apolipoprotein E4 Domain Interaction: Arginine 61 and glutamic acid 255 interact to direct the preference for very low density lipoproteins. J. Biol. Chem. 1996, 271, 19053–19057. [Google Scholar] [CrossRef] [Green Version]
- Raussens, V.; Drury, J.; Forte, T.M.; Choy, N.; Goormaghtigh, E.; Ruysschaert, J.-M.; Narayanaswami, V. Orientation and mode of lipid-binding interaction of human apolipoprotein E C-terminal domain. Biochem. J. 2005, 387, 747–754. [Google Scholar] [CrossRef] [Green Version]
- Narayanaswami, V.; Maiorano, J.N.; Dhanasekaran, P.; Ryan, R.O.; Phillips, M.C.; Lund-Katz, S.; Davidson, W. Helix Orientation of the Functional Domains in Apolipoprotein E in Discoidal High Density Lipoprotein Particles. J. Biol. Chem. 2004, 279, 14273–14279. [Google Scholar] [CrossRef] [Green Version]
- Hatters, D.M.; Voss, J.C.; Budamagunta, M.S.; Newhouse, Y.N.; Weisgraber, K.H. Insight on the Molecular Envelope of Lipid-Bound Apolipoprotein E from Electron Paramagnetic Resonance Spectroscopy. J. Mol. Biol. 2009, 386, 261–271. [Google Scholar] [CrossRef] [Green Version]
- Frieden, C.; Wang, H.; Ho, C.M.W. A mechanism for lipid binding to apoE and the role of intrinsically disordered regions coupled to domain–domain interactions. Proc. Natl. Acad. Sci. USA 2017, 114, 6292–6297. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peters-Libeu, C.A.; Newhouse, Y.; Hatters, D.M.; Weisgraber, K.H. Model of Biologically Active Apolipoprotein E Bound to Dipalmitoylphosphatidylcholine. J. Biol. Chem. 2006, 281, 1073–1079. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clément-Collin, V.; Barbier, A.; Dergunov, A.D.; Visvikis, A.; Siest, G.; Desmadril, M.; Takahashi, M.; Aggerbeck, L.P. The structure of human apolipoprotein E2, E3 and E4 in solution. Multidomain organization correlates with the stability of apoE structure. Biophys. Chem. 2006, 119, 170–185. [Google Scholar] [CrossRef] [PubMed]
- Hatters, D.M.; Zhong, N.; Rutenber, E.; Weisgraber, K.H. Amino-terminal Domain Stability Mediates Apolipoprotein E Aggregation into Neurotoxic Fibrils. J. Mol. Biol. 2006, 361, 932–944. [Google Scholar] [CrossRef] [Green Version]
- Islam, T.; Gharibyan, A.L.; Golchin, S.A.; Pettersson, N.; Brännström, K.; Hedberg, I.; Virta, M.; Olofsson, L.; Olofsson, A. Apolipoprotein E impairs amyloid-β fibril elongation and maturation. FEBS J. 2020, 287, 1208–1219. [Google Scholar] [CrossRef]
- Holtzman, D.M.; Bales, K.R.; Tenkova, T.; Fagan, A.M.; Parsadanian, M.; Sartorius, L.J.; Mackey, B.; Olney, J.; McKeel, D.; Wozniak, D.; et al. Apolipoprotein E isoform-dependent amyloid deposition and neuritic degeneration in a mouse model of Alzheimer’s disease. Proc. Natl. Acad. Sci. USA 2000, 97, 2892–2897. [Google Scholar] [CrossRef] [Green Version]
- Liao, F.; Li, A.; Xiong, M.; Bien-Ly, N.; Jiang, H.; Zhang, Y.; Finn, M.B.; Hoyle, R.; Keyser, J.; Lefton, K.B.; et al. Targeting of nonlipidated, aggregated apoE with antibodies inhibits amyloid accumulation. J. Clin. Investig. 2018, 128, 2144–2155. [Google Scholar] [CrossRef] [Green Version]
- Jiang, Q.; Lee, C.Y.D.; Mandrekar, S.; Wilkinson, B.; Cramer, P.; Zelcer, N.; Mann, K.; Lamb, B.; Willson, T.M.; Collins, J.L.; et al. ApoE Promotes the Proteolytic Degradation of Aβ. Neuron 2008, 58, 681–693. [Google Scholar] [CrossRef] [Green Version]
- Robert, J.; Button, E.B.; Yuen, B.; Gilmour, M.; Kang, K.; Bahrabadi, A.; Stukas, S.; Zhao, W.; Kulic, I.; Wellington, C.L. Clearance of beta-amyloid is facilitated by apolipoprotein E and circulating high-density lipoproteins in bioengineered human vessels. eLife 2017, 6, e29595. [Google Scholar] [CrossRef]
- Wilhelmus, M.M.M.; Otte-Höller, I.; Davis, J.; Van Nostrand, W.E.; De Waal, R.M.W.; Verbeek, M.M. Apolipoprotein E Genotype Regulates Amyloid-β Cytotoxicity. J. Neurosci. 2005, 25, 3621–3627. [Google Scholar] [CrossRef]
- Rasmussen, K.L. Plasma levels of apolipoprotein E, APOE genotype and risk of dementia and ischemic heart disease: A review. Atherosclerosis 2016, 255, 145–155. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rasmussen, K.L.; Tybjaerg-Hansen, A.; Nordestgaard, B.G.; Frikke-Schmidt, R. Plasma apolipoprotein E levels and risk of dementia: A Mendelian randomization study of 106,562 individuals. Alzheimers Dement. 2018, 14, 71–80. [Google Scholar] [CrossRef]
- Gupta, V.B.; Wilson, A.C.; Burnham, S.; Hone, E.; Pedrini, S.; Laws, S.M.; Lim, W.L.F.; Rembach, A.; Rainey-Smith, S.; Ames, D.; et al. Follow-up plasma apolipoprotein E levels in the Australian Imaging, Biomarkers and Lifestyle Flagship Study of Ageing (AIBL) cohort. Alzheimer’s Res. Ther. 2015, 7, 16. [Google Scholar] [CrossRef] [Green Version]
- Wolters, F.J.; Koudstaal, P.J.; Hofman, A.; Van Duijn, C.M.; Ikram, M.A. Serum apolipoprotein E is associated with long-term risk of Alzheimer’s disease: The Rotterdam Study. Neurosci. Lett. 2016, 617, 139–142. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Morillo, E.; Hansson, O.; Atagi, Y.; Bu, G.; Minthon, L.; Diamandis, E.P.; Nielsen, H.M. Total apolipoprotein E levels and specific isoform composition in cerebrospinal fluid and plasma from Alzheimer’s disease patients and controls. Acta Neuropathol. 2014, 127, 633–643. [Google Scholar] [CrossRef] [PubMed]
- Bertrand, P.; Poirier, J.; Oda, T.; Finch, C.E.; Pasinetti, G. Association of apolipoprotein E genotype with brain levels of apolipoprotein E and apolipoprotein J (clusterin) in Alzheimer disease. Brain Res. Mol. Brain Res. 1995, 33, 174–178. [Google Scholar] [CrossRef]
- Talwar, P.; Sinha, J.; Grover, S.; Agarwal, R.; Kushwaha, S.; Srivastava, M.V.P.; Kukreti, R. Meta-analysis of apolipoprotein E levels in the cerebrospinal fluid of patients with Alzheimer’s disease. J. Neurol. Sci. 2016, 360, 179–187. [Google Scholar] [CrossRef]
- Cruchaga, C.; Kauwe, J.S.; Nowotny, P.; Bales, K.; Pickering, E.H.; Mayo, K.; Bertelsen, S.; Hinrichs, A.; Fagan, A.M.; Holtzman, D.M.; et al. Cerebrospinal fluid APOE levels: An endophenotype for genetic studies for Alzheimer’s disease. Hum. Mol. Genet. 2012, 21, 4558–4571. [Google Scholar] [CrossRef] [Green Version]
- Sihlbom, C.; Davidsson, P.; Sjögren, M.; Wahlund, L.-O.; Nilsson, C.L. Structural and Quantitative Comparison of Cerebrospinal Fluid Glycoproteins in Alzheimer’s Disease Patients and Healthy Individuals. Neurochem. Res. 2008, 33, 1332–1340. [Google Scholar] [CrossRef]
- Akram, A.; Schmeidler, J.; Katsel, P.; Hof, P.R.; Haroutunian, V. Association of ApoE and LRP mRNA levels with dementia and AD neuropathology. Neurobiol. Aging 2012, 33, 628.e1–628.e14. [Google Scholar] [CrossRef] [Green Version]
- Baker-Nigh, A.T.; Mawuenyega, K.G.; Bollinger, J.G.; Ovod, V.; Kasten, T.; Franklin, E.E.; Liao, F.; Jiang, H.; Holtzman, D.; Cairns, N.J.; et al. Human Central Nervous System (CNS) ApoE Isoforms Are Increased by Age, Differentially Altered by Amyloidosis, and Relative Amounts Reversed in the CNS Compared with Plasma. J. Biol. Chem. 2016, 291, 27204–27218. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schmidt, C.; Becker, H.; Zerr, I. Cerebrospinal Fluid Apolipoprotein E Concentration and Severity of Cognitive Impairment in Patients With Newly Diagnosed Alzheimer’s Disease. Am. J. Alzheimer’s Dis. Other Dement. 2014, 29, 54–60. [Google Scholar] [CrossRef] [PubMed]
- Arboleda-Velasquez, J.F.; Lopera, F.; O’Hare, M.; Delgado-Tirado, S.; Marino, C.; Chmielewska, N.; Saez-Torres, K.L.; Amarnani, D.; Schultz, A.P.; Sperling, R.A.; et al. Resistance to autosomal dominant Alzheimer’s disease in an APOE3 Christchurch homozygote: A case report. Nat. Med. 2019, 25, 1680–1683. [Google Scholar] [CrossRef] [PubMed]
- Lei, P.; Wu, W.-H.; Li, R.-W.; Ma, J.-W.; Yu, Y.-P.; Cui, W.; Zhao, Y.-F.; Li, Y.-M. Prevention and promotion effects of apolipoprotein E4 on amylin aggregation. Biochem. Biophys. Res. Commun. 2008, 368, 414–418. [Google Scholar] [CrossRef] [PubMed]
- Emamzadeh, F.N.; Aojula, H.; McHugh, P.; Allsop, D. Effects of different isoforms of apoE on aggregation of the α-synuclein protein implicated in Parkinson’s disease. Neurosci. Lett. 2016, 618, 146–151. [Google Scholar] [CrossRef] [Green Version]
- Gharibyan, A.L.; Islam, T.; Pettersson, N.; Golchin, S.A.; Lundgren, J.; Johansson, G.; Genot, M.; Schultz, N.; Wennström, M.; Olofsson, A. Apolipoprotein E Interferes with IAPP Aggregation and Protects Pericytes from IAPP-Induced Toxicity. Biomolecules 2020, 10, 134. [Google Scholar] [CrossRef] [Green Version]
- Gallardo, G.; Schlüter, O.M.; Südhof, T.C. A molecular pathway of neurodegeneration linking α-synuclein to ApoE and Aβ peptides. Nat. Neurosci. 2008, 11, 301–308. [Google Scholar] [CrossRef]
- Williams, T.; Borchelt, D.R.; Chakrabarty, P. Therapeutic approaches targeting Apolipoprotein E function in Alzheimer’s disease. Mol. Neurodegener. 2020, 15, 8. [Google Scholar] [CrossRef] [Green Version]
- Moezzi, S.-M.; Mozafari, N.; Fazel-Hoseini, S.-M.; Nadimi-Parashkoohi, S.; Abbasi, H.; Ashrafi, H.; Azadi, A. Apolipoprotein J in Alzheimer’s Disease: Shedding Light on Its Role with Cell Signaling Pathway Perspective and Possible Therapeutic Approaches. ACS Chem. Neurosci. 2020, 11, 4060–4072. [Google Scholar] [CrossRef]
- Foster, E.M.; Dangla-Valls, A.; Lovestone, S.; Ribe, E.M.; Buckley, N.J. Clusterin in Alzheimer’s Disease: Mechanisms, Genetics, and Lessons from Other Pathologies. Front. Neurosci. 2019, 13, 164. [Google Scholar] [CrossRef] [Green Version]
- De Silva, H.V.; Harmony, J.A.; Stuart, W.D.; Gil, C.M.; Robbins, J. Apolipoprotein J: Structure and tissue distribution. Biochemistry 1990, 29, 5380–5389. [Google Scholar] [CrossRef] [PubMed]
- Calero, M.; Rostagno, A.; Matsubara, E.; Zlokovic, B.; Frangione, B.; Ghiso, J. Apolipoprotein J (clusterin) and Alzheimer’s disease. Microsc. Res. Tech. 2000, 50, 305–315. [Google Scholar] [CrossRef]
- Morrissey, C.; Lakins, J.; Moquin, A.; Hussain, M.; Tenniswood, M. An antigen capture assay for the measurement of serum clusterin concentrations. J. Biochem. Biophys. Methods 2001, 48, 13–21. [Google Scholar] [CrossRef]
- Arranz, A.M.; De Strooper, B. The role of astroglia in Alzheimer’s disease: Pathophysiology and clinical implications. Lancet Neurol. 2019, 18, 406–414. [Google Scholar] [CrossRef]
- Dunker, A.K.; Lawson, J.D.; Brown, C.J.; Williams, R.M.; Romero, P.; Oh, J.S.; Oldfield, C.J.; Campen, A.M.; Ratliff, C.M.; Hipps, K.W.; et al. Intrinsically disordered protein. J. Mol. Graph. Model. 2001, 19, 26–59. [Google Scholar] [CrossRef] [Green Version]
- Jones, S.E.; Jomary, C. Clusterin. Int. J. Biochem. Cell Biol. 2002, 34, 427–431. [Google Scholar] [CrossRef]
- Bartl, M.M.; Luckenbach, T.; Bergner, O.; Ullrich, O.; Koch-Brandt, C. Multiple Receptors Mediate apoJ-Dependent Clearance of Cellular Debris into Nonprofessional Phagocytes. Exp. Cell Res. 2001, 271, 130–141. [Google Scholar] [CrossRef]
- Zhang, H.; Kim, J.K.; Edwards, C.A.; Xu, Z.; Taichman, R.S.; Wang, C.-Y. Clusterin inhibits apoptosis by interacting with activated Bax. Nat. Cell Biol. 2005, 7, 909–915. [Google Scholar] [CrossRef]
- Wilson, M.R.; Easterbrook-Smith, S.B. Clusterin is a secreted mammalian chaperone. Trends Biochem. Sci. 2000, 25, 95–98. [Google Scholar] [CrossRef] [Green Version]
- Humphreys, D.T.; Carver, J.A.; Easterbrook-Smith, S.B.; Wilson, M.R. Clusterin Has Chaperone-like Activity Similar to That of Small Heat Shock Proteins. J. Biol. Chem. 1999, 274, 6875–6881. [Google Scholar] [CrossRef] [Green Version]
- Wyatt, A.; Yerbury, J.; Poon, S.; Dabbs, R.; Wilson, M. Chapter 6: The Chaperone Action of Clusterin and Its Putative Role in Quality Control of Extracellular Protein Folding. Adv. Cancer Res. 2009, 104, 89–114. [Google Scholar] [CrossRef] [PubMed]
- Nuutinen, T.; Suuronen, T.; Kauppinen, A.; Salminen, A. Clusterin: A forgotten player in Alzheimer’s disease. Brain Res. Rev. 2009, 61, 89–104. [Google Scholar] [CrossRef] [PubMed]
- Harold, D.; Abraham, R.; Hollingworth, P.; Sims, R.; Gerrish, A.; Hamshere, M.L.; Pahwa, J.S.; Moskvina, V.; Dowzell, K.; Williams, A.; et al. Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer’s disease. Nat. Genet. 2009, 41, 1088–1093. [Google Scholar] [CrossRef] [Green Version]
- Lambert, J.C.; Ibrahim-Verbaas, C.A.; Harold, D.; Naj, A.C.; Sims, R.; Bellenguez, C.; DeStafano, A.L.; Bis, J.C.; Beecham, G.W.; Grenier-Boley, B.; et al. Meta-Analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat. Genet. 2013, 45, 1452–1458. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tan, L.; Initiative, A.D.N.; Wang, H.-F.; Tan, M.-S.; Tan, C.-C.; Zhu, X.-C.; Miao, D.; Yu, W.-J.; Jiang, T.; Tan, L.; et al. Effect of CLU genetic variants on cerebrospinal fluid and neuroimaging markers in healthy, mild cognitive impairment and Alzheimer’s disease cohorts. Sci. Rep. 2016, 6, 26027. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schrijvers, E.M.; Koudstaal, P.J.; Hofman, A.; Breteler, M.M. Plasma Clusterin and the Risk of Alzheimer Disease. JAMA 2011, 305, 1322–1326. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nilselid, A.-M.; Davidsson, P.; Nägga, K.; Andreasen, N.; Fredman, P.; Blennow, K. Clusterin in cerebrospinal fluid: Analysis of carbohydrates and quantification of native and glycosylated forms. Neurochem. Int. 2006, 48, 718–728. [Google Scholar] [CrossRef]
- Miners, J.S.; Clarke, P.; Love, S. Clusterin levels are increased in Alzheimer’s disease and influence the regional distribution of Aβ. Brain Pathol. 2017, 27, 305–313. [Google Scholar] [CrossRef] [PubMed]
- Lidström, A.-M.; Bogdanovic, N.; Hesse, C.; Volkman, I.; Davidsson, P.; Blennow, K. Clusterin (Apolipoprotein J) Protein Levels Are Increased in Hippocampus and in Frontal Cortex in Alzheimer’s Disease. Exp. Neurol. 1998, 154, 511–521. [Google Scholar] [CrossRef] [PubMed]
- Bell, R.D.; Sagare, A.P.; Friedman, A.E.; Bedi, G.S.; Holtzman, D.M.; Deane, R.; Zlokovic, B.V. Transport Pathways for Clearance of Human Alzheimer’s Amyloid β-Peptide and Apolipoproteins E and J in the Mouse Central Nervous System. J. Cereb. Blood Flow Metab. 2007, 27, 909–918. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Ma, Y.; Wei, X.; Li, Y.; Wu, H.; Zhuang, J.; Zhao, Z. Clusterin in Alzheimer’s disease: A player in the biological behavior of amyloid-beta. Neurosci. Bull. 2014, 30, 162–168. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oda, T.; Wals, P.; Osterburg, H.H.; Johnson, S.A.; Pasinetti, G.; Morgan, T.E.; Rozovsky, I.; Stine, W.B.; Snyder, S.; Holzman, T.F.; et al. Clusterin (apoJ) Alters the Aggregation of Amyloid β-Peptide (Aβ1-42) and Forms Slowly Sedimenting Aβ Complexes That Cause Oxidative Stress. Exp. Neurol. 1995, 136, 22–31. [Google Scholar] [CrossRef] [PubMed]
- Wojtas, A.M.; Kang, S.S.; Olley, B.M.; Gatherer, M.; Shinohara, M.; Lozano, P.A.; Liu, C.-C.; Kurti, A.; Baker, K.E.; Dickson, D.W.; et al. Loss of clusterin shifts amyloid deposition to the cerebrovasculature via disruption of perivascular drainage pathways. Proc. Natl. Acad. Sci. USA 2017, 114, E6962–E6971. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deane, R.; Bell, R.D.; Sagare, A.; Zlokovic, B.V. Clearance of Amyloid-β Peptide Across the Blood-Brain Barrier: Implication for Therapies in Alzheimers Disease. CNS Neurol. Disord. Drug Targets 2009, 8, 16–30. [Google Scholar] [CrossRef] [PubMed]
- Yerbury, J.J.; Poon, S.; Meehan, S.; Thompson, B.; Kumita, J.R.; Dobson, C.M.; Wilson, M.R. The extracellular chaperone clusterin influences amyloid formation and toxicity by interacting with prefibrillar structures. FASEB J. 2007, 21, 2312–2322. [Google Scholar] [CrossRef]
- Scheidt, T.; Łapińska, U.; Kumita, J.R.; Whiten, D.R.; Klenerman, D.; Wilson, M.R.; Cohen, S.I.A.; Linse, S.; Vendruscolo, M.; Dobson, C.M.; et al. Secondary nucleation and elongation occur at different sites on Alzheimer’s amyloid-β aggregates. Sci. Adv. 2019, 5, eaau3112. [Google Scholar] [CrossRef] [Green Version]
- Wojtas, A.M.; Carlomagno, Y.; Sens, J.P.; Kang, S.S.; Jensen, T.D.; Kurti, A.; Baker, K.E.; Berry, T.J.; Phillips, V.R.; Castanedes, M.C.; et al. Clusterin ameliorates tau pathology in vivo by inhibiting fibril formation. Acta Neuropathol. Commun. 2020, 8, 210. [Google Scholar] [CrossRef]
- De Retana, S.F.; Marazuela, P.; Solé, M.; Colell, G.; Pastra, A.B.; Sánchez-Quesada, J.L.; Montaner, J.; Maspoch, D.; Cano-Sarabia, M.; Hernández-Guillamon, M. Peripheral administration of human recombinant ApoJ/clusterin modulates brain beta-amyloid levels in APP23 mice. Alzheimer’s Res. Ther. 2019, 11, 42. [Google Scholar] [CrossRef] [Green Version]
- Hedlund, J.; Johansson, J.; Persson, B. BRICHOS—A superfamily of multidomain proteins with diverse functions. BMC Res. Notes 2009, 2, 180. [Google Scholar] [CrossRef] [Green Version]
- Pulido, L.S.; Devos, D.; Valencia, A. BRICHOS: A conserved domain in proteins associated with dementia, respiratory distress and cancer. Trends Biochem. Sci. 2002, 27, 329–332. [Google Scholar] [CrossRef]
- Willander, H.; Askarieh, G.; Landreh, M.; Westermark, P.; Nordling, K.; Keränen, H.; Hermansson, E.; Hamvas, A.; Nogee, L.M.; Bergman, T.; et al. High-resolution structure of a BRICHOS domain and its implications for anti-amyloid chaperone activity on lung surfactant protein C. Proc. Natl. Acad. Sci. USA 2012, 109, 2325–2329. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martin, L.; Fluhrer, R.; Reiss, K.; Kremmer, E.; Saftig, P.; Haass, C. Regulated Intramembrane Proteolysis of Bri2 (Itm2b) by ADAM10 and SPPL2a/SPPL2b. J. Biol. Chem. 2008, 283, 1644–1652. [Google Scholar] [CrossRef] [Green Version]
- Knight, S.D.; Presto, J.; Linse, S.; Johansson, J. The BRICHOS Domain, Amyloid Fibril Formation, and Their Relationship. Biochemistry 2013, 52, 7523–7531. [Google Scholar] [CrossRef] [PubMed]
- Conkright, J.J.; Na, C.-L.; Weaver, T.E. Overexpression of Surfactant Protein–C Mature Peptide Causes Neonatal Lethality in Transgenic Mice. Am. J. Respir. Cell Mol. Biol. 2002, 26, 85–90. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Poska, H.; Haslbeck, M.; Kurudenkandy, F.R.; Hermansson, E.; Chen, G.; Kostallas, G.; Abelein, A.; Biverstål, H.; Crux, S.; Fisahn, A.; et al. Dementia-related Bri2 BRICHOS is a versatile molecular chaperone that efficiently inhibits Aβ42 toxicity in Drosophila. Biochem. J. 2016, 473, 3683–3704. [Google Scholar] [CrossRef] [Green Version]
- Johansson, J.; Szyperski, T.; Wüthrich, K. Pulmonary surfactant-associated polypeptide SP-C in lipid micelles: CD studies of intact SP-C and NMR secondary structure determination of depalmitoyl-SP-C(1-17). FEBS Lett. 1995, 362, 261–265. [Google Scholar] [CrossRef] [Green Version]
- Johansson, J.; Szyperski, T.; Curstedt, T.; Wuethrich, K. The NMR Structure of the Pulmonary Surfactant-Associated Polypeptide SP-C in an Apolar Solvent Contains a Valyl-Rich.alpha.-Helix. Biochemistry 1994, 33, 6015–6023. [Google Scholar] [CrossRef]
- Mulugeta, S.; Beers, M.F. Surfactant protein C: Its unique properties and emerging immunomodulatory role in the lung. Microbes Infect. 2006, 8, 2317–2323. [Google Scholar] [CrossRef]
- Johansson, H.; Nordling, K.; Weaver, T.E.; Johansson, J. The Brichos Domain-containing C-terminal Part of Pro-surfactant Protein C Binds to an Unfolded Poly-Val Transmembrane Segment. J. Biol. Chem. 2006, 281, 21032–21039. [Google Scholar] [CrossRef] [Green Version]
- Nogee, L.M.; Dunbar, A.E.; Wert, S.; Askin, F.; Hamvas, A.; Whitsett, J.A. Mutations in the Surfactant Protein C Gene Associated With Interstitial Lung Disease. Chest 2002, 121, 20S–21S. [Google Scholar] [CrossRef] [Green Version]
- Nerelius, C.; Martin, E.; Peng, S.; Gustafsson, M.; Nordling, K.; Weaver, T.; Johansson, J. Mutations linked to interstitial lung disease can abrogate anti-amyloid function of prosurfactant protein C. Biochem. J. 2008, 416, 201–209. [Google Scholar] [CrossRef] [PubMed]
- Dolfe, L.; Tambaro, S.; Tigro, H.; Del Campo, M.; Hoozemans, J.J.; Wiehager, B.; Graff, C.; Winblad, B.; Ankarcrona, M.; Kaldmäe, M.; et al. The Bri2 and Bri3 BRICHOS Domains Interact Differently with Aβ 42 and Alzheimer Amyloid Plaques. J. Alzheimer’s Dis. Rep. 2018, 2, 27–39. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Poska, H.; Leppert, A.; Tigro, H.; Zhong, X.; Kaldmäe, M.; Nilsson, H.E.; Hebert, H.; Chen, G.; Johansson, J. Recombinant Bri3 BRICHOS domain is a molecular chaperone with effect against amyloid formation and non-fibrillar protein aggregation. Sci. Rep. 2020, 10, 9817. [Google Scholar] [CrossRef] [PubMed]
- Cantlon, A.; Frigerio, C.S.; Walsh, M.M. Lessons from a Rare Familial Dementia: Amyloid and Beyond. J. Park. Dis. Alzheimer’s Dis. 2015, 2, 12. [Google Scholar] [CrossRef]
- Mead, S.; James-Galton, M.; Revesz, T.; Doshi, R.B.; Harwood, G.; Pan, E.L.; Ghiso, J.; Frangione, B.; Plant, G. Familial British dementia with amyloid angiopathy: Early clinical, neuropsychological and imaging findings. Brain 2000, 123 Pt 5, 975–991. [Google Scholar] [CrossRef] [Green Version]
- Matsuda, S.; Tamayev, R.; D’Adamio, L. Increased AβPP Processing in Familial Danish Dementia Patients. J. Alzheimer’s Dis. 2011, 27, 385–391. [Google Scholar] [CrossRef] [Green Version]
- Fotinopoulou, A.; Tsachaki, M.; Vlavaki, M.; Poulopoulos, A.; Rostagno, A.; Frangione, B.; Ghiso, J.; Efthimiopoulos, S. BRI2 Interacts with Amyloid Precursor Protein (APP) and Regulates Amyloid β (Aβ) Production. J. Biol. Chem. 2005, 280, 30768–30772. [Google Scholar] [CrossRef] [Green Version]
- Matsuda, S.; Matsuda, Y.; Snapp, E.L.; D’Adamio, L. Maturation of BRI2 generates a specific inhibitor that reduces APP processing at the plasma membrane and in endocytic vesicles. Neurobiol. Aging 2011, 32, 1400–1408. [Google Scholar] [CrossRef] [Green Version]
- Tamayev, R.; Matsuda, S.; Fà, M.; Arancio, O.; D’Adamio, L. Danish dementia mice suggest that loss of function and not the amyloid cascade causes synaptic plasticity and memory deficits. Proc. Natl. Acad. Sci. USA 2010, 107, 20822–20827. [Google Scholar] [CrossRef] [Green Version]
- Tamayev, R.; Giliberto, L.; Li, W.; D’Abramo, C.; Arancio, O.; Vidal, R.; D’Adamio, L. Memory Deficits Due to Familial British Dementia BRI2 Mutation Are Caused by Loss of BRI2 Function Rather than Amyloidosis. J. Neurosci. 2010, 30, 14915–14924. [Google Scholar] [CrossRef]
- Kim, J.; Miller, V.M.; Levites, Y.; West, K.J.; Zwizinski, C.W.; Moore, B.D.; Troendle, F.J.; Bann, M.; Verbeeck, C.; Price, R.W.; et al. BRI2 (ITM2b) Inhibits Aβ Deposition In Vivo. J. Neurosci. 2008, 28, 6030–6036. [Google Scholar] [CrossRef] [PubMed]
- Hermansson, E.; Schultz, S.; Crowther, D.; Linse, S.; Winblad, B.; Westermark, G.; Johansson, J.; Presto, J. The chaperone domain BRICHOS prevents CNS toxicity of amyloid-β peptide in Drosophila melanogaster. Dis. Model. Mech. 2014, 7, 659–665. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cohen, S.I.A.; Arosio, P.; Presto, J.; Kurudenkandy, F.R.; Biverstal, H.; Dolfe, L.; Dunning, C.; Yang, X.; Frohm, B.; Vendruscolo, M.; et al. A molecular chaperone breaks the catalytic cycle that generates toxic Aβ oligomers. Nat. Struct. Mol. Biol. 2015, 22, 207–213. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Abelein, A.; Nilsson, H.E.; Leppert, A.; Andrade-Talavera, Y.; Tambaro, S.; Hemmingsson, L.; Roshan, F.; Landreh, M.; Biverstal, H.; et al. Bri2 BRICHOS client specificity and chaperone activity are governed by assembly state. Nat. Commun. 2017, 8, 2081. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arosio, P.; Michaels, T.; Linse, S.; Månsson, C.; Emanuelsson, C.; Presto, J.; Johansson, J.; Vendruscolo, M.; Dobson, C.M.; Knowles, T. Kinetic analysis reveals the diversity of microscopic mechanisms through which molecular chaperones suppress amyloid formation. Nat. Commun. 2016, 7, 10948. [Google Scholar] [CrossRef] [Green Version]
- Tambaro, S.; Galan-Acosta, L.; Leppert, A.; Chen, G.; Biverstål, H.; Presto, J.; Nilsson, P.; Johansson, J. Blood–brain and blood–cerebrospinal fluid passage of BRICHOS domains from two molecular chaperones in mice. J. Biol. Chem. 2019, 294, 2606–5220. [Google Scholar] [CrossRef] [Green Version]
- Galan-Acosta, L.; Sierra, C.; Leppert, A.; Pouliopoulos, A.N.; Kwon, N.; Noel, R.L.; Tambaro, S.; Presto, J.; Nilsson, P.; Konofagou, E.E.; et al. Recombinant BRICHOS chaperone domains delivered to mouse brain parenchyma by focused ultrasound and microbubbles are internalized by hippocampal and cortical neurons. Mol. Cell. Neurosci. 2020, 105, 103498. [Google Scholar] [CrossRef]
- Chen, G.; Andrade-Talavera, Y.; Tambaro, S.; Leppert, A.; Nilsson, H.E.; Zhong, X.; Landreh, M.; Nilsson, P.; Hebert, H.; Biverstål, H.; et al. Augmentation of Bri2 molecular chaperone activity against amyloid-β reduces neurotoxicity in mouse hippocampus in vitro. Commun. Biol. 2020, 3, 32. [Google Scholar] [CrossRef] [Green Version]
- Oskarsson, M.E.; Hermansson, E.; Wang, Y.; Welsh, N.; Presto, J.; Johansson, J.; Westermark, G.T. BRICHOS domain of Bri2 inhibits islet amyloid polypeptide (IAPP) fibril formation and toxicity in human beta cells. Proc. Natl. Acad. Sci. USA 2018, 115, E2752–E2761. [Google Scholar] [CrossRef] [Green Version]
- Akter, K.; Lanza, E.A.; Martin, S.A.; Myronyuk, N.; Rua, M.; Raffa, R.B. Diabetes mellitus and Alzheimer’s disease: Shared pathology and treatment? Br. J. Clin. Pharmacol. 2011, 71, 365–376. [Google Scholar] [CrossRef] [Green Version]
- Uchikado, H.; Lin, W.-L.; DeLucia, M.W.; Dickson, D.W. Alzheimer Disease with Amygdala Lewy Bodies: A distinct form of alpha-synucleinopathy. J. Neuropathol. Exp. Neurol. 2006, 65, 685–697. [Google Scholar] [CrossRef] [PubMed] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gharibyan, A.L.; Wasana Jayaweera, S.; Lehmann, M.; Anan, I.; Olofsson, A. Endogenous Human Proteins Interfering with Amyloid Formation. Biomolecules 2022, 12, 446. https://doi.org/10.3390/biom12030446
Gharibyan AL, Wasana Jayaweera S, Lehmann M, Anan I, Olofsson A. Endogenous Human Proteins Interfering with Amyloid Formation. Biomolecules. 2022; 12(3):446. https://doi.org/10.3390/biom12030446
Chicago/Turabian StyleGharibyan, Anna L., Sanduni Wasana Jayaweera, Manuela Lehmann, Intissar Anan, and Anders Olofsson. 2022. "Endogenous Human Proteins Interfering with Amyloid Formation" Biomolecules 12, no. 3: 446. https://doi.org/10.3390/biom12030446
APA StyleGharibyan, A. L., Wasana Jayaweera, S., Lehmann, M., Anan, I., & Olofsson, A. (2022). Endogenous Human Proteins Interfering with Amyloid Formation. Biomolecules, 12(3), 446. https://doi.org/10.3390/biom12030446





