Endogenous Controls for the Evaluation of Osteoarthritis-Related miRNAs in Extracellular Vesicles from Bone-Marrow-Derived Mesenchymal Stromal Cells and the Impact of Osteoarthritis Synovial Fluid
Abstract
1. Introduction
2. Materials and Methods
2.1. OA-Synovial Fluid Collection
2.2. OA-SF Characterization
2.3. BMSCs Isolation and Expansion
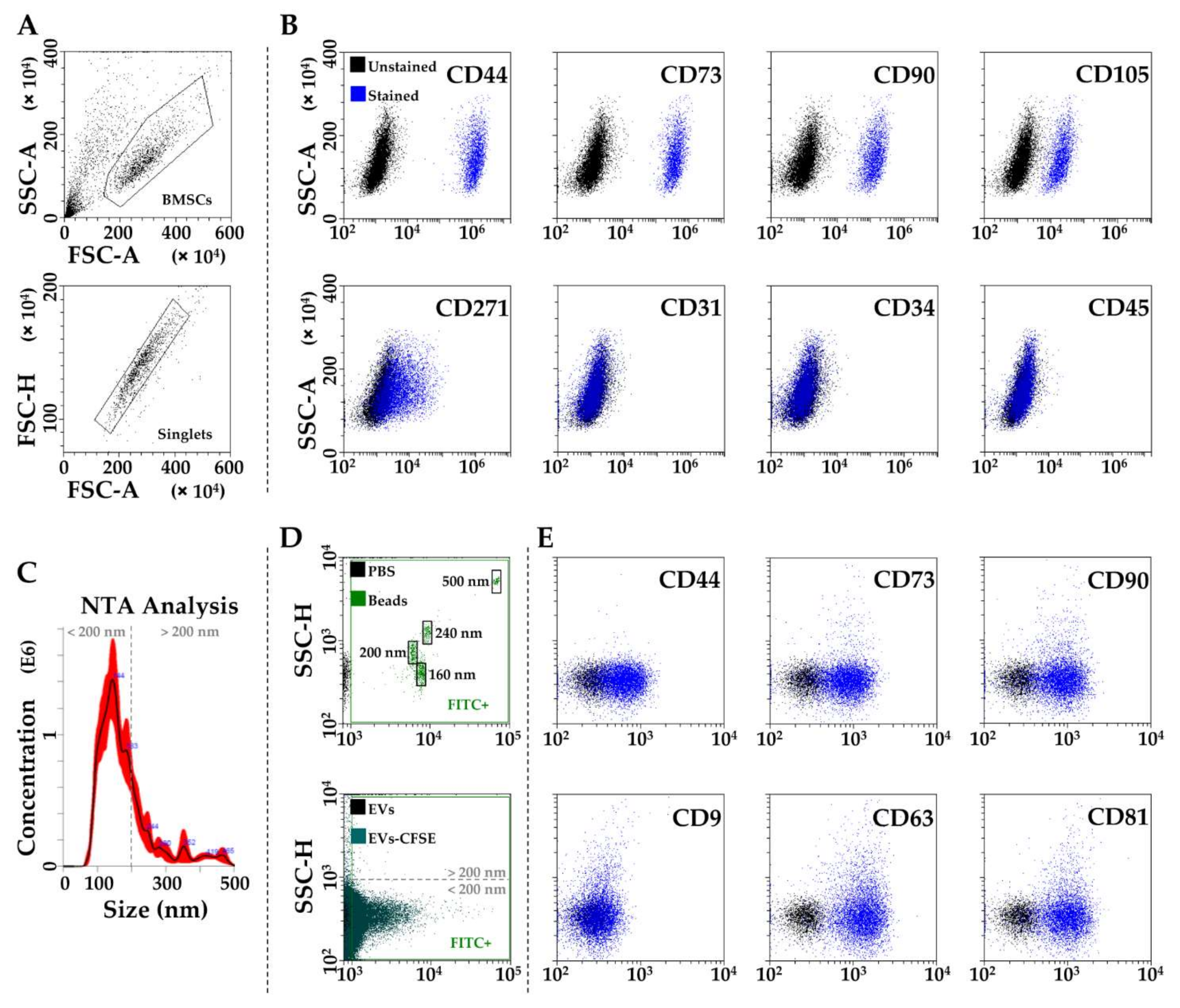
2.4. BMSCs Characterization by Flow Cytometry
2.5. BMSC-EVs Isolation and Characterization
2.6. Total RNA Isolation and miRNA Profiling
2.7. Data Analysis
2.8. Statistical Analysis
3. Results
3.1. BMSCs and EVs Characterization
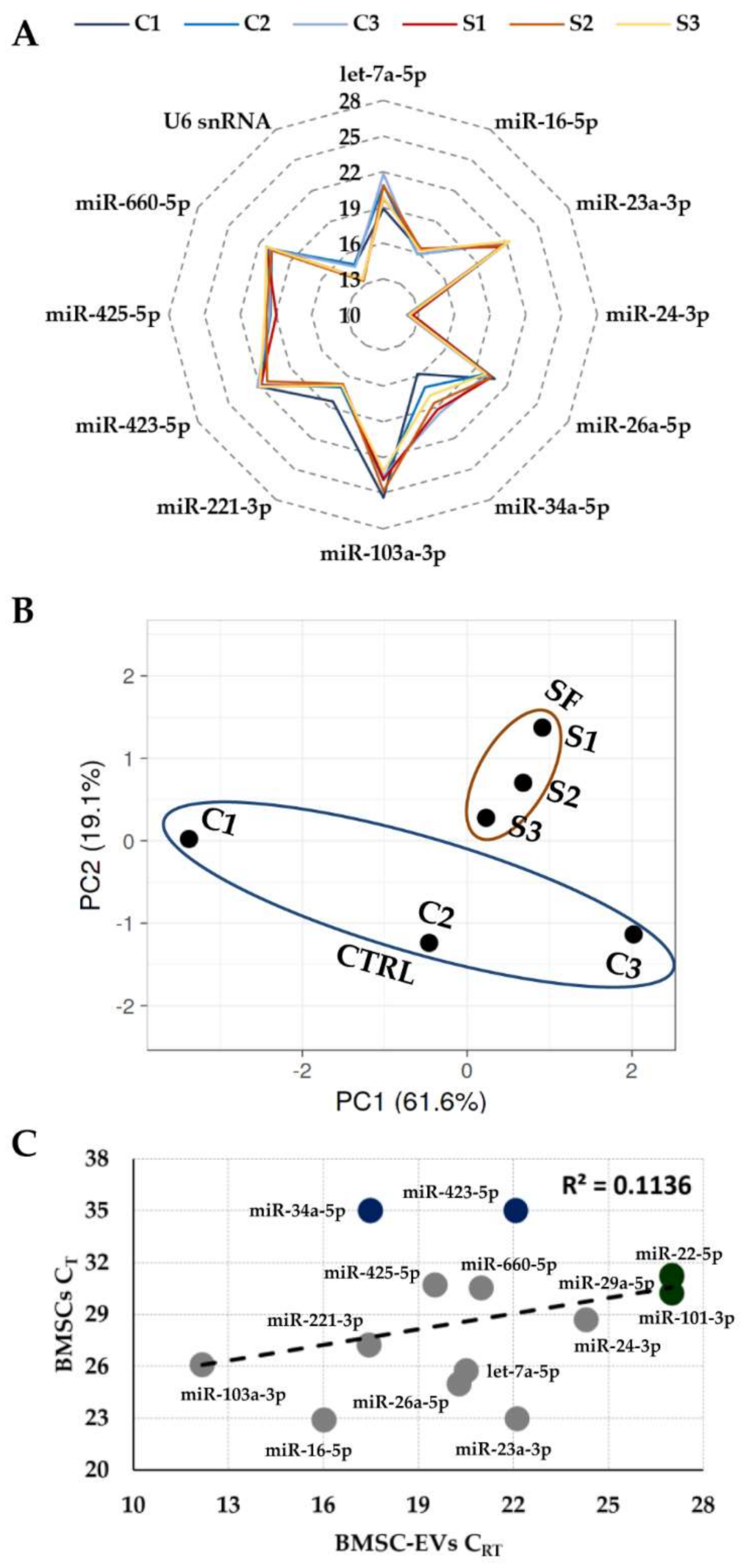
3.2. Expression of Candidate Reference Genes
3.3. RGs Stability Analysis
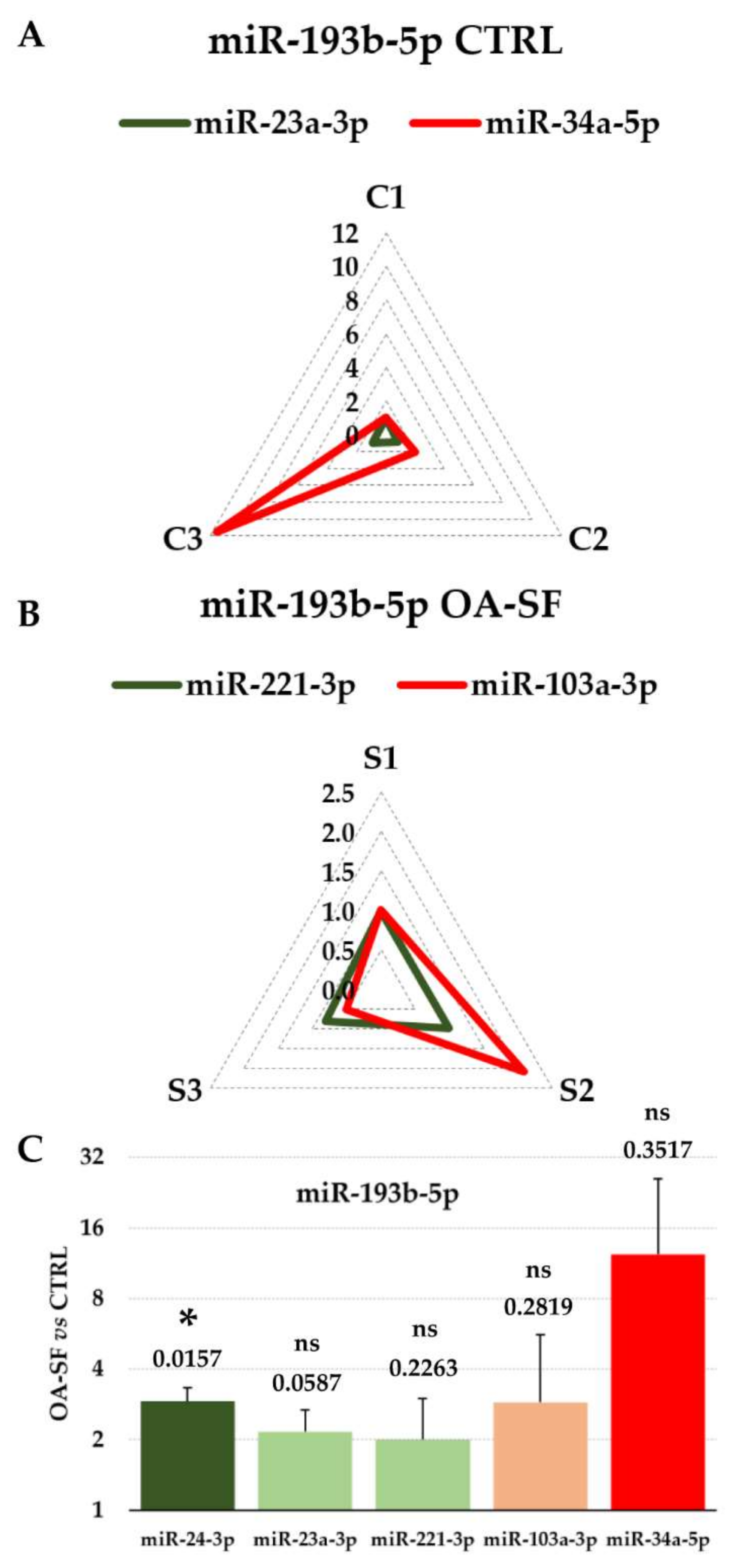
3.4. Impact of RGs Choice on the Quantification of Specific Target miRNAs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cui, A.; Li, H.; Wang, D.; Zhong, J.; Chen, Y.; Lu, H. Global, regional prevalence, incidence and risk factors of knee osteoarthritis in population-based studies. EClinicalMedicine 2020, 29-30, 29–30. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Shen, J.; Zhao, W.; Wang, T.; Han, L.; Hamilton, J.L.; Im, H.-J. Osteoarthritis: Toward a comprehensive understanding of pathological mechanism. Bone Res. 2017, 5, 16044. [Google Scholar] [CrossRef]
- Vos, T.; Allen, C.; Arora, M.; Barber, R.M.; Bhutta, Z.A.; Brown, A.; Carter, A.; Casey, D.C.; Charlson, F.J.; Chen, A.Z.; et al. Global, regional, and national incidence, prevalence, and years lived with disability for 310 diseases and injuries, 1990–2015: A systematic analysis for the Global Burden of Disease Study 2015. Lancet 2016, 388, 1545–1602. [Google Scholar] [CrossRef]
- Felson, D.T. Osteoarthritis of the Knee. N. Engl. J. Med. 2006, 354, 841–848. [Google Scholar] [CrossRef] [PubMed]
- Bannuru, R.R.; Osani, M.C.; Vaysbrot, E.E.; Arden, N.K.; Bennell, K.; Bierma-Zeinstra, S.M.A.; Kraus, V.B.; Lohmander, L.S.; Abbott, J.H.; Bhandari, M.; et al. OARSI guidelines for the non-surgical management of knee, hip, and polyarticular osteoarthritis. Osteoarthr. Cartil. 2019, 27, 1578–1589. [Google Scholar] [CrossRef] [PubMed]
- De Girolamo, L.; Kon, E.; Filardo, G.; Marmotti, A.G.; Soler, F.; Peretti, G.; Vannini, F.; Madry, H.; Chubinskaya, S. Regenerative approaches for the treatment of early OA. Knee Surg. Sports Traumatol. Arthrosc. 2016, 24, 1826–1835. [Google Scholar] [CrossRef]
- Cho, Y.; Jeong, S.; Kim, H.; Kang, D.; Lee, J.; Kang, S.-B.; Kim, J.-H. Disease-modifying therapeutic strategies in osteoarthritis: Current status and future directions. Exp. Mol. Med. 2021, 53, 1689–1696. [Google Scholar] [CrossRef]
- Han, Y.; Li, X.; Zhang, Y.; Han, Y.; Chang, F.; Ding, J. Mesenchymal Stem Cells for Regenerative Medicine. Cells 2019, 8, 886. [Google Scholar] [CrossRef] [PubMed]
- Weiss, A.R.R.; Dahlke, M.H. Immunomodulation by Mesenchymal Stem Cells (MSCs): Mechanisms of Action of Living, Apoptotic, and Dead MSCs. Front. Immunol. 2019, 10, 1191. [Google Scholar] [CrossRef] [PubMed]
- Pers, Y.-M.; Rackwitz, L.; Ferreira, R.; Pullig, O.; Delfour, C.; Barry, F.; Sensebe, L.; Casteilla, L.; Fleury, S.; Bourin, P.; et al. Adipose Mesenchymal Stromal Cell-Based Therapy for Severe Osteoarthritis of the Knee: A Phase I Dose-Escalation Trial. Stem Cells Transl. Med. 2016, 5, 847–856. [Google Scholar] [CrossRef]
- Emadedin, M.; Aghdami, N.; Taghiyar, L.; Fazeli, R.; Moghadasali, R.; Jahangir, S.; Farjad, R.; Eslaminejad, M.B. Intra-articular injection of autologous mesenchymal stem cells in six patients with knee osteoarthritis. Arch. Iran. Med. 2012, 15, 422–428. [Google Scholar] [PubMed]
- Jo, C.H.; Gil Lee, Y.; Shin, W.H.; Kim, H.; Chai, J.W.; Jeong, E.C.; Kim, J.E.; Shim, H.; Shin, J.S.; Shin, I.S.; et al. Intra-Articular Injection of Mesenchymal Stem Cells for the Treatment of Osteoarthritis of the Knee: A Proof-of-Concept Clinical Trial. Stem Cells 2014, 32, 1254–1266. [Google Scholar] [CrossRef] [PubMed]
- Berebichez-Fridman, R.; Montero-Olvera, P.R. Sources and Clinical Applications of Mesenchymal Stem Cells: State-of-the-art review. Sultan Qaboos Univ. Med. J. 2018, 18, e264–e277. [Google Scholar] [CrossRef]
- Afizah, H.; Hui, J.H.P. Mesenchymal stem cell therapy for osteoarthritis. J. Clin. Orthop. Trauma 2016, 7, 177–182. [Google Scholar] [CrossRef][Green Version]
- Jevotovsky, D.S.; Alfonso, A.R.; Einhorn, T.; Chiu, E.S. Osteoarthritis and stem cell therapy in humans: A systematic review. Osteoarthr. Cartil. 2018, 26, 711–729. [Google Scholar] [CrossRef]
- Cotter, E.J.; Wang, K.C.; Yanke, A.B.; Chubinskaya, S. Bone Marrow Aspirate Concentrate for Cartilage Defects of the Knee: From Bench to Bedside Evidence. Cartilage 2018, 9, 161–170. [Google Scholar] [CrossRef]
- Caplan, A.I. Mesenchymal Stem Cells: Time to Change the Name! Stem Cells Transl. Med. 2017, 6, 1445–1451. [Google Scholar] [CrossRef] [PubMed]
- Maas, S.L.N.; Breakefield, X.O.; Weaver, A.M. Extracellular Vesicles: Unique Intercellular Delivery Vehicles. Trends Cell Biol. 2017, 27, 172–188. [Google Scholar] [CrossRef]
- Qiu, G.; Zheng, G.; Ge, M.; Wang, J.; Huang, R.; Shu, Q.; Xu, J. Mesenchymal stem cell-derived extracellular vesicles affect disease outcomes via transfer of microRNAs. Stem Cell Res. Ther. 2018, 9, 320. [Google Scholar] [CrossRef]
- Malemud, C.J. MicroRNAs and Osteoarthritis. Cells 2018, 7, 92. [Google Scholar] [CrossRef]
- Miao, C.; Zhou, W.; Wang, X.; Fang, J. The Research Progress of Exosomes in Osteoarthritis, With Particular Emphasis on the Mediating Roles of miRNAs and lncRNAs. Front. Pharmacol. 2021, 12. [Google Scholar] [CrossRef] [PubMed]
- Toh, W.S.; Lai, R.C.; Hui, J.H.P.; Lim, S.K. MSC exosome as a cell-free MSC therapy for cartilage regeneration: Implications for osteoarthritis treatment. Semin. Cell Dev. Biol. 2017, 67, 56–64. [Google Scholar] [CrossRef]
- Chevillet, J.R.; Kang, Q.; Ruf, I.K.; Briggs, H.A.; Vojtech, L.; Hughes, S.; Cheng, H.H.; Arroyo, J.; Meredith, E.K.; Gallichotte, E.N.; et al. Quantitative and stoichiometric analysis of the microRNA content of exosomes. Proc. Natl. Acad. Sci. USA 2014, 111, 14888–14893. [Google Scholar] [CrossRef]
- Ragni, E.; Palombella, S.; Lopa, S.; Talò, G.; Orfei, C.P.; De Luca, P.; Moretti, M.; De Girolamo, L. Innovative Visualization and Quantification of Extracellular Vesicles Interaction with and Incorporation in Target Cells in 3D Microenvironments. Cells 2020, 9, 1180. [Google Scholar] [CrossRef] [PubMed]
- Ragni, E.; Orfei, C.P.; De Luca, P.; Mondadori, C.; Viganò, M.; Colombini, A.; De Girolamo, L. Inflammatory priming enhances mesenchymal stromal cell secretome potential as a clinical product for regenerative medicine approaches through secreted factors and EV-miRNAs: The example of joint disease. Stem Cell Res. Ther. 2020, 11, 1–19. [Google Scholar] [CrossRef]
- Ragni, E.; Colombini, A.; Viganò, M.; Libonati, F.; Orfei, C.P.; Zagra, L.; de Girolamo, L. Cartilage Protective and Immunomodulatory Features of Osteoarthritis Synovial Fluid-Treated Adipose-Derived Mesenchymal Stem Cells Secreted Factors and Extracellular Vesicles-Embedded miRNAs. Cells 2021, 10, 1072. [Google Scholar] [CrossRef]
- Zhang, Y.; Bi, J.; Huang, J.; Tang, Y.; Du, S.; Li, P. Exosome: A Review of Its Classification, Isolation Techniques, Storage, Diagnostic and Targeted Therapy Applications. Int. J. Nanomed. 2020, ume 15, 6917–6934. [Google Scholar] [CrossRef]
- Xia, X.; Wang, Y.; Huang, Y.; Zhang, H.; Lu, H.; Zheng, J.C. Exosomal miRNAs in central nervous system diseases: Biomarkers, pathological mediators, protective factors and therapeutic agents. Prog. Neurobiol. 2019, 183, 101694. [Google Scholar] [CrossRef] [PubMed]
- Bellingham, S.A.; Guo, B.B.; Coleman, B.M.; Hill, A.F. Exosomes: Vehicles for the Transfer of Toxic Proteins Associated with Neurodegenerative Diseases? Front. Physiol. 2012, 3, 124. [Google Scholar] [CrossRef]
- Ragni, E.; De Luca, P.; Orfei, C.P.; Colombini, A.; Viganò, M.; Lugano, G.; Bollati, V.; De Girolamo, L.; Luca, D.; Orfei, P.; et al. Insights into Inflammatory Priming of Adipose-Derived Mesenchymal Stem Cells: Validation of Extracellular Vesicles-Embedded miRNA Reference Genes as A Crucial Step for Donor Selection. Cells 2019, 8, 369. [Google Scholar] [CrossRef] [PubMed]
- Cavalleri, T.; Angelici, L.; Favero, C.; Dioni, L.; Mensi, C.; Bareggi, C.; Palleschi, A.; Rimessi, A.; Consonni, D.; Bordini, L.; et al. Plasmatic extracellular vesicle microRNAs in malignant pleural mesothelioma and asbestos-exposed subjects suggest a 2-miRNA signature as potential biomarker of disease. PLoS ONE 2017, 12, e0176680. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, L.; Liu, F.; Xiang, G.; Jiang, D.; Pu, X. Identification of Endogenous Controls for Analyzing Serum Exosomal miRNA in Patients with Hepatitis B or Hepatocellular Carcinoma. Dis. Markers 2015, 2015, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Xiang, G.-M.; Liu, L.-L.; Liu, C.; Liu, F.; Jiang, D.-N.; Pu, X.-Y. Assessment of endogenous reference gene suitability for serum exosomal microRNA expression analysis in liver carcinoma resection studies. Mol. Med. Rep. 2015, 12, 4683–4691. [Google Scholar] [CrossRef] [PubMed]
- Cazzoli, R.; Buttitta, F.; DI Nicola, M.; Malatesta, S.; Marchetti, A.; Rom, W.; Pass, H.I. microRNAs Derived from Circulating Exosomes as Noninvasive Biomarkers for Screening and Diagnosing Lung Cancer. J. Thorac. Oncol. 2013, 8, 1156–1162. [Google Scholar] [CrossRef] [PubMed]
- Ge, Q.; Zhou, Y.; Lu, J.; Bai, Y.; Xie, X.; Lu, Z. miRNA in Plasma Exosome is Stable under Different Storage Conditions. Molecules 2014, 19, 1568–1575. [Google Scholar] [CrossRef]
- Lange, T.; Stracke, S.; Rettig, R.; Lendeckel, U.; Kuhn, J.; Schlüter, R.; Rippe, V.; Endlich, K.; Endlich, N. Identification of miR-16 as an endogenous reference gene for the normalization of urinary exosomal miRNA expression data from CKD patients. PLoS ONE 2017, 12, e0183435. [Google Scholar] [CrossRef]
- Ragni, E.; Perucca Orfei, C.; De Luca, P.; Colombini, A.; Viganò, M.; Lugano, G.; Bollati, V.; De Girolamo, L. Identification of miRNA Reference Genes in Extracellular Vesicles from Adipose Derived Mesenchymal Stem Cells for Studying Osteoarthritis. Int. J. Mol. Sci. 2019, 20, 1108. [Google Scholar] [CrossRef]
- Ragni, E.; Orfei, C.P.; De Luca, P.; Viganò, M.; Colombini, A.; Lugano, G.; Bollati, V.; De Girolamo, L. miR-22-5p and miR-29a-5p Are Reliable Reference Genes for Analyzing Extracellular Vesicle-Associated miRNAs in Adipose-Derived Mesenchymal Stem Cells and Are Stable under Inflammatory Priming Mimicking Osteoarthritis Condition. Stem Cell Rev. Rep. 2019, 15, 743–754. [Google Scholar] [CrossRef]
- Ragni, E.; Orfei, C.P.; Silini, A.R.; Colombini, A.; Viganò, M.; Parolini, O.; De Girolamo, L. miRNA Reference Genes in Extracellular Vesicles Released from Amniotic Membrane-Derived Mesenchymal Stromal Cells. Pharmaceutics 2020, 12, 347. [Google Scholar] [CrossRef]
- Gouin, K.; Peck, K.; Antes, T.; Johnson, J.L.; Li, C.; Vaturi, S.D.; Middleton, R.; de Couto, G.; Walravens, A.S.; Rodriguez-Borlado, L.; et al. A comprehensive method for identification of suitable reference genes in extracellular vesicles. J. Extracell. Vesicles 2017, 6, 1347019. [Google Scholar] [CrossRef]
- Ragni, E.; Orfei, C.P.; Papait, A.; de Girolamo, L. Comparison of miRNA cargo in human adipose-tissue vs. amniotic-membrane derived mesenchymal stromal cells extracellular vesicles for osteoarthritis treatment. Extracell. Vesicles Circ. Nucleic Acids 2021, 2, 202–221. [Google Scholar] [CrossRef]
- Santovito, D.; De Nardis, V.; Marcantonio, P.; Mandolini, C.; Paganelli, C.; Vitale, E.; Buttitta, F.; Bucci, M.; Mezzetti, A.; Consoli, A.; et al. Plasma Exosome MicroRNA Profiling Unravels a New Potential Modulator of Adiponectin Pathway in Diabetes: Effect of Glycemic Control. J. Clin. Endocrinol. Metab. 2014, 99, E1681–E1685. [Google Scholar] [CrossRef]
- Lv, C.Y.; Yang, T.L. Effective enrichment of urinary exosomes by polyethylene glycol for RNA detection. Biomed. Res. 2018, 29, 29. [Google Scholar] [CrossRef]
- Zhang, C.; Zhang, Z.; Chang, Z.; Mao, G.; Hu, S.; Zeng, A.; Fu, M. miR-193b-5p regulates chondrocytes metabolism by directly targeting histone deacetylase 7 in interleukin-1β-induced osteoarthritis. J. Cell. Biochem. 2019, 120, 12775–12784. [Google Scholar] [CrossRef]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, research0034. [Google Scholar] [CrossRef] [PubMed]
- Andersen, C.L.; Jensen, J.L.; Ørntoft, T.F. Normalization of Real-Time Quantitative Reverse Transcription-PCR Data: A Model-Based Variance Estimation Approach to Identify Genes Suited for Normalization, Applied to Bladder and Colon Cancer Data Sets. Cancer Res. 2004, 64, 5245–5250. [Google Scholar] [CrossRef]
- Pfaffl, M.W.; Tichopad, A.; Prgomet, C.; Neuvians, T.P. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper – Excel-based tool using pair-wise correlations. Biotechnol. Lett. 2004, 26, 509–515. [Google Scholar] [CrossRef]
- Silver, N.; Best, S.; Jiang, J.; Thein, S.L. Selection of housekeeping genes for gene expression studies in human reticulocytes using real-time PCR. BMC Mol. Biol. 2006, 7, 33. [Google Scholar] [CrossRef] [PubMed]
- Xie, F.; Xiao, P.; Chen, D.; Xu, L.; Zhang, B. miRDeepFinder: A miRNA analysis tool for deep sequencing of plant small RNAs. Plant Mol. Biol. 2012, 80, 75–84. [Google Scholar] [CrossRef]
- Metsalu, T.; Vilo, J. ClustVis: A web tool for visualizing clustering of multivariate data using Principal Component Analysis and heatmap. Nucleic Acids Res. 2015, 43, W566–W570. [Google Scholar] [CrossRef] [PubMed]
- Dominici, M.; Le Blanc, K.; Mueller, I.; Slaper-Cortenbach, I.; Marini, F.C.; Krause, D.S.; Deans, R.J.; Keating, A.; Prockop, D.J.; Horwitz, E.M. Minimal criteria for defining multipotent mesenchymal stromal cells. The International Society for Cellular Therapy position statement. Cytotherapy 2006, 8, 315–317. [Google Scholar] [CrossRef] [PubMed]
- Barilani, M.; Banfi, F.; Sironi, S.; Ragni, E.; Guillaumin, S.; Polveraccio, F.; Rosso, L.; Moro, M.; Astori, G.; Pozzobon, M.; et al. Low-affinity Nerve Growth Factor Receptor (CD271) Heterogeneous Expression in Adult and Fetal Mesenchymal Stromal Cells. Sci. Rep. 2018, 8, 1–11. [Google Scholar] [CrossRef]
- Barilani, M.; Peli, V.; Cherubini, A.; Dossena, M.; Dolo, V.; Lazzari, L. NG2 as an Identity and Quality Marker of Mesenchymal Stem Cell Extracellular Vesicles. Cells 2019, 8, 1524. [Google Scholar] [CrossRef]
- Ramos, T.L.; Sánchez-Abarca, L.I.; Muntión, S.; Preciado, S.; Puig, N.; López-Ruano, G.; Hernández-Hernández, Á.; Redondo, A.; Ortega, R.; Rodríguez, C.; et al. MSC surface markers (CD44, CD73, and CD90) can identify human MSC-derived extracellular vesicles by conventional flow cytometry. Cell Commun. Signal. 2016, 14, 2. [Google Scholar] [CrossRef] [PubMed]
- Megraw, M.; Sethupathy, P.; Corda, B.; Hatzigeorgiou, A.G. miRGen: A database for the study of animal microRNA genomic organization and function. Nucleic Acids Res. 2007, 35, D149–D155. [Google Scholar] [CrossRef] [PubMed]
- Ragni, E.; Montemurro, T.; Montelatici, E.; Lavazza, C.; Viganò, M.; Rebulla, P.; Giordano, R.; Lazzari, L. Differential microRNA signature of human mesenchymal stem cells from different sources reveals an “environmental-niche memory” for bone marrow stem cells. Exp. Cell Res. 2013, 319, 1562–1574. [Google Scholar] [CrossRef]
- Ni, Z.; Zhou, S.; Li, S.; Kuang, L.; Chen, H.; Luo, X.; Ouyang, J.; He, M.; Du, X.; Chen, L. Exosomes: Roles and therapeutic potential in osteoarthritis. Bone Res. 2020, 8, 25. [Google Scholar] [CrossRef]
- Sánchez, G.B.; Bunn, K.E.; Pua, H.H.; Rafat, M. Extracellular vesicles: Mediators of intercellular communication in tissue injury and disease. Cell Commun. Signal. 2021, 19, 1–18. [Google Scholar] [CrossRef]
- Wiklander, O.P.B.; Brennan, M.Á.; Lötvall, J.; Breakefield, X.O.; EL Andaloussi, S. Advances in therapeutic applications of extracellular vesicles. Sci. Transl. Med. 2019, 11, 492. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, K.; Breyne, K.; Ughetto, S.; Laurent, L.C.; Breakefield, X.O. RNA delivery by extracellular vesicles in mammalian cells and its applications. Nat. Rev. Mol. Cell Biol. 2020, 21, 585–606. [Google Scholar] [CrossRef]
- Mao, G.; Zhang, Z.; Hu, S.; Zhang, Z.; Chang, Z.; Huang, Z.; Liao, W.; Kang, Y. Exosomes derived from miR-92a-3p-overexpressing human mesenchymal stem cells enhance chondrogenesis and suppress cartilage degradation via targeting WNT5A. Stem Cell Res. Ther. 2018, 9, 247. [Google Scholar] [CrossRef]
- Tao, S.-C.; Yuan, T.; Zhang, Y.-L.; Yin, W.-J.; Guo, S.-C.; Zhang, C.-Q. Exosomes derived from miR-140-5p-overexpressing human synovial mesenchymal stem cells enhance cartilage tissue regeneration and prevent osteoarthritis of the knee in a rat model. Theranostics 2017, 7, 180–195. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Hu, S.; Zhang, Z.; Lun, J.; Liao, W.; Zhang, Z. Expression of exosomal microRNAs during chondrogenic differentiation of human bone mesenchymal stem cells. J. Cell. Biochem. 2019, 120, 171–181. [Google Scholar] [CrossRef]
- De Abreu, R.C.; Ramos, C.V.; Becher, C.; Lino, M.; Jesus, C.; Martins, P.A.D.C.; Martins, P.A.T.; Moreno, M.J.; Fernandes, H.; Ferreira, L. Exogenous loading of miRNAs into small extracellular vesicles. J. Extracell. Vesicles 2021, 10, e12111. [Google Scholar] [CrossRef] [PubMed]
- Mestdagh, P.; Van Vlierberghe, P.; De Weer, A.; Muth, D.; Westermann, F.; Speleman, F.; Vandesompele, J. A novel and universal method for microRNA RT-qPCR data normalization. Genome Biol. 2009, 10, R64. [Google Scholar] [CrossRef] [PubMed]
- De Cássia Noronha, N.; Mizukami, A.; Caliári-Oliveira, C.; Cominal, J.G.; Rocha, J.L.M.; Covas, D.T.; Swiech, K.; Malmegrim, K.C.R. Priming approaches to improve the efficacy of mesenchymal stromal cell-based therapies. Stem Cell Res. Ther. 2019, 10, 131. [Google Scholar] [CrossRef]
- Lee, Y.; Ahn, C.; Han, J.; Choi, H.; Kim, J.; Yim, J.; Lee, J.; Provost, P.; Rådmark, O.; Kim, S.; et al. The nuclear RNase III Drosha initiates microRNA processing. Nature 2003, 425, 415–419. [Google Scholar] [CrossRef] [PubMed]
- Fu, S.; Wang, Y.; Xia, X.; Zheng, J.C. Exosome engineering: Current progress in cargo loading and targeted delivery. NanoImpact 2020, 20, 100261. [Google Scholar] [CrossRef]
- Ragni, E.; Banfi, F.; Barilani, M.; Cherubini, A.; Parazzi, V.; Larghi, P.; Dolo, V.; Bollati, V.; Lazzari, L. Extracellular Vesicle-Shuttled mRNA in Mesenchymal Stem Cell Communication. Stem Cells 2017, 35, 1093–1105. [Google Scholar] [CrossRef]
- Zheng, X.; Zhao, F.-C.; Pang, Y.; Li, D.-Y.; Yao, S.-C.; Sun, S.-S.; Guo, K.-J. Downregulation of miR-221-3p contributes to IL-1β-induced cartilage degradation by directly targeting the SDF1/CXCR4 signaling pathway. Klin. Wochenschr. 2017, 95, 615–627. [Google Scholar] [CrossRef]
- Xu, J.; Liu, Y.; Deng, M.; Li, J.; Cai, H.; Meng, Q.; Fang, W.; Long, X.; Ke, J. MicroRNA221-3p modulates Ets-1 expression in synovial fibroblasts from patients with osteoarthritis of temporomandibular joint. Osteoarthr. Cartil. 2016, 24, 2003–2011. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Qian, X.; Ding, R. MiR-24-3p attenuates IL-1β-induced chondrocyte injury associated with osteoarthritis by targeting BCL2L12. J. Orthop. Surg. Res. 2021, 16, 1–10. [Google Scholar] [CrossRef]
- Naqvi, J.B.F.A.R.; Nares, S.; Fordham, J.B. miR-24 Regulates Macrophage Polarization and Plasticity. J. Clin. Cell. Immunol. 2015, 6, 1–9. [Google Scholar] [CrossRef]
- Li, L.; Jia, J.; Liu, X.; Yang, S.; Ye, S.; Yang, W.; Zhang, Y. MicroRNA-16-5p Controls Development of Osteoarthritis by Targeting SMAD3 in Chondrocytes. Curr. Pharm. Des. 2015, 21, 5160–5167. [Google Scholar] [CrossRef] [PubMed]
| Accession Number | Gene Name | Target Sequence (5′-3′) | Reference |
|---|---|---|---|
| MIMAT0000062 | let-7a-5p | UGAGGUAGUAGGUUGUAUAGUU | [32,33,34] |
| MIMAT0000069 | miR-16-5p | UAGCAGCACGUAAAUAUUGGCG | [30,35,36,37] |
| MIMAT0004495 | miR-22-5p | AGUUCUUCAGUGGCAAGCUUUA | [38,39] |
| MIMAT0000078 | miR-23a-3p | AUCACAUUGCCAGGGAUUUCC | [37,40] |
| MIMAT0000080 | miR-24-3p | UGGCUCAGUUCAGCAGGAACAG | [41] |
| MIMAT0000082 | miR-26a-5p | UUCAAGUAAUCCAGGAUAGGCU | [30,33,40] |
| MIMAT0004503 | miR-29a-5p | ACUGAUUUCUUUUGGUGUUCAG | [38] |
| MIMAT0000255 | miR-34a-5p | UGGCAGUGUCUUAGCUGGUUGU | [41] |
| MIMAT0000099 | miR-101-3p | UACAGUACUGUGAUAACUGAA | [39,40] |
| MIMAT0000101 | miR-103a-3p | AGCAGCAUUGUACAGGGCUAUGA | [32] |
| MIMAT0000278 | miR-221-3p | AGCUACAUUGUCUGCUGGGUUUC | [32,33] |
| MIMAT0004748 | miR-423-5p | UGAGGGGCAGAGAGCGAGACUUU | [42] |
| MIMAT0003393 | miR-425-5p | AAUGACACGAUCACUCCCGUUGA | [42] |
| MIMAT0003338 | miR-660-5p | UACCCAUUGCAUAUCGGAGUUG | [38] |
| NR_004394.1 | U6 snRNA | GUGCUCGCUUCGGCAGCACAUAUACUAAAAU UGGAACGATACAGAGAAGAUUAGCAUGGCCC CUGCGCAAGGAUGACACGCAAAUUCGUGAAG CGUUCCAUAUUUU | [43] |
| miRNA target | |||
| MIMAT0004767 | miR-193b-5p | CGGGGUUUUGAGGGCGAGAUGA | [44] |
| (A) CTRL | ||||||||||
| Ranking Order | Gene Name | Geomean | Genorm M-Value | Normfinder SV | BestKeeper SD | Delta CT SD | ||||
| 1 | miR-23a-3p | 1.19 | 0.074 | (1) | 0.037 | (1) | 0.07 | (2) | 0.64 | (1) |
| 2 | miR-425-5p | 1.57 | 0.074 | (1) | 0.037 | (2) | 0.05 | (1) | 0.65 | (3) |
| 3 | miR-660-5p | 3.98 | 0.122 | (3) | 0.055 | (3) | 0.10 | (3) | 0.69 | (7) |
| 4 | U6 snRNA | 4.16 | 0.156 | (4) | 0.106 | (5) | 0.10 | (4) | 0.66 | (5) |
| 5 | miR-16-5p | 4.36 | 0.219 | (5) | 0.203 | (6) | 0.19 | (6) | 0.64 | (2) |
| 6 | miR-423-5p | 5.38 | 0.250 | (7) | 0.081 | (4) | 0.16 | (5) | 0.67 | (6) |
| 7 | miR-24-3p | 5.86 | 0.235 | (6) | 0.246 | (7) | 0.20 | (7) | 0.65 | (4) |
| 8 | miR-26a-5p | 8.00 | 0.303 | (8) | 0.501 | (8) | 0.38 | (8) | 0.77 | (8) |
| 9 | miR-221-3p | 9.00 | 0.402 | (9) | 0.935 | (9) | 0.65 | (9) | 1.02 | (9) |
| 10 | miR-103a-3p | 10.00 | 0.485 | (10) | 1.109 | (10) | 0.75 | (10) | 1.15 | (10) |
| 11 | let-7a-5p | 11.00 | 0.719 | (11) | 1.568 | (11) | 1.10 | (11) | 1.68 | (11) |
| 12 | miR-34a-5p | 12.00 | 0.940 | (12) | 2.006 | (12) | 1.41 | (12) | 2.05 | (12) |
| (B) OA-SF | ||||||||||
| Ranking Order | Gene Name | Geomean | Genorm M-Value | Normfinder SV | BestKeeper SD | Delta CT SD | ||||
| 1 | miR-221-3p | 1.00 | 0.049 | (1) | 0.025 | (1) | 0.03 | (1) | 0.41 | (1) |
| 2 | U6 snRNA | 2.06 | 0.049 | (1) | 0.101 | (3) | 0.07 | (2) | 0.42 | (3) |
| 3 | miR-16-5p | 2.45 | 0.116 | (3) | 0.063 | (2) | 0.08 | (3) | 0.42 | (2) |
| 4 | miR-24-3p | 4.00 | 0.139 | (4) | 0.169 | (4) | 0.15 | (4) | 0.44 | (4) |
| 5 | miR-660-5p | 5.23 | 0.183 | (5) | 0.356 | (6) | 0.22 | (5) | 0.51 | (5) |
| 6 | miR-26a-5p | 5.96 | 0.289 | (7) | 0.277 | (5) | 0.27 | (6) | 0.52 | (6) |
| 7 | miR-423-5p | 6.74 | 0.233 | (6) | 0.51 | (7) | 0.32 | (7) | 0.61 | (7) |
| 8 | miR-23a-3p | 8.00 | 0.350 | (8) | 0.541 | (8) | 0.34 | (8) | 0.65 | (8) |
| 9 | let-7a-5p | 9.49 | 0.474 | (10) | 0.55 | (9) | 0.44 | (10) | 0.70 | (9) |
| 10 | miR-425-5p | 9.49 | 0.417 | (9) | 0.602 | (10) | 0.42 | (9) | 0.72 | (10) |
| 11 | miR-34a-5p | 11.00 | 0.528 | (11) | 0.677 | (11) | 0.47 | (11) | 0.77 | (11) |
| 12 | miR-103a-3p | 12.00 | 0.590 | (12) | 0.831 | (12) | 0.59 | (12) | 0.90 | (12) |
| (C) ALL | ||||||||||
| Ranking Order | Gene Name | Geomean | Genorm M-Value | Normfinder SV | BestKeeper SD | Delta CT SD | ||||
| 1 | miR-24-3p | 1.00 | 0.135 | (1) | 0.213 | (1) | 0.18 | (1) | 0.64 | (1) |
| 2 | miR-16-5p | 1.68 | 0.135 | (1) | 0.263 | (2) | 0.20 | (2) | 0.66 | (2) |
| 3 | miR-660-5p | 3.22 | 0.300 | (4) | 0.291 | (3) | 0.20 | (3) | 0.70 | (3) |
| 4 | miR-423-5p | 4.47 | 0.368 | (5) | 0.296 | (4) | 0.24 | (4) | 0.70 | (4) |
| 5 | miR-26a-5p | 5.45 | 0.218 | (3) | 0.365 | (7) | 0.33 | (7) | 0.71 | (6) |
| 6 | miR-23a-3p | 5.48 | 0.408 | (6) | 0.306 | (5) | 0.28 | (6) | 0.70 | (5) |
| 7 | miR-425-5p | 5.86 | 0.433 | (7) | 0.354 | (6) | 0.24 | (5) | 0.74 | (7) |
| 8 | miR-221-3p | 8.00 | 0.484 | (8) | 0.643 | (8) | 0.42 | (8) | 0.84 | (8) |
| 9 | U6 snRNA | 9.24 | 0.557 | (9) | 0.766 | (9) | 0.71 | (10) | 0.97 | (9) |
| 10 | miR-103a-3p | 9.74 | 0.617 | (10) | 0.881 | (10) | 0.67 | (9) | 1.03 | (10) |
| 11 | let-7a-5p | 11.00 | 0.731 | (11) | 1.054 | (11) | 0.83 | (11) | 1.23 | (11) |
| 12 | miR-34a-5p | 12.00 | 0.876 | (12) | 1.521 | (12) | 1.12 | (12) | 1.60 | (12) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ragni, E.; Perucca Orfei, C.; Viganò, M.; Valli, F.; de Girolamo, L. Endogenous Controls for the Evaluation of Osteoarthritis-Related miRNAs in Extracellular Vesicles from Bone-Marrow-Derived Mesenchymal Stromal Cells and the Impact of Osteoarthritis Synovial Fluid. Biomolecules 2022, 12, 316. https://doi.org/10.3390/biom12020316
Ragni E, Perucca Orfei C, Viganò M, Valli F, de Girolamo L. Endogenous Controls for the Evaluation of Osteoarthritis-Related miRNAs in Extracellular Vesicles from Bone-Marrow-Derived Mesenchymal Stromal Cells and the Impact of Osteoarthritis Synovial Fluid. Biomolecules. 2022; 12(2):316. https://doi.org/10.3390/biom12020316
Chicago/Turabian StyleRagni, Enrico, Carlotta Perucca Orfei, Marco Viganò, Federico Valli, and Laura de Girolamo. 2022. "Endogenous Controls for the Evaluation of Osteoarthritis-Related miRNAs in Extracellular Vesicles from Bone-Marrow-Derived Mesenchymal Stromal Cells and the Impact of Osteoarthritis Synovial Fluid" Biomolecules 12, no. 2: 316. https://doi.org/10.3390/biom12020316
APA StyleRagni, E., Perucca Orfei, C., Viganò, M., Valli, F., & de Girolamo, L. (2022). Endogenous Controls for the Evaluation of Osteoarthritis-Related miRNAs in Extracellular Vesicles from Bone-Marrow-Derived Mesenchymal Stromal Cells and the Impact of Osteoarthritis Synovial Fluid. Biomolecules, 12(2), 316. https://doi.org/10.3390/biom12020316








