Searching for the Metabolic Signature of Cancer: A Review from Warburg’s Time to Now
Abstract
1. Introduction on Metabolism
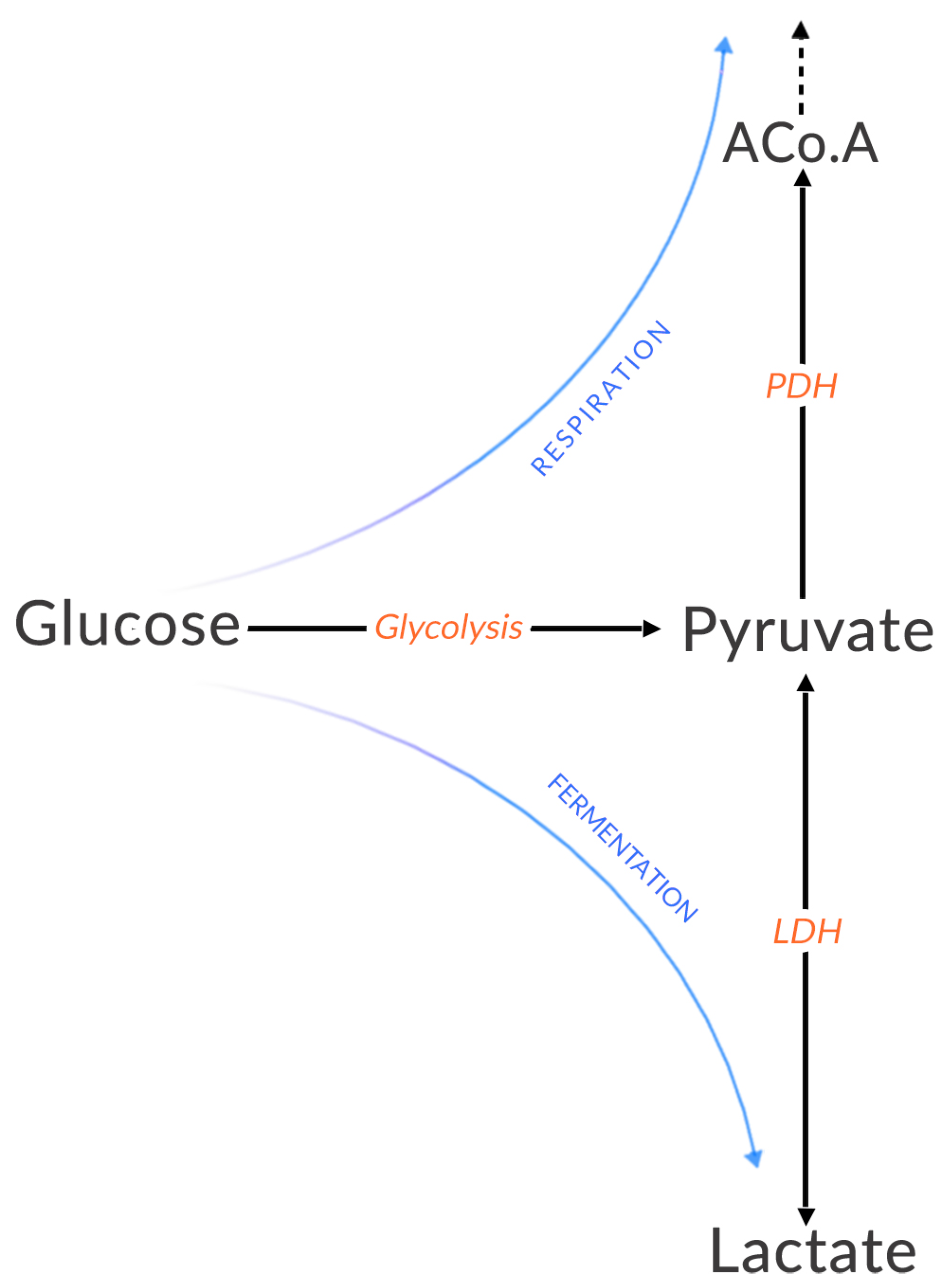
1.1. Energy Production Pathways
1.2. Delineations of Metabolic Pathways
2. The Tumor Singularity
2.1. General Characteristics of Cancers
- independence from proliferation signals,
- insensitivity to anti-growth signals,
- escape from programmed cell death,
- unlimited replication potential,
- persistent angiogenesis,
- tissue invasion and metastasis,
- evasion of the immune system,
- inflammation,
- genetic instability,
- dysregulated metabolism.
2.2. About Genetic Mutations
“Two decades from now, having fully charted the wiring diagrams of every cellular signaling pathway, it will be possible to lay out the complete “integrated circuit of the cell” upon its current outline. We will then be able to apply the tools of mathematical modeling to explain how specific genetic lesions serve to reprogram this integrated circuit in each of the constituent cell types so as to manifest cancer.”
2.3. The Warburg Effect
2.4. The Concept of Metabolic Switch
2.5. Is Warburg’s Phenotype Universal?
3. Differences around the Concepts Associated with the Warburg Effect between Its Discovery and Now
3.1. Warburg’s Observations in 1956
- Healthy cells make little use of fermentation in the presence of oxygen because ATP is sufficiently produced by respiration [55].
- If lactate production is high in the absence of oxygen, this is normal since the cell must produce ATP to survive.
- If lactate production is high and oxygen is available, it is abnormal.
- Cells favor a particular mode of energy production.
- The reprogramming of energy metabolism is a dysregulation [56].
“I shall not consider aerobic fermentation, which is a result of the interaction of respiration and fermentation, because aerobic fermentation is too labile and too dependent on external conditions. Of importance for the considerations that follow are only the two stable independent metabolic processes, respiration and anaerobic fermentation-respiration, which is measured by the oxygen consumption of cells that are saturated with oxygen, and fermentation, which is measured by the formation of lactic acid in the absence of oxygen.”
“The mysterious latency period of the production of cancer is, therefore, nothing more than the time in which the fermentation increases after a damaging of the respiration. This time differs in various animals; it is especially long in man and here often amounts to several decades, as can be determined in the cases in which the time of the respiratory damage is known for example, in arsenic cancer and irradiation cancer....Since the increase in fermentation in the development of cancer cells takes place gradually, there must be a transitional phase between normal body cells and fully formed cancer cells.”
“The Warburg effect is instead a crucial component of the malignant phenotype and a central feature of the ‘selfish’ metabolic reprogramming of cancer cells which is considered a “hallmark of cancer” (Hanahan & Weinberg, 2011). The switch to aerobic glycolysis (i.e., the conversion of glucose to pyruvate) followed by lactate formation is acquired very early in carcinogenesis (oncogenesis), even before tumor cells are exposed to hypoxic conditions” (Vander Heiden et al. 2009).
3.2. “Aerobic Glycolysis”
3.3. The Reverse Warburg Effect and the Questioning of a Universal Phenotype
4. Metabolic Landscape
4.1. On the Importance of Heterogeneity
4.2. Transient States, Stationary States and Equilibrium States
4.3. Homeostasis and Reachability
- the robustness of the cellular phenotype: i.e., the conservation of functions despite environmental variations, by multiple redundancies at the level of the genetic code, in the role of amino acids or metabolic pathways and checkpoints,
- the system evolution: the ability to mutate and authorize new behaviors/functionings of the biological system,
- the viability: all the environmental constraints of a physico-chemical, radiative, mechanical, social nature, etc. These constraints contribute to test and challenge/experience the phenotype.
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Rich, P.R. The molecular machinery of Keilin’s respiratory chain. Biochem. Soc. Trans. 2003, 31, 1095–1105. [Google Scholar] [CrossRef] [PubMed]
- Hinkle, P.C. P/O ratios of mitochondrial oxidative phosphorylation. Biochim. Biophys. Acta (BBA)-Bioenerg. 2005, 1706, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Nakhleh, L. Properties of metabolic graphs: Biological organization or representation artifacts? BMC Bioinform. 2011, 12, 132. [Google Scholar] [CrossRef] [PubMed]
- Beguerisse-Díaz, M.; Bosque, G.; Oyarzún, D.; Picó, J.; Barahona, M. Flux-dependent graphs for metabolic networks. NPJ Syst. Biol. Appl. 2018, 4, 32. [Google Scholar] [CrossRef] [PubMed]
- Larhlimi, A.; Blachon, S.; Selbig, J.; Nikoloski, Z. Robustness of metabolic networks: A review of existing definitions. Biosystems 2011, 106, 1–8. [Google Scholar] [CrossRef]
- Caspi, R.; Dreher, K.; Karp, P.D. The challenge of constructing, classifying, and representing metabolic pathways. FEMS Microbiol. Lett. 2013, 345, 85–93. [Google Scholar] [CrossRef]
- Jose, C.; Bellance, N.; Rossignol, R. Choosing between glycolysis and oxidative phosphorylation: A tumor’s dilemma? Biochim. Biophys. Acta (BBA)-Bioenerg. 2011, 1807, 552–561. [Google Scholar] [CrossRef]
- Zheng, J. Energy metabolism of cancer: Glycolysis versus oxidative phosphorylation (review). Oncol. Lett. 2012, 4, 1151–1157. [Google Scholar] [CrossRef]
- Basu, H.S.; Wilganowski, N.; Robertson, S.; Ramachandran, S.; Zurita-Saavedra, A.; Titus, M.; Cohen, E.; Reuben, J.; Wilding, G. Metabolic switch from glycolysis to oxidative phosphorylation (ox-phos) provides survival advantage to anti-androgen-treated prostate cancer cells and make them vulnerable to mitochondrial metabolism inhibitors IACS-010759 and CB-839. Cancer Res. 2020, 80, 4791. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. The Hallmarks of Cancer. Cell 2000, 100, 57–70. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed]
- Petrova, V.; Annicchiarico-Petruzzelli, M.; Melino, G.; Amelio, I. The hypoxic tumour microenvironment. Oncogenesis 2018, 7, 10. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhao, L.; Li, X.F. Hypoxia and the tumor microenvironment. Technol. Cancer Res. Treat. 2021, 20, 153303382110363. [Google Scholar] [CrossRef] [PubMed]
- Krock, B.L.; Skuli, N.; Simon, M.C. Hypoxia-Induced Angiogenesis: Good and Evil. Genes Cancer 2011, 2, 1117–1133. [Google Scholar] [CrossRef]
- Roy, S.; Kumaravel, S.; Sharma, A.; Duran, C.L.; Bayless, K.J.; Chakraborty, S. Hypoxic tumor microenvironment: Implications for cancer therapy. Exp. Biol. Med. 2020, 245, 1073–1086. [Google Scholar] [CrossRef]
- Zhu, G.; Pan, C.; Bei, J.X.; Li, B.; Liang, C.; Xu, Y.; Fu, X. Mutant p53 in Cancer Progression and Targeted Therapies. Front. Oncol. 2020, 10, 595187. [Google Scholar] [CrossRef]
- Mendiratta, G.; Ke, E.; Aziz, M.; Liarakos, D.; Tong, M.; Stites, E.C. Cancer gene mutation frequencies for the U.S. population. Nat. Commun. 2021, 12, 5961. [Google Scholar] [CrossRef]
- Chen, J. The Cell-Cycle Arrest and Apoptotic Functions of p53 in Tumor Initiation and Progression. Cold Spring Harb. Perspect. Med. 2016, 6, a026104. [Google Scholar] [CrossRef]
- Feroz, W.; Sheikh, A.M.A. Exploring the multiple roles of guardian of the genome: P53. Egypt. J. Med. Hum. Genet. 2020, 21, 49. [Google Scholar] [CrossRef]
- Olivier, M.; Hollstein, M.; Hainaut, P. TP53 Mutations in Human Cancers: Origins, Consequences, and Clinical Use. Cold Spring Harb. Perspect. Biol. 2009, 2, a001008. [Google Scholar] [CrossRef]
- Gerstung, M.; Jolly, C.; Leshchiner, I.; Dentro, S.C. The evolutionary history of 2658 cancers. Nature 2020, 578, 122–128. [Google Scholar] [CrossRef] [PubMed]
- Krewski, D.; Bird, M.; Al-Zoughool, M.; Birkett, N.; Billard, M.; Milton, B.; Rice, J.M.; Grosse, Y.; Cogliano, V.J.; Hill, M.A.; et al. Key characteristics of 86 agents known to cause cancer in humans. J. Toxicol. Environ. Health Part B 2019, 22, 244–263. [Google Scholar] [CrossRef] [PubMed]
- Poschl, G. Alcohol and Cancer. Alcohol Alcohol. 2004, 39, 155–165. [Google Scholar] [CrossRef] [PubMed]
- Cannataro, V.L.; Mandell, J.D.; Townsend, J.P. Attribution of Cancer Origins to Endogenous, Exogenous, and Preventable Mutational Processes. Mol. Biol. Evol. 2022, 39. [Google Scholar] [CrossRef] [PubMed]
- Ewald, P.W.; Ewald, H.A.S. Infection, mutation, and cancer evolution. J. Mol. Med. 2012, 90, 535–541. [Google Scholar] [CrossRef] [PubMed]
- Hasin, Y.; Seldin, M.; Lusis, A. Multi-omics approaches to disease. Genome Biol. 2017, 18, 155–165. [Google Scholar] [CrossRef]
- Subramanian, I.; Verma, S.; Kumar, S.; Jere, A.; Anamika, K. Multi-omics Data Integration, Interpretation, and Its Application. Bioinform. Biol. Insights 2020, 14, 117793221989905. [Google Scholar] [CrossRef]
- Potter, M.; Newport, E.; Morten, K.J. The Warburg effect: 80 years on. Biochem. Soc. Trans. 2016, 44, 1499–1505. [Google Scholar] [CrossRef]
- Warburg, O. On the Origin of Cancer Cells. Science 1956, 123, 309–314. [Google Scholar] [CrossRef]
- Senyilmaz, D.; Teleman, A.A. Chicken or the egg: Warburg effect and mitochondrial dysfunction. F1000Prime Rep. 2015, 7, 41. [Google Scholar] [CrossRef]
- Medina, M.A. Metabolic Reprogramming is a Hallmark of Metabolism Itself. BioEssays 2020, 42, 2000058. [Google Scholar] [CrossRef]
- Chen, J.; Cao, X.; Li, B.; Zhao, Z.; Chen, S.; Lai, S.W.T.; Muend, S.A.; Nossa, G.K.; Wang, L.; Guo, W.; et al. Warburg Effect Is a Cancer Immune Evasion Mechanism against Macrophage Immunosurveillance. Front. Immunol. 2021, 11, 621757. [Google Scholar] [CrossRef] [PubMed]
- Liberti, M.V.; Locasale, J.W.; Biology, C.C.; Biology, C.C. The Warburg Effect: How Does it Benefit Cancer Cells? Trends Biochem. Sci. 2016, 41, 211–218. [Google Scholar] [CrossRef] [PubMed]
- Xie, J.; Wu, H.; Dai, C.; Pan, Q.; Ding, Z.; Hu, D.; Ji, B.; Luo, Y.; Hu, X. Beyond Warburg effect—Dual metabolic nature of cancer cells. Sci. Rep. 2014, 4, 4927. [Google Scholar] [CrossRef]
- Frezza, C. Metabolism and cancer: The future is now. Br. J. Cancer 2020, 122, 133–135. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.T.; Huang, C.H.; Wei, Y.Q. The metabolic switch and its regulation in cancer cells. Sci. China Life Sci. 2010, 53, 942–958. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Chen, X.; Sun, X.; Wang, L.; Chen, S. The Glycolytic Switch in Tumors: How Many Players Are Involved? J. Cancer 2017, 8, 3430–3440. [Google Scholar] [CrossRef]
- DeBerardinis, R.J.; Chandel, N.S. We need to talk about the Warburg effect. Nat. Metab. 2020, 2, 127–129. [Google Scholar] [CrossRef]
- Murphy, M.P.; O’Neill, L.A.J. How should we talk about metabolism? Nat. Immunol. 2020, 21, 713–715. [Google Scholar] [CrossRef]
- Xiao, Z.; Dai, Z.; Locasale, J.W. Metabolic landscape of the tumor microenvironment at single cell resolution. Nat. Commun. 2019, 10, 3763. [Google Scholar] [CrossRef]
- Almuhaideb, A.; Papathanasiou, N.; Bomanji, J. F-FDG PET/CT Imaging In Oncology. Ann. Saudi Med. 2011, 31, 3–13. [Google Scholar] [CrossRef]
- Faubert, B.; Solmonson, A.; DeBerardinis, R.J. Metabolic reprogramming and cancer progression. Science 2020, 368, eaaw5473. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, G.J. Metabolic reprogramming: The emerging concept and associated therapeutic strategies. J. Exp. Clin. Cancer Res. 2015, 34, 111. [Google Scholar] [CrossRef] [PubMed]
- Dienel, G.A.; Cruz, N.F. Aerobic glycolysis during brain activation: Adrenergic regulation and influence of norepinephrine on astrocytic metabolism. J. Neurochem. 2016, 138, 14–52. [Google Scholar] [CrossRef] [PubMed]
- Jones, W.; Bianchi, K. Aerobic glycolysis: Beyond proliferation. Front. Immunol. 2015, 6, 227. [Google Scholar] [CrossRef] [PubMed]
- Pacheco, M.P.; Bintener, T.; Ternes, D.; Kulms, D.; Haan, S.; Letellier, E.; Sauter, T. Identifying and targeting cancer-specific metabolism with network-based drug target prediction. EBioMedicine 2019, 43, 98–106. [Google Scholar] [CrossRef]
- Persi, E.; Duran-Frigola, M.; Damaghi, M.; Roush, W.R.; Aloy, P.; Cleveland, J.L.; Gillies, R.J.; Ruppin, E. Systems analysis of intracellular pH vulnerabilities for cancer therapy. Nat. Commun. 2018, 9, 2997. [Google Scholar] [CrossRef]
- Damaghi, M.; Wojtkowiak, J.W.; Gillies, R.J. pH sensing and regulation in cancer. Front. Physiol. 2013, 4, 370. [Google Scholar] [CrossRef]
- White, K.A.; Grillo-Hill, B.K.; Barber, D.L. Cancer cell behaviors mediated by dysregulated pH dynamics at a glance. J. Cell Sci. 2017, 130, 663–669. [Google Scholar] [CrossRef]
- Luo, X.; Yang, H.; Wang, H.; Ye, Z.; Zhou, Z.; Gu, L.; Chen, J.; Xiao, Y.; Liang, X.; Qian, X.; et al. Highly Sensitive Hill-Type Small-Molecule pH Probe That Recognizes the Reversed pH Gradient of Cancer Cells. Anal. Chem. 2018, 90, 5803–5809. [Google Scholar] [CrossRef]
- Jin, M.Z.; Jin, W.L. The updated landscape of tumor microenvironment and drug repurposing. Signal Transduct. Target. Ther. 2020, 5, 166. [Google Scholar] [CrossRef] [PubMed]
- Spencer, N.Y.; Stanton, R.C. The Warburg Effect, Lactate, and Nearly a Century of Trying to Cure Cancer. Semin. Nephrol. 2019, 39, 380–393. [Google Scholar] [CrossRef] [PubMed]
- Sies, H. Oxidative Stress Eustress and Distress; Elsevier Science and Technology: Amsterdam, The Netherlands, 2019. [Google Scholar]
- Cazzaniga, M.; Bonanni, B. Relationship between metabolic reprogramming and mitochondrial activity in cancer cells. Understanding the anticancer effect of metformin and its clinical implications. Anticancer Res. 2015, 35, 5789–5796. [Google Scholar] [PubMed]
- Kubicka, A.; Matczak, K.; Łabieniec Watała, M. More Than Meets the Eye Regarding Cancer Metabolism. Int. J. Mol. Sci. 2021, 22, 9507. [Google Scholar] [CrossRef] [PubMed]
- Robey, R.; Weisz, J.; Kuemmerle, N.B.; Salzberg, A.C.; Berg, A.; Brown, D.G.; Kubik, L.; Palorini, R.; Al-Mulla, F.; Al-Temaimi, R.; et al. Metabolic reprogramming and dysregulated metabolism: Cause, consequence and/or enabler of environmental carcinogenesis? Carcinogenesis 2015, 36, S203–S231. [Google Scholar] [CrossRef]
- Vaupel, P.; Multhoff, G. Revisiting the Warburg effect: Historical dogma versus current understanding. J. Physiol. 2021, 599, 1745–1757. [Google Scholar] [CrossRef]
- Bouchez, C.L.; Hammad, N.; Cuvellier, S.; Ransac, S.; Rigoulet, M.; Devin, A. The Warburg Effect in Yeast: Repression of Mitochondrial Metabolism Is Not a Prerequisite to Promote Cell Proliferation. Front. Oncol. 2020, 10, 1333. [Google Scholar] [CrossRef]
- Srinivasan, S.; Guha, M.; Avadhani, N.G. Mitochondrial respiratory defects promote the Warburg effect and cancer progression. Mol. Cell. Oncol. 2015, 3, e1085120. [Google Scholar] [CrossRef]
- Boland, M.L.; Chourasia, A.H.; Macleod, K.F. Mitochondrial Dysfunction in Cancer. Front. Oncol. 2013, 3, 292. [Google Scholar] [CrossRef]
- Jia, D.; Lu, M.; Jung, K.H.; Park, J.H.; Yu, L.; Onuchic, J.N.; Kaipparettu, B.A.; Levine, H. Elucidating cancer metabolic plasticity by coupling gene regulation with metabolic pathways. Proc. Natl. Acad. Sci. USA 2019, 116, 3909–3918. [Google Scholar] [CrossRef]
- Wang, D.; Green, M.F.; McDonnell, E.; Hirschey, M.D. Oxygen Flux Analysis to Understand the Biological Function of Sirtuins. In Sirtuins; Humana Press: Totowa, NJ, USA, 2013; pp. 241–258. [Google Scholar] [CrossRef]
- Muz, B.; de la Puente, P.; Azab, F.; Azab, A.K. The role of hypoxia in cancer progression, angiogenesis, metastasis, and resistance to therapy. Hypoxia 2015, 2015, 83–92. [Google Scholar] [CrossRef] [PubMed]
- Emami Nejad, A.; Najafgholian, S.; Rostami, A.; Sistani, A.; Shojaeifar, S.; Esparvarinha, M.; Nedaeinia, R.; Haghjooy Javanmard, S.; Taherian, M.; Ahmadlou, M.; et al. The role of hypoxia in the tumor microenvironment and development of cancer stem cell: A novel approach to developing treatment. Cancer Cell Int. 2021, 21, 62. [Google Scholar] [CrossRef]
- Cassim, S.; Vučetić, M.; Ždralević, M.; Pouyssegur, J. Warburg and Beyond: The Power of Mitochondrial Metabolism to Collaborate or Replace Fermentative Glycolysis in Cancer. Cancers 2020, 12, 1119. [Google Scholar] [CrossRef] [PubMed]
- Pavlides, S.; Whitaker-Menezes, D.; Castello-Cros, R.; Flomenberg, N.; Witkiewicz, A.K.; Frank, P.G.; Casimiro, M.C.; Wang, C.; Fortina, P.; Addya, S.; et al. The reverse Warburg effect: Aerobic glycolysis in cancer associated fibroblasts and the tumor stroma. Cell Cycle 2009, 8, 3984–4001. [Google Scholar] [CrossRef]
- Gladden, L.B.; Goodwin, M.L.; McDonald, J.R.; Nijsten, M.W.N. Fuel for cancer cells? Cell Cycle 2011, 10, 2422. [Google Scholar] [CrossRef][Green Version]
- Schiliro, C.; Firestein, B.L. Mechanisms of Metabolic Reprogramming in Cancer Cells Supporting Enhanced Growth and Proliferation. Cells 2021, 10, 1056. [Google Scholar] [CrossRef]
- Jacquet, P.; Stéphanou, A. Metabolic Reprogramming, Questioning, and Implications for Cancer. Biology 2021, 10, 129. [Google Scholar] [CrossRef]
- Goetz, L.H.; Schork, N.J. Personalized medicine: Motivation, challenges, and progress. Fertil. Steril. 2018, 109, 952–963. [Google Scholar] [CrossRef]
- Niida, A.; Hasegawa, T.; Innan, H.; Shibata, T.; Mimori, K.; Miyano, S. A unified simulation model for understanding the diversity of cancer evolution. PeerJ 2020, 8, e8842. [Google Scholar] [CrossRef]
- Hausser, J.; Szekely, P.; Bar, N.; Zimmer, A.; Sheftel, H.; Caldas, C.; Alon, U. Tumor diversity and the trade-off between universal cancer tasks. Nat. Commun. 2019, 10, 5423. [Google Scholar] [CrossRef]
- Sonveaux, P.; Végran, F.; Schroeder, T.; Wergin, M.C.; Verrax, J.; Rabbani, Z.N.; De Saedeleer, C.J.; Kennedy, K.M.; Diepart, C.; Jordan, B.F.; et al. Targeting lactate-fueled respiration selectively kills hypoxic tumor cells in mice. J. Clin. Investig. 2008, 118, 3930–3942. [Google Scholar] [CrossRef]
- Strickaert, A.; Saiselet, M.; Dom, G.; De Deken, X.; Dumont, J.E.; Feron, O.; Sonveaux, P.; Maenhaut, C. Cancer heterogeneity is not compatible with one unique cancer cell metabolic map. Oncogene 2017, 36, 2637–2642. [Google Scholar] [CrossRef] [PubMed]
- Damiani, C.; Maspero, D.; Di Filippo, M.; Colombo, R.; Pescini, D.; Graudenzi, A.; Westerhoff, H.V.; Alberghina, L.; Vanoni, M.; Mauri, G. Integration of single-cell RNA-seq data into population models to characterize cancer metabolism. PLoS Comput. Biol. 2019, 15, e1006733. [Google Scholar] [CrossRef] [PubMed]
- Dewhirst, M.W.; Cao, Y.; Moeller, B. Cycling hypoxia and free radicals regulate angiogenesis and radiotherapy response. Nat. Rev. Cancer 2008, 8, 425–437. [Google Scholar] [CrossRef]
- Dewhirst, M.W. Relationships between Cycling Hypoxia, HIF-1, Angiogenesis and Oxidative Stress. Radiat. Res. 2009, 172, 653–665. [Google Scholar] [CrossRef]
- Wolfe, J. Cellular Thermodynamics: The Molecular and Macroscopic Views; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2015. [Google Scholar] [CrossRef]
- Casciari, J.J.; Sotirchos, S.V.; Sutherland, R.M. Mathematical modelling of microenvironment and growth in EMT6/Ro multicellular tumour spheroids. Cell Prolif. 1992, 25, 1–22. [Google Scholar] [CrossRef]
- DuBois, C.; Farnham, J.; Aaron, E.; Radunskaya, A. A multiple time-scale computational model of a tumor and its micro environment. Math. Biosci. Eng. 2012, 10, 121–150. [Google Scholar] [CrossRef]
- Phipps, C.; Molavian, H.; Kohandel, M. A microscale mathematical model for metabolic symbiosis: Investigating the effects of metabolic inhibition on ATP turnover in tumors. J. Theor. Biol. 2015, 366, 103–114. [Google Scholar] [CrossRef]
- Jagiella, N.; Müller, B.; Müller, M.; Vignon-Clementel, I.E.; Drasdo, D. Inferring Growth Control Mechanisms in Growing Multi-cellular Spheroids of NSCLC Cells from Spatial-Temporal Image Data. PLoS Comput. Biol. 2016, 12, e1004412. [Google Scholar] [CrossRef]
- Shamsi, M.; Saghafian, M.; Dejam, M.; Sanati-Nezhad, A. Mathematical Modeling of the Function of Warburg Effect in Tumor Microenvironment. Sci. Rep. 2018, 8, 8903. [Google Scholar] [CrossRef]
- Pourhasanzade, F.; Sabzpoushan, S.H. A New Mathematical Model for Controlling Tumor Growth Based on Microenvironment Acidity and Oxygen Concentration. BioMed Res. Int. 2021, 2021, 8886050. [Google Scholar] [CrossRef]
- Li, W.; Wang, J. Uncovering the Underlying Mechanisms of Cancer Metabolism through the Landscapes and Probability Flux Quantifications. iScience 2020, 23, 101002. [Google Scholar] [CrossRef]
- Billman, G.E. Homeostasis: The Underappreciated and Far Too Often Ignored Central Organizing Principle of Physiology. Front. Physiol. 2020, 11, 200. [Google Scholar] [CrossRef]
- Glade, N. Le Vivant Rare, Faible et Amorphe. Evolution depuis les Origines jusqu’à la Vie telle qu’elle nous Apparaît. In Habilitation à Diriger des Recherches; Université Grenoble Alpes: Grenoble, France, 2022. (In French) [Google Scholar]






Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jacquet, P.; Stéphanou, A. Searching for the Metabolic Signature of Cancer: A Review from Warburg’s Time to Now. Biomolecules 2022, 12, 1412. https://doi.org/10.3390/biom12101412
Jacquet P, Stéphanou A. Searching for the Metabolic Signature of Cancer: A Review from Warburg’s Time to Now. Biomolecules. 2022; 12(10):1412. https://doi.org/10.3390/biom12101412
Chicago/Turabian StyleJacquet, Pierre, and Angélique Stéphanou. 2022. "Searching for the Metabolic Signature of Cancer: A Review from Warburg’s Time to Now" Biomolecules 12, no. 10: 1412. https://doi.org/10.3390/biom12101412
APA StyleJacquet, P., & Stéphanou, A. (2022). Searching for the Metabolic Signature of Cancer: A Review from Warburg’s Time to Now. Biomolecules, 12(10), 1412. https://doi.org/10.3390/biom12101412





