PAQR6 Upregulation Is Associated with AR Signaling and Unfavorite Prognosis in Prostate Cancers
Abstract
:1. Introduction
2. Materials and Methods
2.1. Gene Expression Profiles in Benign Prostate Tissues
2.2. Gene Expression Profiles in Prostate Cancer LNCaP Cells
2.3. Gene Expression Profiles in Primary Prostate Cancers
2.4. Differences of Gene Expression in Relation to Clinical Parameters
2.5. Assessment of Patient Survival Outcomes
2.6. Gene Expression Profiles and Correlation Analysis in Metastatic Late-Stage Prostate Cancers
2.7. Data Presentation and Statistical Analysis
3. Results
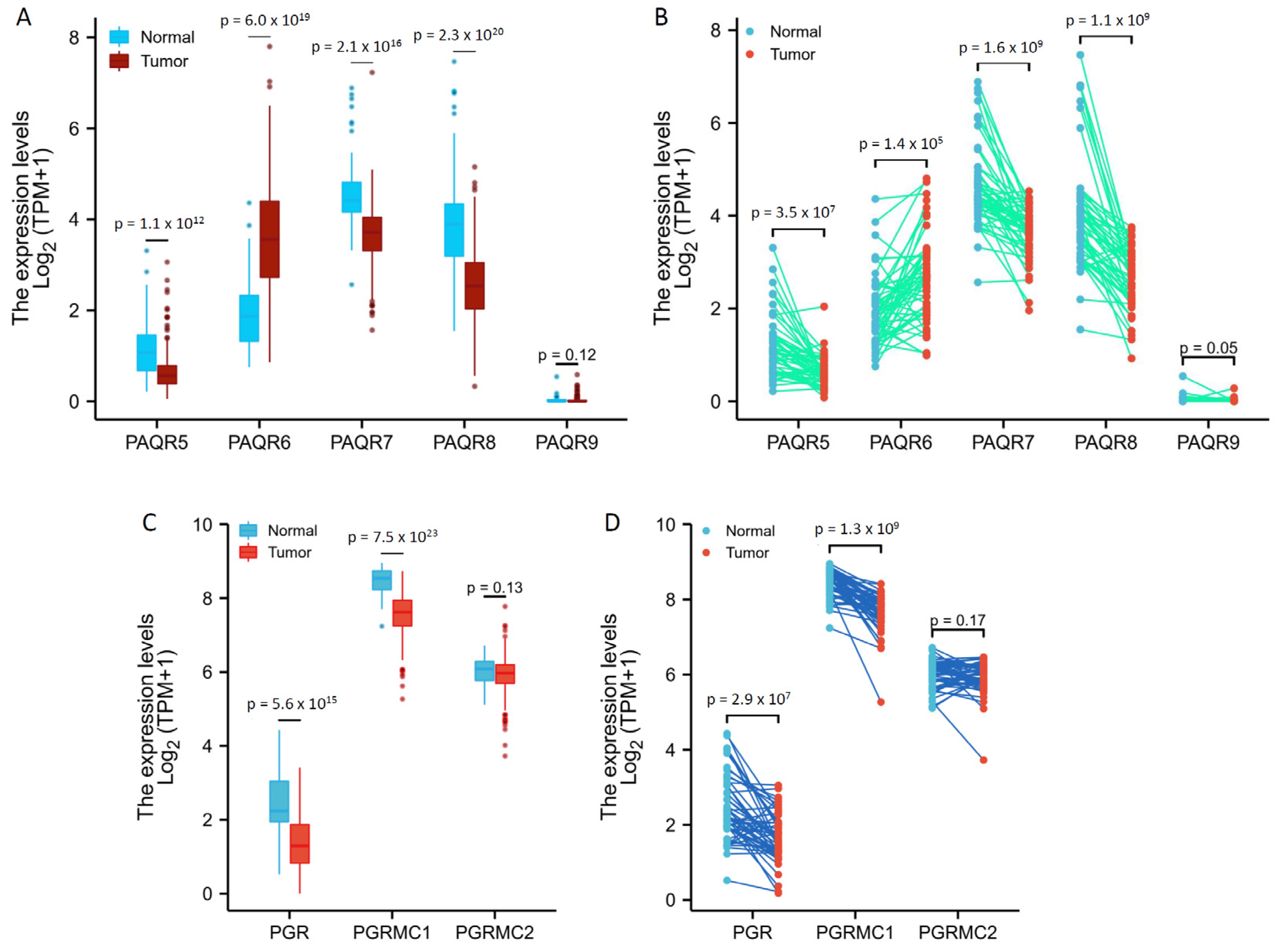
3.1. Expression Levels of mPR Genes Were Aberrantly Altered in Prostate Cancers
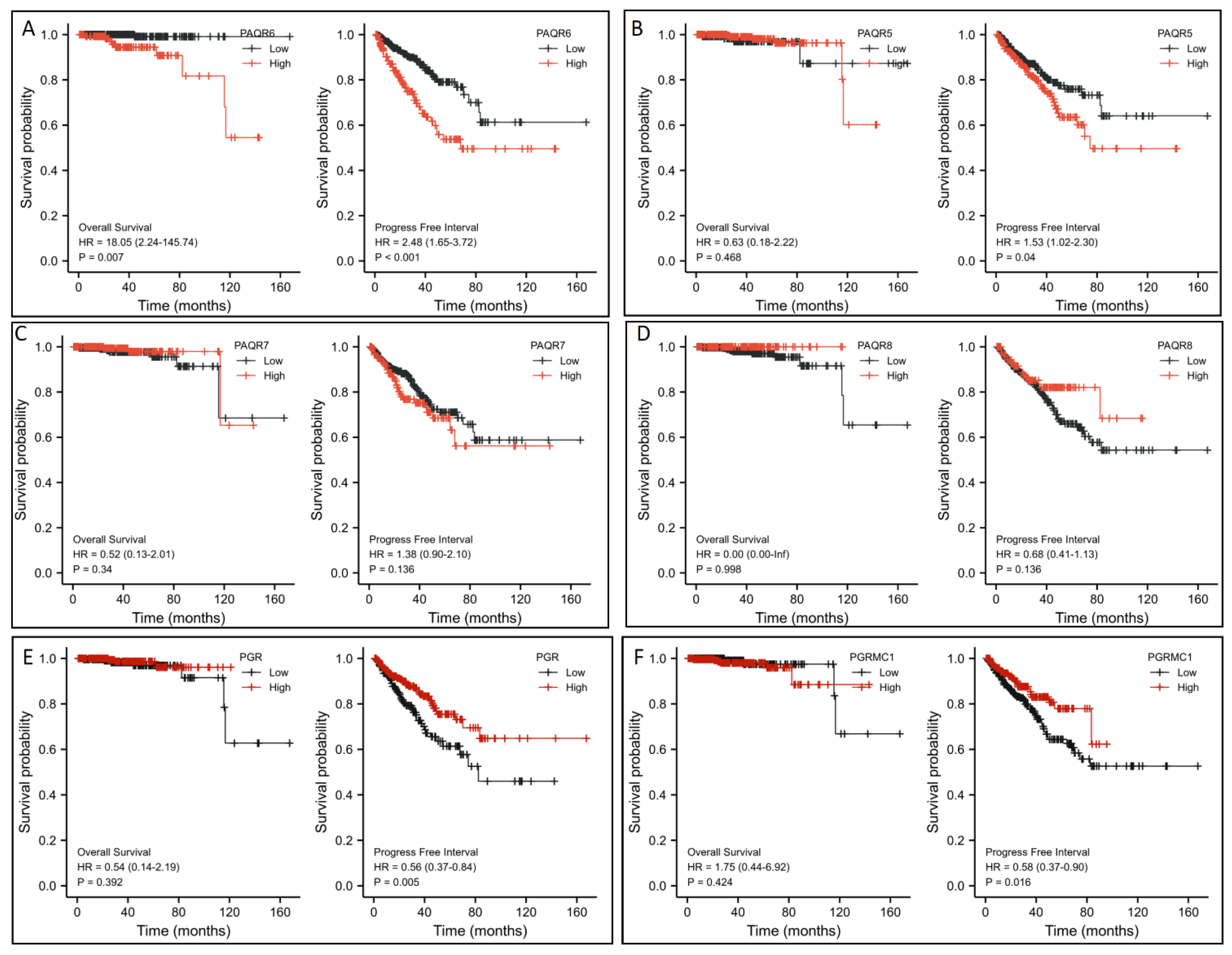
3.2. PAQR6 Upregulation Is a Strong Prognosis Factor for Disease Progression and Patient Survival
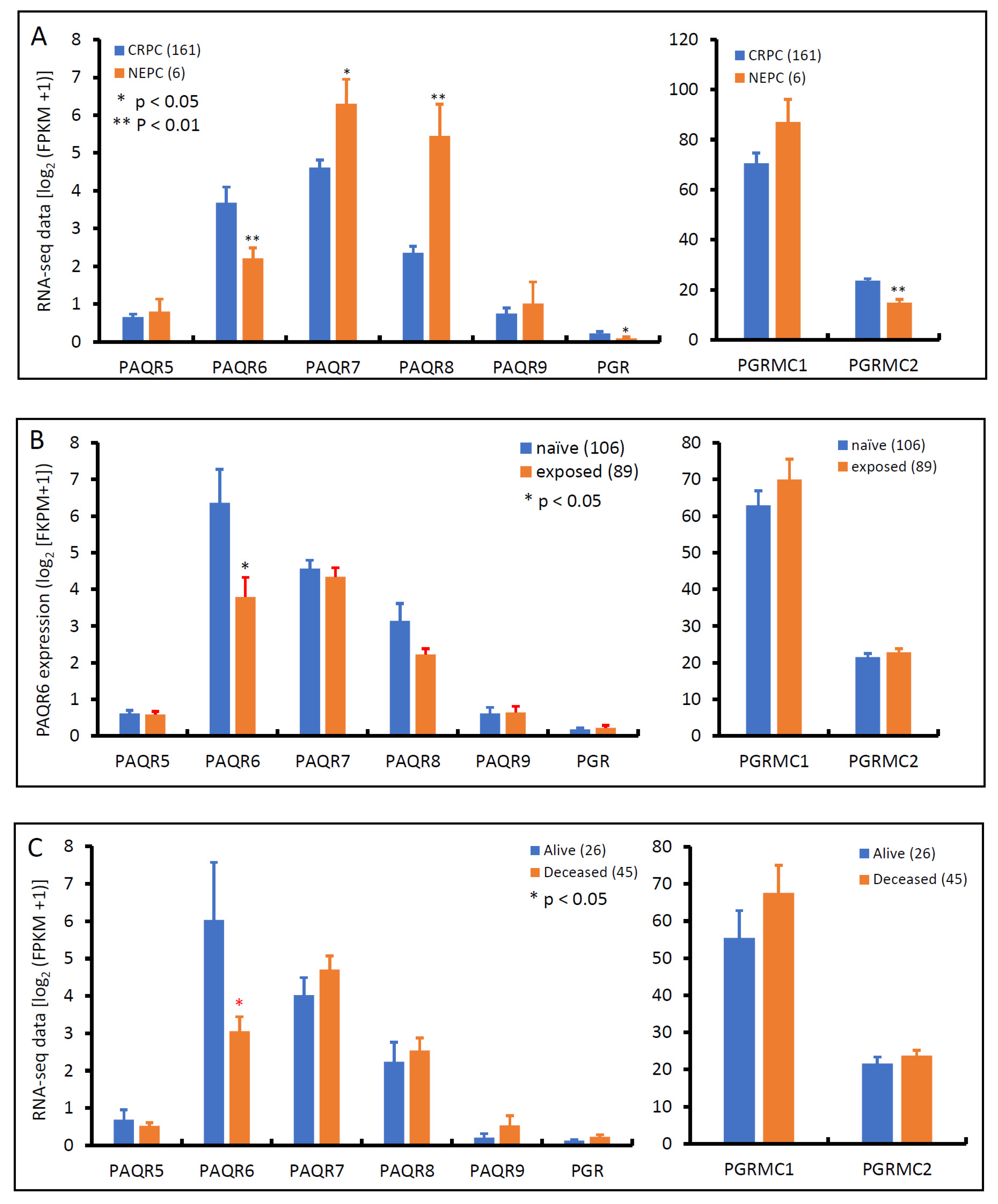
3.3. PAQR6 Expression Is Reduced in Neuroendocrinal Prostate Cancers
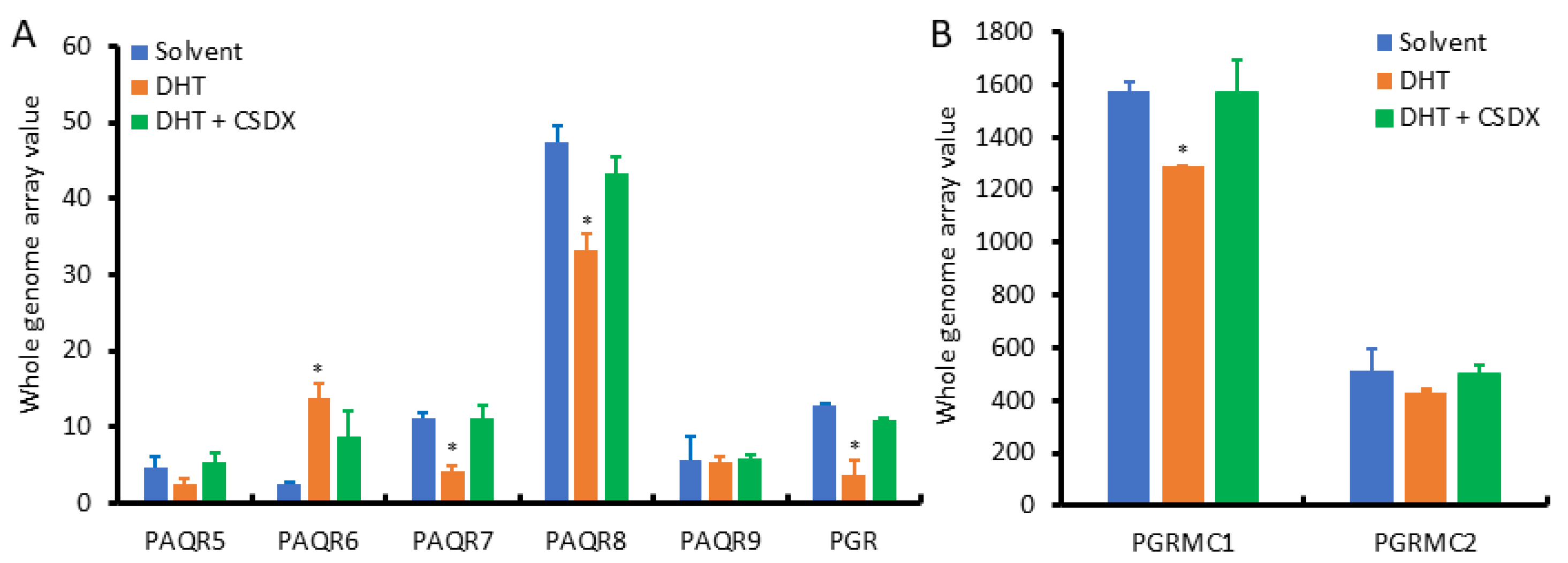
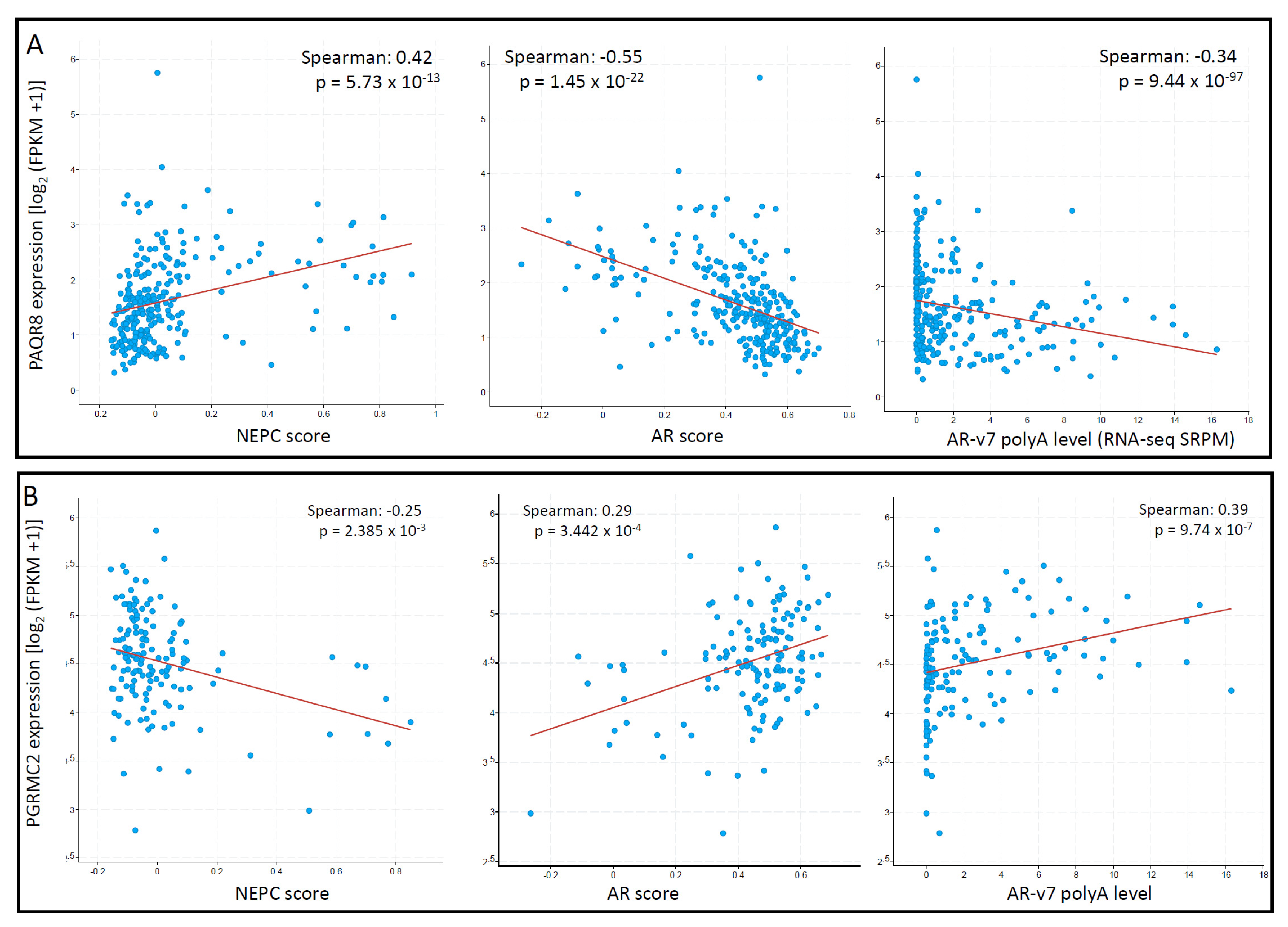
3.4. PAQR8 and PGRMC2 Oppositely Correlated with AR Signaling and NE Features
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| AR | androgen receptor |
| BCR | Biochemical Relapse |
| BPH | benign prostate hyperplasia |
| CRPC | castration-resistant prostate cancer |
| DHT | dihydrotestosterone |
| FPKM | fragments per kilobase per million |
| HR | hazard ratio |
| mPR | membrane progesterone receptor |
| NEPC | neuroendocrinal prostate cancer |
| PGR | progesterone receptor |
| PGRMC1/2 | progesterone receptor membrane component 1/2 |
| PAQR | progestin and adipoQ receptor |
| PSA | prostate-specific antigen |
| TCGA | the Cancer Genome Atlas |
| TPM | transcript per million reads |
| SEM | standard error of the mean |
References
- Trabert, B.; Sherman, M.E.; Kannan, N.; Stanczyk, F.Z. Progesterone and Breast Cancer. Endocr. Rev. 2019, 41, 320–344. [Google Scholar] [CrossRef] [PubMed]
- Winkelmann, B.R.; März, W.; Boehm, B.O.; Zotz, R.; Hager, J.; Hellstern, P.; Senges, J. Rationale and design of the LURIC study-a resource for functional genomics, pharmacogenomics and long-term prognosis of cardiovascular disease. Pharmacogenomics 2001, 2, S1–S73. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Liu, Y.; Cao, J.-M. G Protein-Coupled Receptors: Extranuclear Mediators for the Non-Genomic Actions of Steroids. Int. J. Mol. Sci. 2014, 15, 15412–15425. [Google Scholar] [CrossRef] [Green Version]
- Petersen, S.L.P.; Intlekofer, K.A.P.; Moura-Conlon, P.J.P.; Brewer, D.N.P.; Sans, J.P.D.P.; Lopez, J.A. Novel progesterone receptors: Neural localization and possible functions. Front. Neurosci. 2013, 7, 164. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaluka, D.; Batabyal, D.; Chiang, B.-Y.; Poulos, T.L.; Yeh, S.-R. Spectroscopic and Mutagenesis Studies of Human PGRMC1. Biochemistry 2015, 54, 1638–1647. [Google Scholar] [CrossRef]
- Roberto, D.; Selvarajah, S.; Park, P.C.; Berman, D.; Venkateswaran, V. Functional validation of metabolic genes that distinguish Gleason 3 from Gleason 4 prostate cancer foci. Prostate 2019, 79, 1777–1788. [Google Scholar] [CrossRef]
- Paul, M.R.; Pan, T.-C.; Pant, D.K.; Shih, N.N.; Chen, Y.; Harvey, K.L.; Solomon, A.; Lieberman, D.; Morrissette, J.J.; Soucier-Ernst, D.; et al. Genomic landscape of metastatic breast cancer identifies preferentially dysregulated pathways and targets. J. Clin. Investig. 2020, 130, 4252–4265. [Google Scholar] [CrossRef]
- Charles, N.J.; Thomas, P.; Lange, C.A. Expression of Membrane Progesterone Receptors (mPR/PAQR) in Ovarian Cancer Cells: Implications for Progesterone-Induced Signaling Events. Horm. Cancer 2010, 1, 167–176. [Google Scholar] [CrossRef] [Green Version]
- Cai, Z.; Chen, H.; Bai, J.; Zheng, Y.; Ma, J.; Cai, X.; Liu, Y.; Zhang, K.; Shou, J.; Gao, Y. Copy Number Variations of CEP63, FOSL2 and PAQR6 Serve as Novel Signatures for the Prognosis of Bladder Cancer. Front. Oncol. 2021, 11, 674933. [Google Scholar] [CrossRef]
- Kunitomi, H.; Kobayashi, Y.; Wu, R.-C.; Takeda, T.; Tominaga, E.; Banno, K.; Aoki, D. LAMC1 is a prognostic factor and a potential therapeutic target in endometrial cancer. J. Gynecol. Oncol. 2020, 31, e11. [Google Scholar] [CrossRef]
- Sinreih, M.; Knific, T.; Thomas, P.; Grazio, S.F.; Rižner, T.L. Membrane progesterone receptors β and γ have potential as prognostic biomarkers of endometrial cancer. J. Steroid Biochem. Mol. Biol. 2018, 178, 303–311. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Lin, Z.; Liang, Q.; Hu, Y.; Xu, W.-F. PAQR6 expression enhancement suggests a worse prognosis in prostate cancer patients. Open Life Sci. 2018, 13, 511–517. [Google Scholar] [CrossRef] [PubMed]
- Takezawa, Y.; Ito, K.; Suzuki, K.; Fukabori, Y.; Yamanaka, H.; Honma, S.; Mieda, M.; Hamataki, N.; Kushitani, M. Effects of a new steroidal antiandrogen, TZP-4238 (17 α-acetoxy-6-chloro-2-oxa-4, 6-pregnadiene-3, 20-dione), on spontaneously developed canine benign prostatic hyperplasia. Prostate 1995, 27, 321–328. [Google Scholar] [CrossRef]
- Murakoshi, M.; Ikeda, R.; Fukui, N.; Tagawa, M. Histopathological and immunocytochemical studies of chlormadinone acetate on the rat prostate. Tokai J. Exp. Clin. Med. 2002, 27, 91–95. [Google Scholar]
- Yu, Y.; Liu, L.; Xie, N.; Xue, H.; Fazli, L.; Buttyan, R.; Wang, Y.; Gleave, M.; Dong, X. Expression and Function of the Progesterone Receptor in Human Prostate Stroma Provide Novel Insights to Cell Proliferation Control. J. Clin. Endocrinol. Metab. 2013, 98, 2887–2896. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, Y.; Yang, O.; Fazli, L.; Rennie, P.S.; Gleave, M.E.; Dong, X. Progesterone receptor expression during prostate cancer progression suggests a role of this receptor in stromal cell differentiation. Prostate 2015, 75, 1043–1050. [Google Scholar] [CrossRef] [PubMed]
- Grindstad, T.; Richardsen, E.; Andersen, S.; Skjefstad, K.; Khanehkenari, M.R.; Donnem, T.; Ness, N.; Nordby, Y.; Bremnes, R.M.; Al-Saad, S.; et al. Progesterone Receptors in Prostate Cancer: Progesterone receptor B is the isoform associated with disease progression. Sci. Rep. 2018, 8, 11358. [Google Scholar] [CrossRef]
- Latil, A.; Bieche, I.; Vidaud, D.; Lidereau, R.; Berthon, P.; Cussenot, O.; Vidaud, M. Evaluation of androgen, estrogen (ER alpha and ER beta), and progesterone receptor expression in human prostate cancer by real-time quantitative reverse transcription-polymerase chain reaction assays. Cancer Res. 2001, 61, 1919–1926. [Google Scholar] [PubMed]
- Oudes, A.J.; Campbell, D.S.; Sorensen, C.M.; Walashek, L.S.; True, L.D.; Liu, A.Y. Transcriptomes of human prostate cells. BMC Genom. 2006, 7, 92. [Google Scholar] [CrossRef] [Green Version]
- Nickols, N.G.; Dervan, P.B. Suppression of androgen receptor-mediated gene expression by a sequence-specific DNA-binding polyamide. Proc. Natl. Acad. Sci. USA 2007, 104, 10418–10423. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Canc Genome Atlas Res The Cancer Genome Atlas Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nat. Cell Biol. 2008, 455, 1061–1068. [Google Scholar] [CrossRef]
- Grasso, C.S.; Wu, Y.-M.; Robinson, D.R.; Cao, X.; Dhanasekaran, S.M.; Khan, A.P.; Quist, M.J.; Jing, X.; Lonigro, R.J.; Brenner, J.C.; et al. The mutational landscape of lethal castration-resistant prostate cancer. Nature 2012, 487, 239–243. [Google Scholar] [CrossRef] [Green Version]
- Taylor, B.S.; Schultz, N.; Hieronymus, H.; Gopalan, A.; Xiao, Y.; Carver, B.S.; Arora, V.K.; Kaushik, P.; Cerami, E.; Reva, B.; et al. Integrative Genomic Profiling of Human Prostate Cancer. Cancer Cell 2010, 18, 11–22. [Google Scholar] [CrossRef] [Green Version]
- Gerhauser, C.; Favero, F.; Risch, T.; Simon, R.; Feuerbach, L.; Assenov, Y.; Heckmann, D.; Sidiropoulos, N.; Waszak, S.M.; Hübschmann, D.; et al. Molecular Evolution of Early-Onset Prostate Cancer Identifies Molecular Risk Markers and Clinical Trajectories. Cancer Cell 2018, 34, 996–1011.e8,. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, J.; Lichtenberg, T.; Hoadley, K.A.; Poisson, L.M.; Lazar, A.J.; Cherniack, A.D.; Kovatich, A.J.; Benz, C.C.; Levine, D.A.; Lee, A.V.; et al. An Integrated TCGA Pan-Cancer Clinical Data Resource to Drive High-Quality Survival Outcome Analytics. Cell 2018, 173, 400–416.e11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abida, W.; Cyrta, J.; Heller, G.; Prandi, D.; Armenia, J.; Coleman, I.; Cieslik, M.; Benelli, M.; Robinson, D.; Van Allen, E.M.; et al. Genomic correlates of clinical outcome in advanced prostate cancer. Proc. Natl. Acad. Sci. USA 2019, 116, 11428–11436. [Google Scholar] [CrossRef] [Green Version]
- Hieronymus, H.; Lamb, J.; Ross, K.N.; Peng, X.P.; Clement, C.; Rodina, A.; Nieto, M.; Du, J.; Stegmaier, K.; Raj, S.M.; et al. Gene expression signature-based chemical genomic prediction identifies a novel class of HSP90 pathway modulators. Cancer Cell 2006, 10, 321–330. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beltran, H.; Prandi, D.; Mosquera, J.M.; Benelli, M.; Puca, L.; Cyrta, J.; Marotz, C.; Giannopoulou, E.; Chakravarthi, B.V.; Varambally, S.; et al. Divergent clonal evolution of castration-resistant neuroendocrine prostate cancer. Nat. Med. 2016, 22, 298–305. [Google Scholar] [CrossRef] [Green Version]
- Ren, S.; Wei, G.-H.; Liu, D.; Wang, L.; Hou, Y.; Zhu, S.; Peng, L.; Zhang, Q.; Cheng, Y.; Su, H.; et al. Whole-genome and Transcriptome Sequencing of Prostate Cancer Identify New Genetic Alterations Driving Disease Progression. Eur. Urol. 2018, 73, 322–339. [Google Scholar] [CrossRef] [PubMed]
- Schiewer, M.J.; Knudsen, K.E. Basic Science and Molecular Genetics of Prostate Cancer Aggressiveness. Urol. Clin. North Am. 2021, 48, 339–347. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, Y.; Ci, X.; Choi, S.Y.C.; Crea, F.; Lin, D.; Wang, Y. Molecular events in neuroendocrine prostate cancer development. Nat. Rev. Urol. 2021, 1–16. [Google Scholar] [CrossRef]
- Dressing, G.E.; Goldberg, J.E.; Charles, N.J.; Schwertfeger, K.L.; Lange, C.A. Membrane progesterone receptor expression in mammalian tissues: A review of regulation and physiological implications. Steroids 2011, 76, 11–17. [Google Scholar] [CrossRef] [Green Version]
- Desai, K.; McManus, J.M.; Sharifi, N. Hormonal Therapy for Prostate Cancer. Endocr. Rev. 2021, 42, 354–373. [Google Scholar] [CrossRef] [PubMed]
- Thakur, A.; Roy, A.; Ghosh, A.; Chhabra, M.; Banerjee, S. Abiraterone acetate in the treatment of prostate cancer. Biomed. Pharmacother. 2018, 101, 211–218. [Google Scholar] [CrossRef]
- Tassinari, D.; Fochessati, F.; Panzini, I.; Poggi, B.; Sartori, S.; Ravaioli, A. Rapid Progression of Advanced “Hormone-Resistant” Prostate Cancer During Palliative Treatment with Progestins for Cancer Cachexia. J. Pain Symptom Manag. 2003, 25, 481–484. [Google Scholar] [CrossRef]



| Characteristic | Low PAQR6 | High PAQR6 | p | Statistic | Method |
|---|---|---|---|---|---|
| n | 249 | 250 | |||
| T stage, n (%) | 0.027 | 7.23 | Chisq.test | ||
| T2 | 109 (22.2%) | 80 (16.3%) | |||
| T3 | 132 (26.8%) | 160 (32.5%) | |||
| T4 | 5 (1%) | 6 (1.2%) | |||
| N stage, n (%) | 0.014 | 6.08 | Chisq.test | ||
| N0 | 179 (42%) | 168 (39.4%) | |||
| N1 | 28 (6.6%) | 51 (12%) | |||
| M stage, n (%) | 0.249 | Fisher.test | |||
| M0 | 224 (48.9%) | 231 (50.4%) | |||
| M1 | 0 (0%) | 3 (0.7%) | |||
| Age, median (IQR) | 62 (56, 66) | 61 (56, 65) | 0.327 | 32701 | Wilcoxon |
| Characteristic | Low PAQR5 | High PAQR5 | p | Statistic | Method |
|---|---|---|---|---|---|
| n | 249 | 250 | |||
| T stage, n (%) | <0.001 | 19.6 | Chisq.test | ||
| T2 | 117 (23.8%) | 72 (14.6%) | |||
| T3 | 128 (26%) | 164 (33.3%) | |||
| T4 | 2 (0.4%) | 9 (1.8%) | |||
| N stage, n (%) | 0.021 | 5.31 | Chisq.test | ||
| N0 | 171 (40.1%) | 176 (41.3%) | |||
| N1 | 27 (6.3%) | 52 (12.2%) | |||
| M stage, n (%) | 0.623 | Fisher.test | |||
| M0 | 226 (49.3%) | 229 (50%) | |||
| M1 | 2 (0.4%) | 1 (0.2%) | |||
| Age, mean ± SD | 60.84 ± 7 | 61.22 ± 6.64 | 0.529 | −0.63 | T test |
| Characteristics | Total (N) | Odds Ratio (OR) | p Value |
|---|---|---|---|
| PAQR5 | |||
| T stage (T3 & T4 vs. T2) | 492 | 2.162 (1.495–3.144) | <0.001 |
| N stage (N1 vs. N0) | 426 | 1.871 (1.132–3.152) | 0.016 |
| PAQR6 | |||
| T stage (T3 & T4 vs. T2) | 492 | 1.651 (1.146–2.387) | 0.007 |
| N stage (N1 vs. N0) | 426 | 1.941 (1.177–3.255) | 0.01 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, M.; Li, J.C.; Tao, C.; Wu, S.; Liu, B.; Shu, Q.; Li, B.; Zhu, R. PAQR6 Upregulation Is Associated with AR Signaling and Unfavorite Prognosis in Prostate Cancers. Biomolecules 2021, 11, 1383. https://doi.org/10.3390/biom11091383
Yang M, Li JC, Tao C, Wu S, Liu B, Shu Q, Li B, Zhu R. PAQR6 Upregulation Is Associated with AR Signaling and Unfavorite Prognosis in Prostate Cancers. Biomolecules. 2021; 11(9):1383. https://doi.org/10.3390/biom11091383
Chicago/Turabian StyleYang, Min, Jean Chong Li, Chang Tao, Sa Wu, Bin Liu, Qiang Shu, Benyi Li, and Runzhi Zhu. 2021. "PAQR6 Upregulation Is Associated with AR Signaling and Unfavorite Prognosis in Prostate Cancers" Biomolecules 11, no. 9: 1383. https://doi.org/10.3390/biom11091383
APA StyleYang, M., Li, J. C., Tao, C., Wu, S., Liu, B., Shu, Q., Li, B., & Zhu, R. (2021). PAQR6 Upregulation Is Associated with AR Signaling and Unfavorite Prognosis in Prostate Cancers. Biomolecules, 11(9), 1383. https://doi.org/10.3390/biom11091383






