In Vitro Secretome Analysis Suggests Differential Pathogenic Mechanisms between Fusarium oxysporum f. sp. cubense Race 1 and Race 4
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals
2.2. Plant Materials and Growth Conditions
2.3. Fungi and Growth Conditions
2.4. Extraction of Secreted Proteins
2.5. Identification of Proteins by LC-MS/MS
2.6. Bioinformatics Analyses of the Secreted Proteins
2.7. Functional Annotation of Secreted Proteins
2.8. Prediction of Pathogenicity-Associated Secreted Proteins
2.9. Prediction of Effectors
2.10. Quantitative Real-Time PCR (RT-qPCR) Analysis
2.11. Statistical Analysis
3. Results
3.1. Shotgun Proteomic Analysis of Foc Secretomes
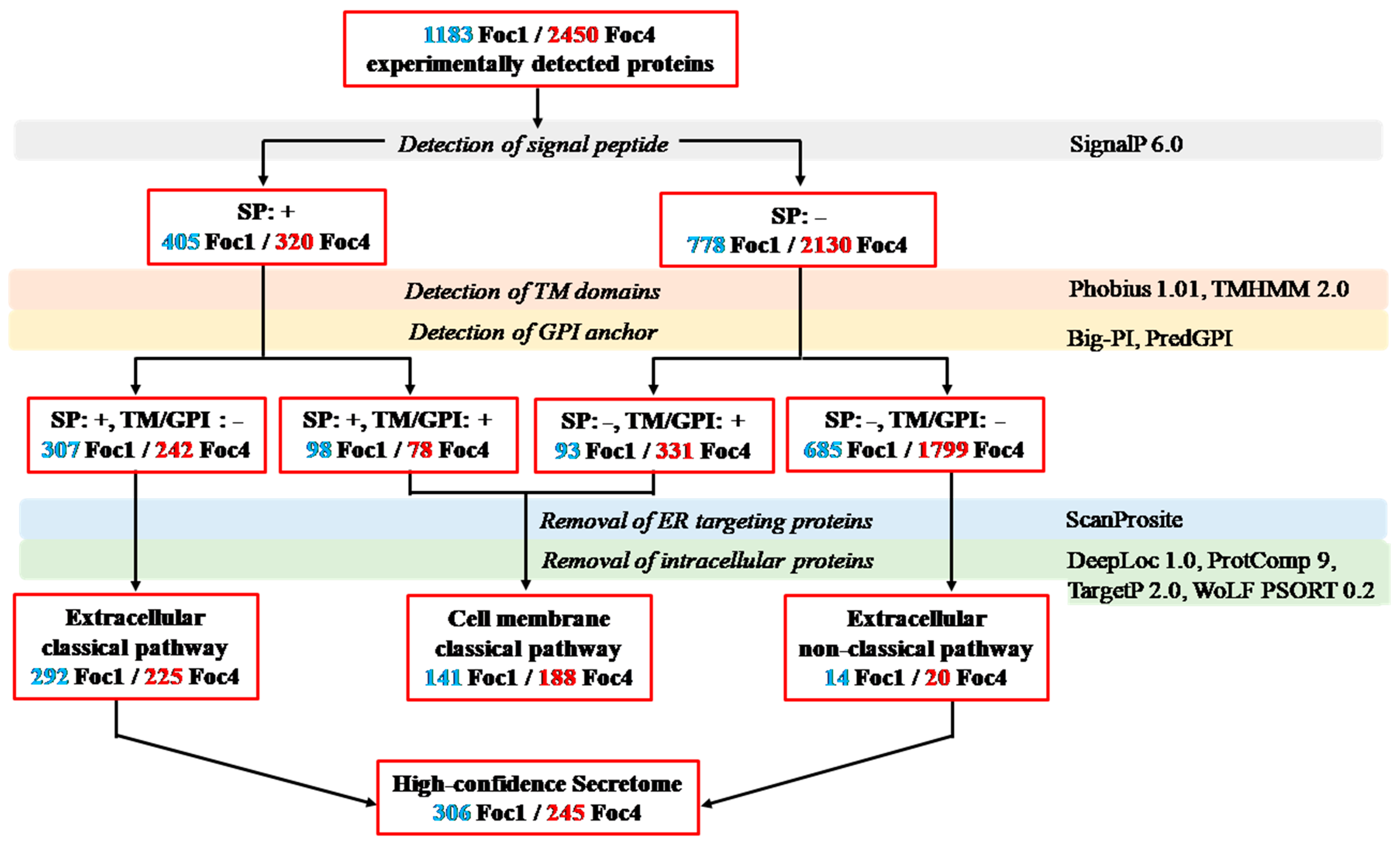
3.2. In silico Analysis of Foc Secretomes
3.3. Functional Annotation and Classification of the Secreted Proteins
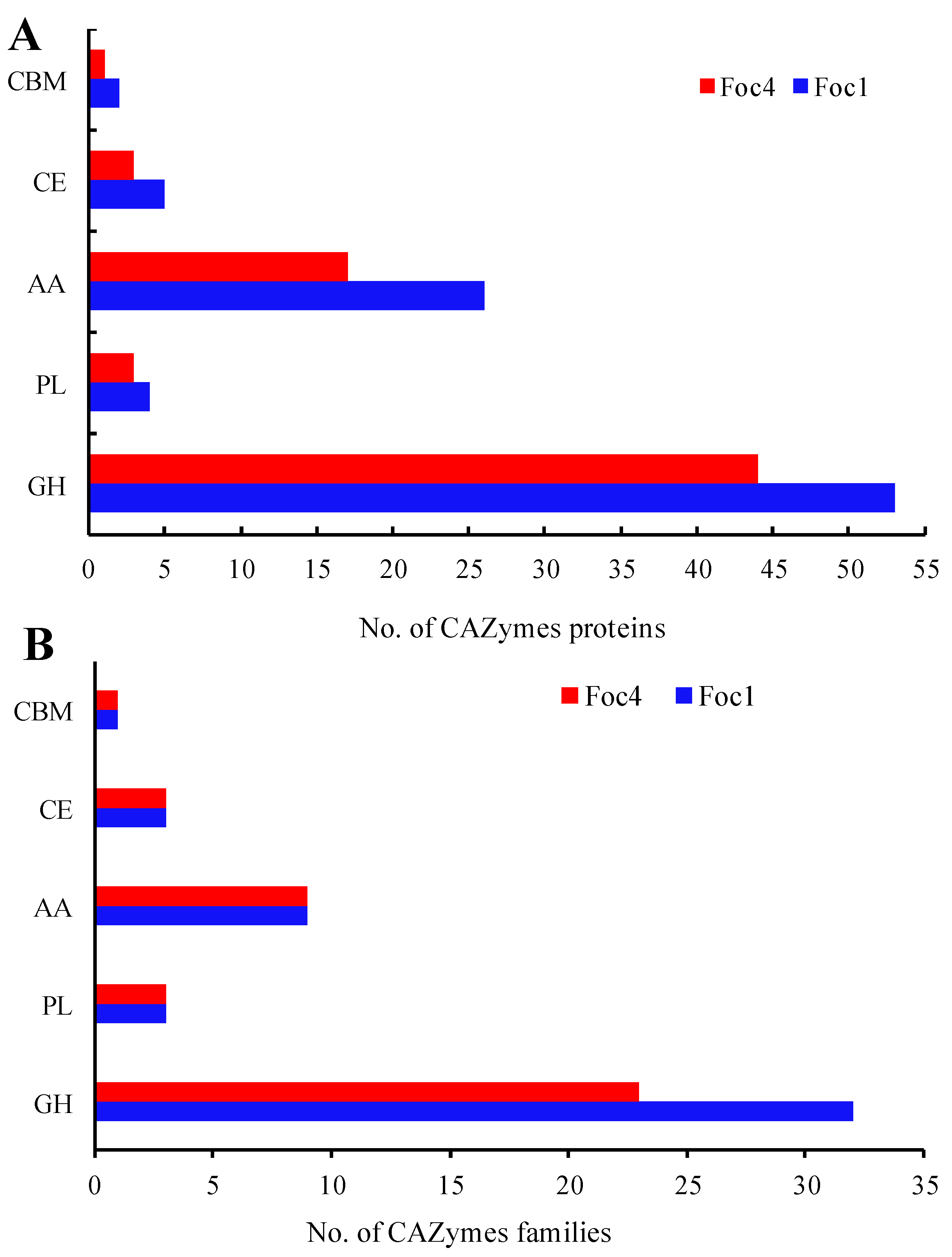
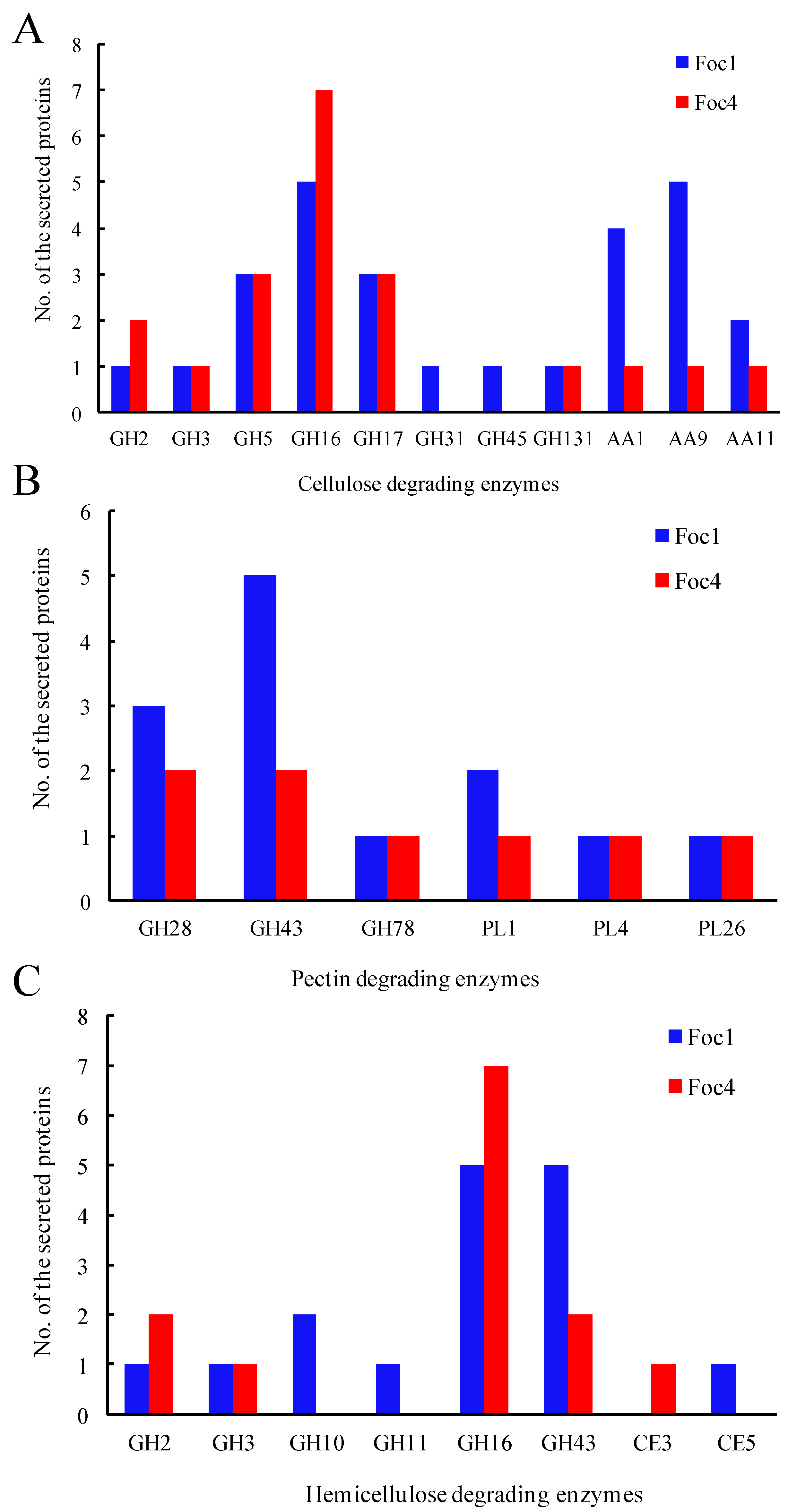
3.4. CAZymes Analysis of the Secreted Proteins
3.5. Pathogenicity-Associated Secreted Proteins
3.6. Effector Analysis of Foc Secretome
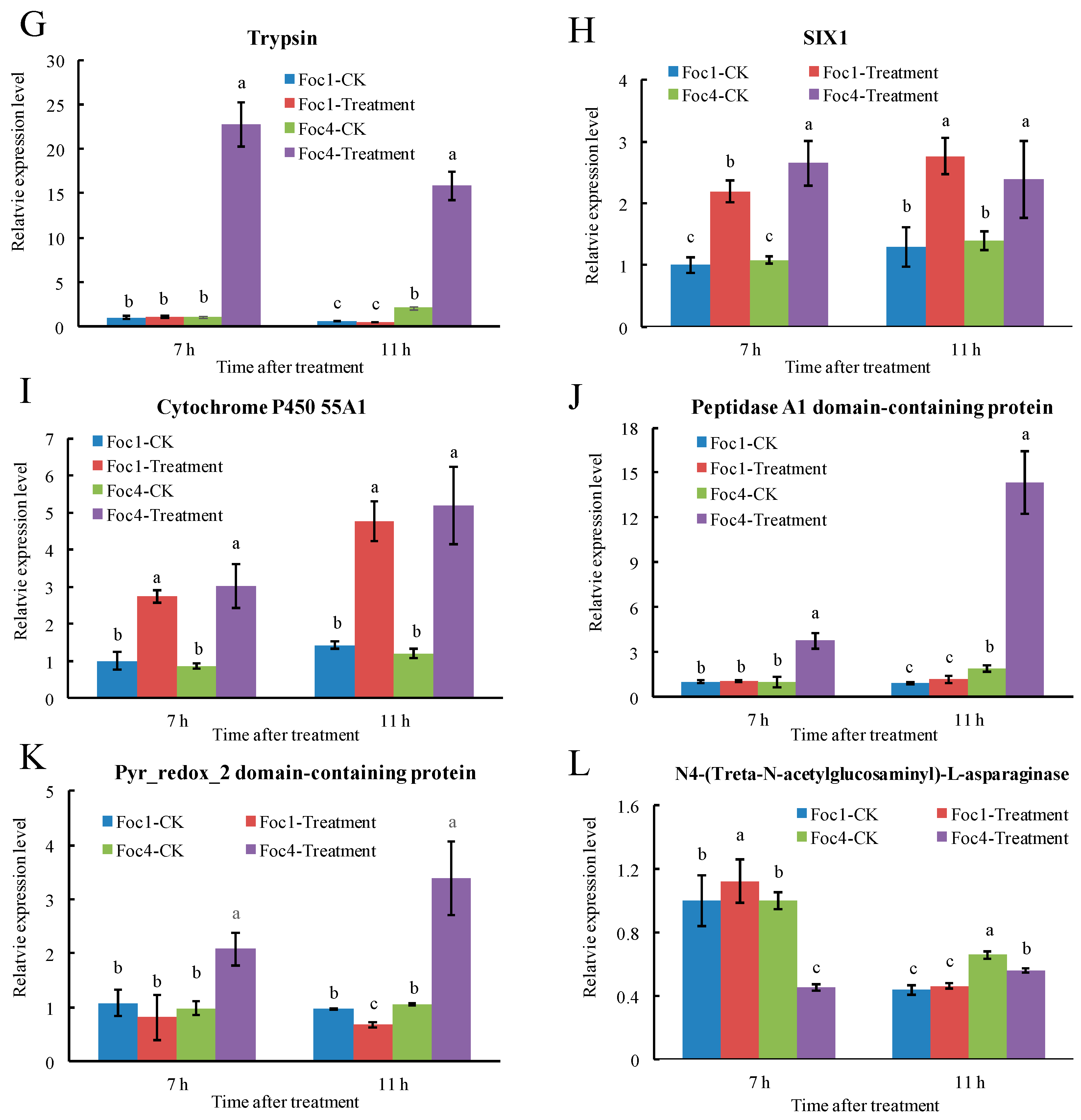
3.7. RT-qPCR Analysis of Foc Secreted Proteins
3.8. Functional Characteristics of Other Experimentally Detected Proteins
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bubici, G.; Kaushal, M.; Prigigallo, M.I.; Cabanas, C.G.L.; Mercado-Blanco, J. Biological control agents against Fusarium wilt of banana. Front. Microbiol. 2019, 10, 616. [Google Scholar] [CrossRef] [Green Version]
- Liu, S.; Li, J.; Zhang, Y.; Liu, N.; Altus, V.; Diane, M.; Zuo, C.; Hu, C.; Bi, F.; Gao, H. Fusaric acid instigates the invasion of banana by Fusarium oxysporum f. sp. cubense TR4. New Phytol. 2020, 228, 2004. [Google Scholar] [CrossRef] [PubMed]
- Thangavelu, R.; Edwin Raj, E.; Pushpakanth, P.; Loganathan, M.; Uma, S. Draft genome of Fusarium oxysporum f. sp. cubense strain tropical race-4 infecting Cavendish (AAA) group of banana in India. Plant. Dis. 2021, 105, 481–483. [Google Scholar] [PubMed]
- Guo, L.; Yang, L.; Liang, C.; Wang, G.; Dai, Q.; Huang, J. Differential colonization patterns of bananas (Musa spp.) by physiological race 1 and race 4 isolates of Fusarium oxysporum f.sp. cubense. J. Phytopathol. 2015, 163, 807–817. [Google Scholar] [CrossRef]
- Dong, H.; Ye, Y.; Guo, Y.; Li, H. Comparative transcriptome analysis revealed resistance differences of Cavendish bananas to Fusarium oxysporum f.sp. cubense race 1 and race 4. BMC Genet. 2020, 21, 122. [Google Scholar]
- Fan, H.; Dong, H.; Xu, C.; Liu, J.; Hu, B.; Ye, J.; Mai, G.; Li, H. Pectin methylesterases contribute the pathogenic differences between races 1 and 4 of Fusarium oxysporum f. sp. cubense. Sci. Rep. 2017, 7, 13140. [Google Scholar] [CrossRef] [Green Version]
- Dong, H.; Fan, H.; Lei, Z.; Wu, C.; Zhou, D.; Li, H. Histological and gene expression analyses in banana reveals the pathogenic differences between races 1 and 4 of banana Fusarium wilt pathogen. Phytopathology 2019, 109, 1029–1042. [Google Scholar] [CrossRef] [PubMed]
- Dong, H.; Li, Y.; Fan, H.; Zhou, D.; Li, H. Quantitative proteomics analysis reveals resistance differences of banana cultivar ‘Brazilian’ to Fusarium oxysporum f. sp. cubense races 1 and 4. J. Proteomics 2019, 203, 103376. [Google Scholar] [CrossRef]
- Ploetz, R.C. Management of Fusarium wilt of banana: A review with special reference to tropical race 4. Crop. Prot. 2015, 73, 7–15. [Google Scholar] [CrossRef]
- Czislowski, E.; Fraser-Smith, S.; Zander, M.; O’neill, W.T.; Meldrum, R.A.; Tran-Nguyen, L.T.T.; Batley, J.; Aitken, E.a.B. Investigation of the diversity of effector genes in the banana pathogen, Fusarium oxysporum f. sp. cubense, reveals evidence of horizontal gene transfer. Mol. Plant. Pathol. 2018, 19, 1155–1171. [Google Scholar] [CrossRef] [Green Version]
- Pandey, V.; Singh, M.; Pandey, D.; Marla, S.; Kumar, A. Secretome analysis identifies potential pathogenicity/virulence factors of Tilletia indica, a quarantined fungal pathogen inciting Karnal bunt disease in wheat. Proteomics 2018, 18, 1700473. [Google Scholar] [CrossRef]
- Dong, Z.; Wang, Z. Isolation and heterologous expression of a polygalacturonase produced by Fusarium oxysporum f. sp. cubense race 1 and 4. Int. J. Mol. Sci. 2015, 16, 7595–7607. [Google Scholar] [CrossRef] [Green Version]
- Qin, S.; Ji, C.; Li, Y.; Wang, Z. Comparative transcriptomic analysis of race 1 and race 4 of Fusarium oxysporum f. sp. cubense induced with different carbon sources. G3 2017, 7, 2125–2138. [Google Scholar]
- Lorrain, C.; Hecker, A.; Duplessis, S. Effector-Mining in the Poplar rust fungus Melampsora larici-populina secretome. Front. Plant. Sci. 2015, 6, 1051. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, D.; Peng, C.; Zheng, X.; Chang, L.; Xu, B.; Tong, Z. Secretome analysis of the banana Fusarium wilt fungi Foc R1 and Foc TR4 reveals a new effector OASTL required for full pathogenicity of Foc TR4 in banana. Biomolecules 2020, 10, 1430. [Google Scholar] [CrossRef] [PubMed]
- Chang, W.; Li, H.; Chen, H.; Qiao, F.; Zeng, H. Identification of mimp-associated effector genes in Fusarium oxysporum f. sp. cubense race 1 and race 4 and virulence confirmation of a candidate effector gene. Microbiol. Res. 2020, 232, 126375. [Google Scholar] [PubMed]
- Qiao, F.; Yang, X.; Xu, F.; Huang, Y.; Zhang, J.; Song, M.; Zhou, S.; Zhang, M.; He, D. TMT-based quantitative proteomic analysis reveals defense mechanism of wheat against the crown rot pathogen Fusarium pseudograminearum. BMC Plant. Biol. 2021, 21, 82. [Google Scholar] [CrossRef]
- Gonzalez-Fernandez, R.; Valero-Galvan, J.; Gomez-Galvez, F.J.; Jorrin-Novo, J.V. Unraveling the in vitro secretome of the phytopathogen Botrytis cinerea to understand the interaction with its hosts. Front. Plant. Sci. 2015, 6, 839. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, L.; Han, L.; Yang, L.; Zeng, H.; Fan, D.; Zhu, Y.; Feng, Y.; Wang, G.; Peng, C.; Jiang, X.; et al. Genome and transcriptome analysis of the fungal pathogen Fusarium oxysporum f. sp. cubense causing banana vascular wilt disease. PLoS ONE 2014, 9, e95543. [Google Scholar] [CrossRef]
- Zhao, S.; An, B.; Guo, Y.; Hou, X.; Luo, H.; He, C.; Wang, Q. Label free proteomics and systematic analysis of secretome reveals effector candidates regulated by SGE1 and FTF1 in the plant pathogen Fusarium oxysporum f. sp. cubense tropical race 4. BMC Genom. 2020, 21, 275. [Google Scholar] [CrossRef]
- Espino, J.J.; Gutierrez-Sanchez, G.; Brito, N.; Shah, P.; Orlando, R.; Gonzalez, C. The Botrytis cinerea early secretome. Proteomics 2010, 10, 3020–3034. [Google Scholar] [CrossRef] [Green Version]
- Schwarz, K.; Fiedler, T.; Fischer, R.J.; Bahl, H. A standard operating procedure (SOP) for the preparation of intra- and extracellular proteins of Clostridium acetobutylicum for proteome analysis. J. Microbiol. Methods 2007, 68, 396–402. [Google Scholar] [CrossRef]
- Lowry, O.H.; Rosebrough, N.J.; Farr, A.L.; Randall, R.J. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 1951, 193, 265–275. [Google Scholar] [CrossRef]
- Wisniewski, J.R.; Zougman, A.; Mann, M. Combination of FASP and StageTip-based fractionation allows in-depth analysis of the hippocampal membrane proteome. J. Proteome Res. 2009, 8, 5674–5678. [Google Scholar] [CrossRef]
- Teufel, F.; Armenteros, J.A.; Johansen, A.R.; Gislason, M.H.; Nielsen, H. SignalP 6.0 achieves signal peptide prediction across all types using protein language models. bioRxiv 2021. [Google Scholar] [CrossRef]
- Käll, L.; Krogh, A.; Sonnhammer, E. A combined transmembrane topology and signal peptide prediction method. J. Mol. Biol. 2004, 338, 1027–1036. [Google Scholar] [CrossRef]
- Sonnhammer, E.; Heijne, G.V.; Krogh, A.V. A hidden markov model for predicting transmembrane helices in protein sequences. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1998, 6, 175–182. [Google Scholar] [PubMed]
- Eisenhaber, B.; Bork, P.; Eisenhaber, F. Post-translational GPI lipid anchor modification of proteins in kingdoms of life: Analysis of protein sequence data from complete genomes. Protein Eng. 2001, 14, 17–25. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pierleoni, A.; Martelli, P.L.; Casadio, R. PredGPI: A GPI-anchor predictor. BMC Bioinform. 2008, 9, 392. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gattiker, A.; Gasteiger, E.; Bairoch, A. ScanProsite: A reference implementation of a PROSITE scanning tool. Appl. Bioinform. 2002, 1, 107. [Google Scholar]
- Almagro Armenteros, J.J.; Sønderby, C.K.; Sønderby, S.K.; Nielsen, H.; Winther, O. DeepLoc: Prediction of protein subcellular localization using deep learning. Bioinformatics 2017, 33, 3387–3395. [Google Scholar] [CrossRef] [PubMed]
- Armenteros, J.; Salvatore, M.; Emanuelsson, O.; Winther, O.; Nielsen, H. Detecting sequence signals in targeting peptides using deep learning. Life Sci. Alliance 2019, 2, e201900429. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Horton, P.; Park, K.-J.; Obayashi, T.; Fujita, N.; Harada, H.; Adams-Collier, C.J.; Nakai, K. WoLF PSORT: Protein localization predictor. Nucleic Acids Res. 2007, 35, W585–W587. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cantalapiedra, C.P.; Hernández-Plaza, A.; Letunic, I.; Bork, P.; Huerta-Cepas, J. eggNOG-mapper v2: Functional annotation, orthology assignments, and domain prediction at the metagenomic scale. bioRxiv 2021. [Google Scholar] [CrossRef]
- Zhang, H.; Tanner, Y.; Huang, L.; Sarah, E.; Wu, P.; Yang, Z.; Busk, P.K.; Xu, Y.; Yin, Y. dbCAN2: A meta server for automated carbohydrate-active enzyme annotation. Nucleic Acids Res. 2018, W95–W101. [Google Scholar] [CrossRef] [Green Version]
- Winnenburg, R.; Baldwin, T.K.; Urban, M.; Rawlings, C.; Koehler, J.; Hammond-Kosack, K.E. PHI-base: A new database for pathogen host interactions. Nucleic Acids Res. 2006, 34, D459–D464. [Google Scholar] [CrossRef] [PubMed]
- Sperschneider, J.; Dodds, P.N. EffectorP 3.0: Prediction of apoplastic and cytoplasmic effectors in fungi and oomycetes. bioRxiv 2021. [Google Scholar] [CrossRef]
- Jones, D.A.; Bertazzoni, S.; Turo, C.J.; Syme, R.A.; Hane, J.K. Bioinformatic prediction of plant-pathogenicity effector proteins of fungi. Curr. Opin. Microbiol. 2018, 46, 43–49. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(T)(-Delta Delta C) method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Deng, G.M.; Yang, Q.S.; He, W.D.; Li, C.Y.; Yang, J.; Zuo, C.W.; Gao, J.; Sheng, O.; Lu, S.Y.; Zhang, S.; et al. Proteomic analysis of conidia germination in Fusarium oxysporum f. sp. cubense tropical race 4 reveals new targets in ergosterol biosynthesis pathway for controlling Fusarium wilt of banana. Appl. Microbiol. Biotechnol. 2015, 99, 7189–7207. [Google Scholar]
- Zhao, Z.; Liu, H.; Wang, C.; Xu, J.R. Comparative analysis of fungal genomes reveals different plant cell wall degrading capacity in fungi. BMC Genom. 2013, 14, 274. [Google Scholar] [CrossRef] [Green Version]
- Roy, A.; Jayaprakash, A.; Rajeswary, R.; Annamalai, A.; Lakshmi, P.T.V. Genome-wide annotation, comparison and functional genomics of carbohydrate-active enzymes in legumes infecting Fusarium oxysporum formae speciales. Mycol. Inter. J. Fungal Biol. 2020, 11, 56–70. [Google Scholar] [CrossRef] [PubMed]
- Sperschneider, J.; Dodds, P.N.; Gardiner, D.M.; Singh, K.B.; Taylor, J.M. Improved prediction of fungal effector proteins from secretomes with EffectorP 2.0. Mol. Plant. Pathol. 2018, 19, 2094–2110. [Google Scholar] [CrossRef] [Green Version]
- Sharpee, W.; Oh, Y.; Yi, M.; Franck, W.; Eyre, A.; Okagaki, L.H.; Valent, B.; Dean, R.A. Identification and characterization of suppressors of plant cell death (SPD) effectors from Magnaporthe oryzae. Mol. Plant. Pathol. 2017, 18, 850–863. [Google Scholar] [CrossRef]
- Zeng, R.; Gao, S.; Xu, L.; Liu, X.; Dai, F. Prediction of pathogenesis-related secreted proteins from Stemphylium lycopersici. BMC Microbiol. 2018, 18, 191. [Google Scholar] [CrossRef]
- Xu, J.; Zhao, Y.; Zhang, X.; Zhang, L.; Hou, Y.; Dong, W. Transcriptome analysis and ultrastructure observation reveal that hawthorn fruit softening is due to cellulose/hemicellulose degradation. Front. Plant. Sci. 2016, 7, 1524. [Google Scholar] [CrossRef] [Green Version]
- He, Y.; Yan, R.; Meng, G.; Yang, W.; Wang, Z.; Li, Y.; Nie, Y. Genome-scale prediction and analysis of secreted proteins and effectors in Fusarium oxysporum f. sp. cubense tropical race 1. Acta Phytopathol. Sinaca 2020, 50, 129–140. (In Chinese) [Google Scholar]
- Nie, Y.; Huang, J.; Zhou, L.; Tu, X.; Chen, H.; Wang, Z.; Li, Y. Genome-scale prediction and analysis of secreted proteins of Fusarium oxysporum f. sp. cubense tropical race 4. Jiangsu J. Agric. Sci. 2017, 33, 288–294. (In Chinese) [Google Scholar]
- Ji, X.L.; Yan, M.; Yang, Z.D.; Li, A.F.; Kong, L.R. Shotgun analysis of the secretome of Fusarium graminearum. Indian J. Microbiol. 2013, 53, 400–409. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.; Chung, K.P.; Lin, W.; Jiang, L. Protein secretion in plants: Conventional and unconventional pathways and new techniques. J. Exp. Bot. 2018, 69, 21–37. [Google Scholar] [CrossRef] [Green Version]
- Kim, S.G.; Wang, Y.; Lee, K.H.; Park, Z.Y.; Park, J.; Wu, J.; Kwon, S.J.; Lee, Y.H.; Agrawal, G.K.; Rakwal, R.; et al. In-depth insight into in vivo apoplastic secretome of rice-Magnaporthe oryzae interaction. J. Proteomics 2013, 78, 58–71. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; McMahon, M.B.; Ramachandran, S.R.; Garrett, W.M.; LeBlanc, N.; Crouch, J.A.; Shishkoff, N.; Luster, D.G. Comparative analysis of extracellular proteomes reveals putative effectors of the boxwood blight pathogens, Calonectria henricotiae and C. pseudonaviculata. Biosci. Rep. 2021, 41, BSR20203544. [Google Scholar] [CrossRef]
- Vincent, D.; Rafiqi, M.; Job, D. The multiple facets of plant-fungal interactions revealed through plant and fungal secretomics. Front. Plant. Sci. 2020, 10, 1626. [Google Scholar] [CrossRef]
- Agrawal, G.K.; Jwa, N.S.; Lebrun, M.H.; Job, D.; Rakwal, R. Plant secretome: Unlocking secrets of the secreted proteins. Proteomics 2010, 10, 799–827. [Google Scholar] [CrossRef]
- Wang, Y.; Wu, J.; Park, Z.Y.; Kim, S.G.; Rakwal, R.; Agrawal, G.K.; Kim, S.T.; Kang, K.Y. Comparative secretome investigation of Magnaporthe oryzae proteins responsive to nitrogen starvation. J. Proteome Res. 2011, 10, 3136–3148. [Google Scholar] [CrossRef] [PubMed]
- Barrett, K.; Jensen, K.; Meyer, A.S.; Frisvad, J.C.; Lange, L. Fungal secretome profile categorization of CAZymes by function and family corresponds to fungal phylogeny and taxonomy: Example Aspergillus and Penicillium. Sci. Rep. 2020, 10, 5158. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, Y.J.; Kong, W.S. Genome-wide comparison of carbohydrate-active enzymes (CAZymes) repertoire of Flammulina ononidis. Mycobiology 2018, 46, 349–360. [Google Scholar] [CrossRef] [Green Version]
- De Assis, L.J.; Silva, L.P.; Bayram, O.; Dowling, P.; Kniemeyer, O.; Krueger, T.; Brakhage, A.A.; Chen, Y.; Dong, L.; Tan, K.; et al. Carbon catabolite repression in filamentous fungi is regulated by phosphorylation of the transcription factor CreA. mBio 2021, 12, e03146-20. [Google Scholar] [CrossRef] [PubMed]
- Singh, J.; Aggarwal, R.; Gurjar, M.S.; Sharma, S.; Saharan, M.S. Identification of carbohydrate active enzymes from whole genome sequence of Tilletia indica and sporulation analysis. Indian J. Agr. Sci. 2019, 89, 1023–1026. [Google Scholar]
- Li, M.H.; Xie, X.L.; Lin, X.F.; Shi, J.X.; Ding, Z.J.; Ling, J.F.; Xi, P.G.; Zhou, J.N.; Leng, Y.; Zhong, S.; et al. Functional characterization of the gene FoOCH1 encoding a putative alpha-1,6-mannosyltransferase in Fusarium oxysporum f. sp. cubense. Fungal Genet. Biol. 2014, 65, 1–13. [Google Scholar] [CrossRef]
- Neu, E.; Debener, T. Prediction of the Diplocarpon rosae secretome reveals candidate genes for effectors and virulence factors. Fungal Biol. 2019, 123, 231–239. [Google Scholar] [CrossRef]
- Fan, H.; Lei, Z.; Dong, H.; Zhou, D.; Li, H. Immune responses in Brazilian banana determining the pathogenic differences between the physiological races 1 and 4 of Fusarium oxysporum f. sp. cubense. J. Plant. Pathol. 2019, 101, 225–234. [Google Scholar] [CrossRef]
- Urban, M.; Cuzick, A.; Seager, J.; Wood, V.; Rutherford, K.; Venkatesh, S.Y.; De Silva, N.; Martinez, M.C.; Pedro, H.; Yates, A.D.; et al. PHI-base: The pathogen-host interactions database. Nucleic Acids Res. 2020, 48, D613–D620. [Google Scholar] [CrossRef]
- Lv, W.; Wang, C.; Yang, N.; Que, Y.; Talbot, N.J.; Wang, Z. Genome-wide functional analysis reveals that autophagy is necessary for growth, sporulation, deoxynivalenol production and virulence in Fusarium graminearum. Sci. Rep. 2017, 7, 11062. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Breakspear, A.; Pasquali, M.; Broz, K.; Dong, Y.; Kistler, H.C. Npc1 is involved in sterol trafficking in the filamentous fungus Fusarium graminearum. Fungal Genet. Biol. 2011, 48, 725–730. [Google Scholar] [CrossRef] [PubMed]
- Subramaniam, R.; Narayanan, S.; Walkowiak, S.; Wang, L.; Joshi, M.; Rocheleau, H.; Ouellet, T.; Harris, L.J. Leucine metabolism regulates TRI6 expression and affects deoxynivalenol production and virulence in Fusarium graminearum. Mol. Microbiol. 2015, 98, 760–769. [Google Scholar] [CrossRef] [PubMed]
- Imazaki, I.; Kurahashi, M.; Iida, Y.; Tsuge, T. Fow2, a Zn(II)2Cys6-type transcription regulator, controls plant infection of the vascular wilt fungus Fusarium oxysporum. Mol. Microbiol. 2007, 63, 737–753. [Google Scholar] [CrossRef] [PubMed]
- Krol, P.; Igielski, R.; Pollmann, S.; Kepczynska, E. Priming of seeds with methyl jasmonate induced resistance to hemi-biotroph Fusarium oxysporum f. sp lycopersici in tomato via 12-oxo-phytodienoic acid, salicylic acid, and flavonol accumulation. J. Plant. Physiol. 2015, 179, 122–132. [Google Scholar] [CrossRef] [PubMed]
- Delgado-Jarana, J.S.; Martinez-Rocha, A.L.; Roldan-Rodriguez, R.; Roncero, M.I.G.; Di Pietro, A. Fusarium oxysporum G-protein beta subunit Fgb1 regulates hyphal growth, development, and virulence through multiple signalling pathways. Fungal Genet. Biol. 2005, 42, 61–72. [Google Scholar] [CrossRef]
- Guo, L.; Yang, L.; Liang, C.; Wang, J.; Liu, L.; Huang, J. The G-protein subunits FGA2 and FGB1 play distinct roles in development and pathogenicity in the banana fungal pathogen Fusarium oxysporum f. sp cubense. Physiol. Mol. Plant. Pathol. 2016, 93, 29–38. [Google Scholar] [CrossRef]
- Tanaka, S.; Djamei, A.; Lo Presti, L.; Schipper, K.; Winterberg, S.; Amati, S.; Becker, D.; Buechner, H.; Kumlehn, J.; Reissmann, S.; et al. Experimental approaches to investigate effector translocation into host cells in the Ustilago maydis/maize pathosystem. Eur. J. Cell Biol. 2015, 94, 349–358. [Google Scholar] [CrossRef] [PubMed]
- Deng, C.H.; Plummer, K.M.; Jones, D.a.B.; Mesarich, C.H.; Shiller, J.; Taranto, A.P.; Robinson, A.J.; Kastner, P.; Hall, N.E.; Templeton, M.D.; et al. Comparative analysis of the predicted secretomes of Rosaceae scab pathogens Venturia inaequalis and V. pirina reveals expanded effector families and putative determinants of host range. BMC Genom. 2017, 18, 339. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Batson, A.M.; Fokkens, L.; Rep, M.; Du Toit, L.J. Putative effector genes distinguish two pathogenicity groups of Fusarium oxysporum f. sp. spinaciae. Mol. Plant. Microbe Interact. 2021, 34, 141–156. [Google Scholar] [CrossRef]
- Oome, S.; Van Den Ackerveken, G. Comparative and functional analysis of the widely occurring family of Nep1-Like proteins. Mol. Plant. Microbe Interact. 2014, 27, 1081–1094. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, S.; Wu, B.; Yang, J.; Bi, F.; Dong, T.; Yang, Q.; Hu, C.; Xiang, D.; Chen, H.; Huang, H.; et al. A Cerato-Platanin family protein FocCP1 is essential for the penetration and virulence of Fusarium oxysporum f. sp. cubense tropical race 4. Int. J. Mol. Sci. 2019, 20, 3785. [Google Scholar] [CrossRef] [Green Version]
- Casarrubia, S.; Daghino, S.; Kohler, A.; Morin, E.; Khouja, H.R.; Daguerre, Y.; Veneault-Fourrey, C.; Martin, F.M.; Perotto, S.; Martino, E. The Hydrophobin-Like OmSSP1 may be an effector in the ericoid Mycorrhizal Symbiosis. Front. Plant. Sci. 2018, 9, 546. [Google Scholar] [CrossRef] [Green Version]
- Wrobel, K.; Wrobel, K.; Garcia Lara, B.; Guerrero Esperanza, M.; Gonzalez Roncero, M.I.; Corrales Escobosa, A.R. Comparative evaluation of two Fusarium oxysporum f. sp. lycopersici strains grown on two different carbon sources: LC-MS-based secretome study after in vivo N-15 metabolic labeling. Int. J. Mass Spectrom. 2020, 449, 116288. [Google Scholar] [CrossRef]
- Rafiqi, M.; Ellis, J.G.; Ludowici, V.A.; Hardham, A.R.; Dodds, P.N. Challenges and progress towards understanding the role of effectors in plant-fungal interactions. Curr. Opin. Plant. Biol. 2012, 15, 477–482. [Google Scholar] [CrossRef] [PubMed]
- Vivek-Ananth, R.P.; Mohanraj, K.; Vandanashree, M.; Jhingran, A.; Craig, J.P.; Samal, A. Comparative systems analysis of the secretome of the opportunistic pathogen Aspergillus fumigatus and other Aspergillus species. Sci. Rep. 2018, 8, 6617. [Google Scholar] [CrossRef] [PubMed]


| Phenotype | No. of PHI-Base Matches | Fraction of the Secretome (%) | ||
|---|---|---|---|---|
| Foc1 | Foc4 | Foc1 | Foc4 | |
| Loss of pathogenicity | 0 | 3 | 0 | 1.22 |
| Reduced virulence | 85 | 76 | 27.78 | 31.02 |
| Unaffected pathogenicity | 53 | 35 | 17.32 | 14.29 |
| Effector (plant avirulence determinant) | 7 | 3 | 2.29 | 1.22 |
| Increased virulence | 2 | 1 | 0.65 | 0.41 |
| Lethal | 1 | 1 | 0.33 | 0.41 |
| Mixed outcome | 11 | 11 | 3.59 | 4.49 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
He, Y.; Zhou, X.; Li, J.; Li, H.; Li, Y.; Nie, Y. In Vitro Secretome Analysis Suggests Differential Pathogenic Mechanisms between Fusarium oxysporum f. sp. cubense Race 1 and Race 4. Biomolecules 2021, 11, 1353. https://doi.org/10.3390/biom11091353
He Y, Zhou X, Li J, Li H, Li Y, Nie Y. In Vitro Secretome Analysis Suggests Differential Pathogenic Mechanisms between Fusarium oxysporum f. sp. cubense Race 1 and Race 4. Biomolecules. 2021; 11(9):1353. https://doi.org/10.3390/biom11091353
Chicago/Turabian StyleHe, Yanqiu, Xiaofan Zhou, Jieling Li, Huaping Li, Yunfeng Li, and Yanfang Nie. 2021. "In Vitro Secretome Analysis Suggests Differential Pathogenic Mechanisms between Fusarium oxysporum f. sp. cubense Race 1 and Race 4" Biomolecules 11, no. 9: 1353. https://doi.org/10.3390/biom11091353
APA StyleHe, Y., Zhou, X., Li, J., Li, H., Li, Y., & Nie, Y. (2021). In Vitro Secretome Analysis Suggests Differential Pathogenic Mechanisms between Fusarium oxysporum f. sp. cubense Race 1 and Race 4. Biomolecules, 11(9), 1353. https://doi.org/10.3390/biom11091353






