TFEB Signalling-Related MicroRNAs and Autophagy
Abstract
1. Introduction
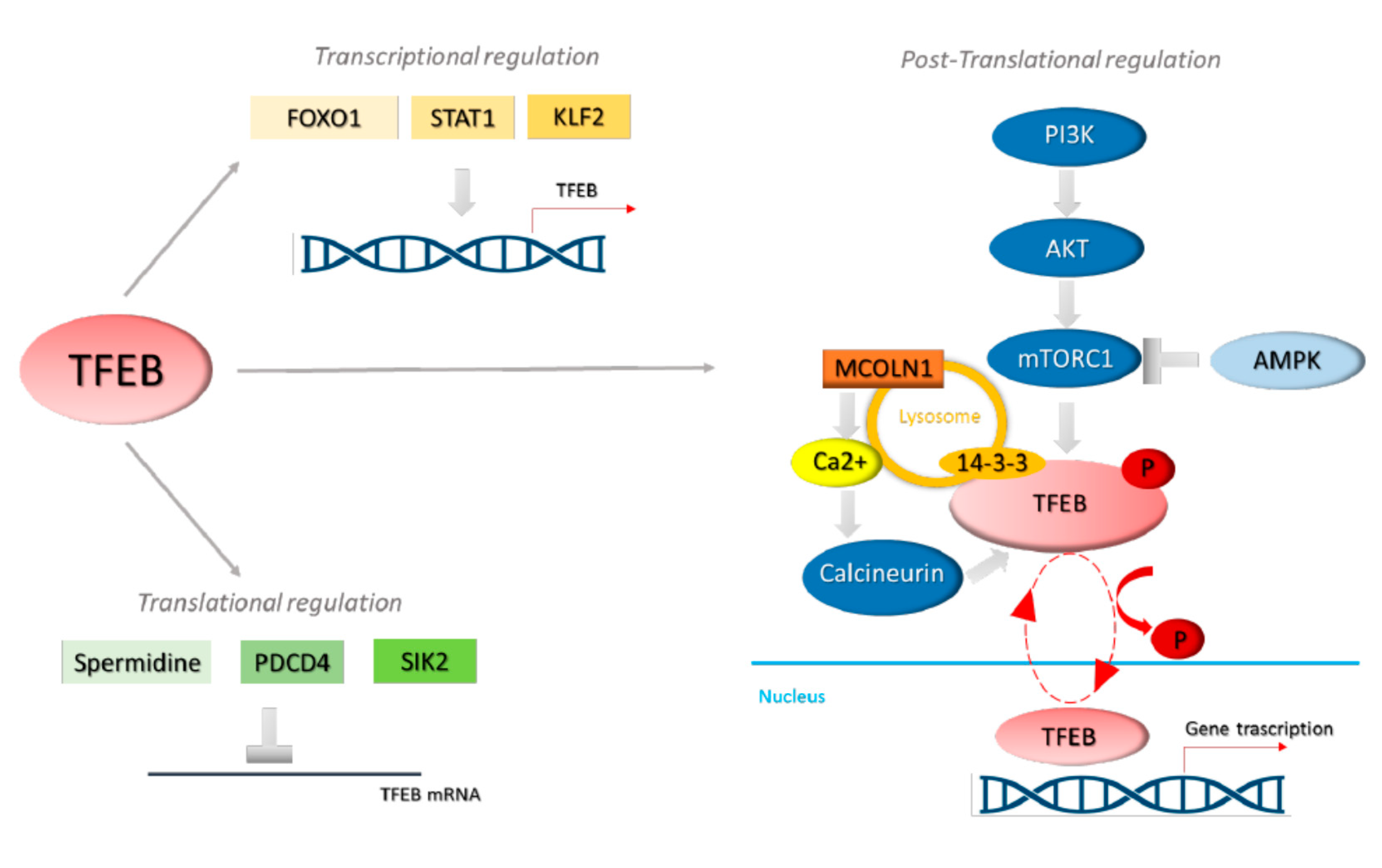
2. miRNAs and TFEB
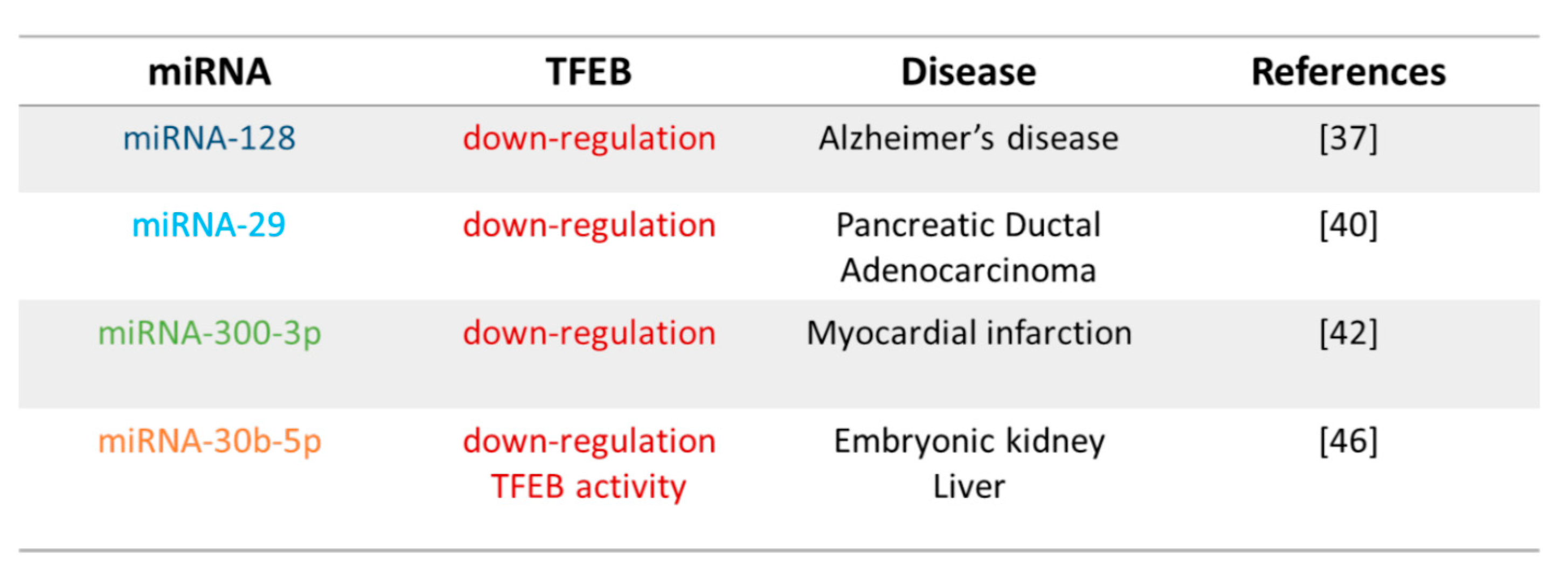
3. Post-Transcriptional Regulation of TFEB via miRNAs
3.1. miRNA-128
3.2. miRNA-29
3.3. miRNA-300
3.4. miRNA-30
4. miRNAs Control of TFEB Activation
4.1. miRNA-211 and mTOR
4.2. miRNA-21 and PTEN
4.3. miRNA-33 and AMPK
4.4. miRNA-211 and Ezrin

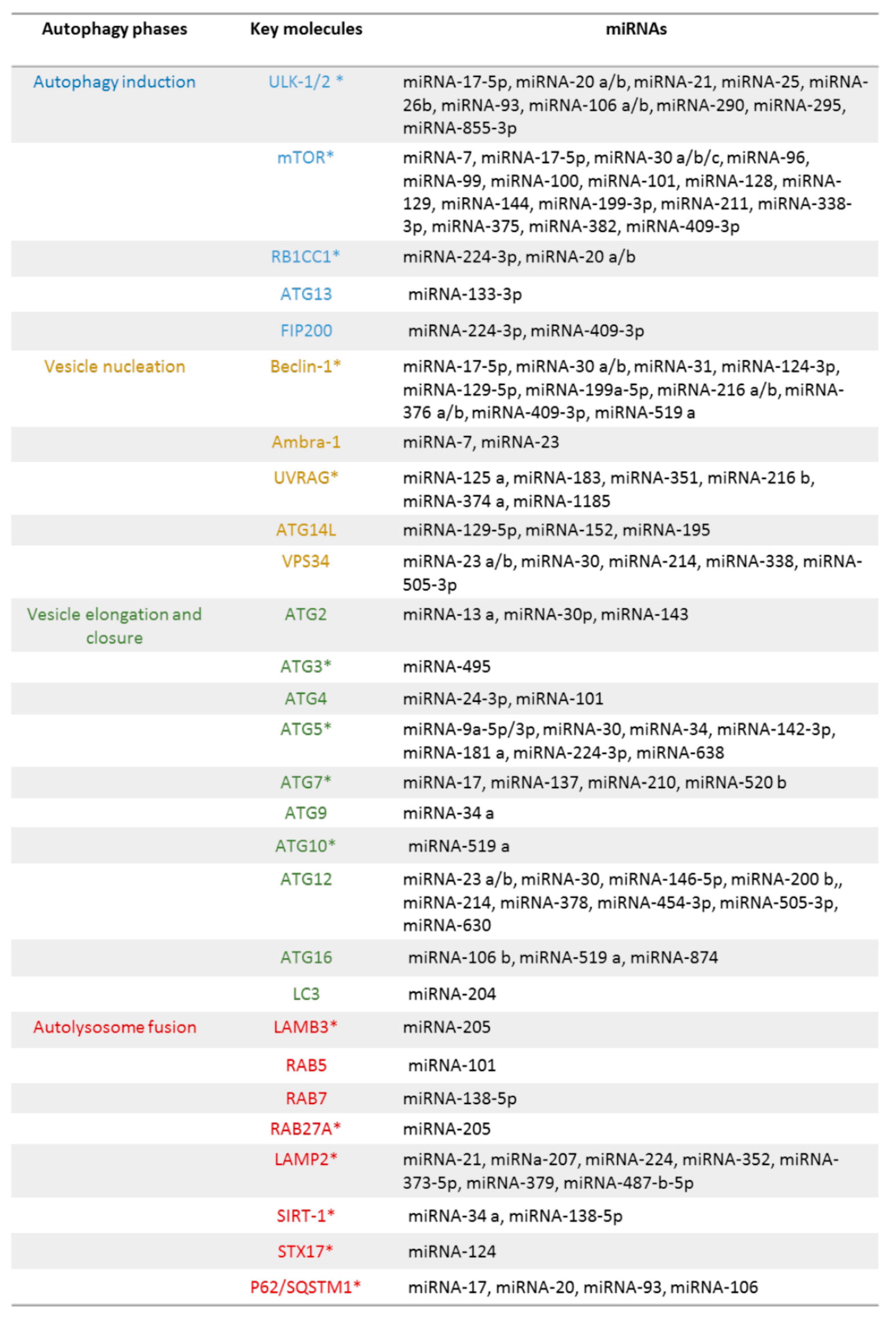
5. Autophagic Genes and miRNAs
5.1. Autophagy initiation
5.2. Vesicle Nucleation
5.3. Vesicle Elongation
5.4. Autolysosome Fusion
6. TFEB Regulation of miRNA Expression
6.1. TFEB and IRS: miRNA-335-3p, miRNA-495-3p, and miRNA-548o-3p
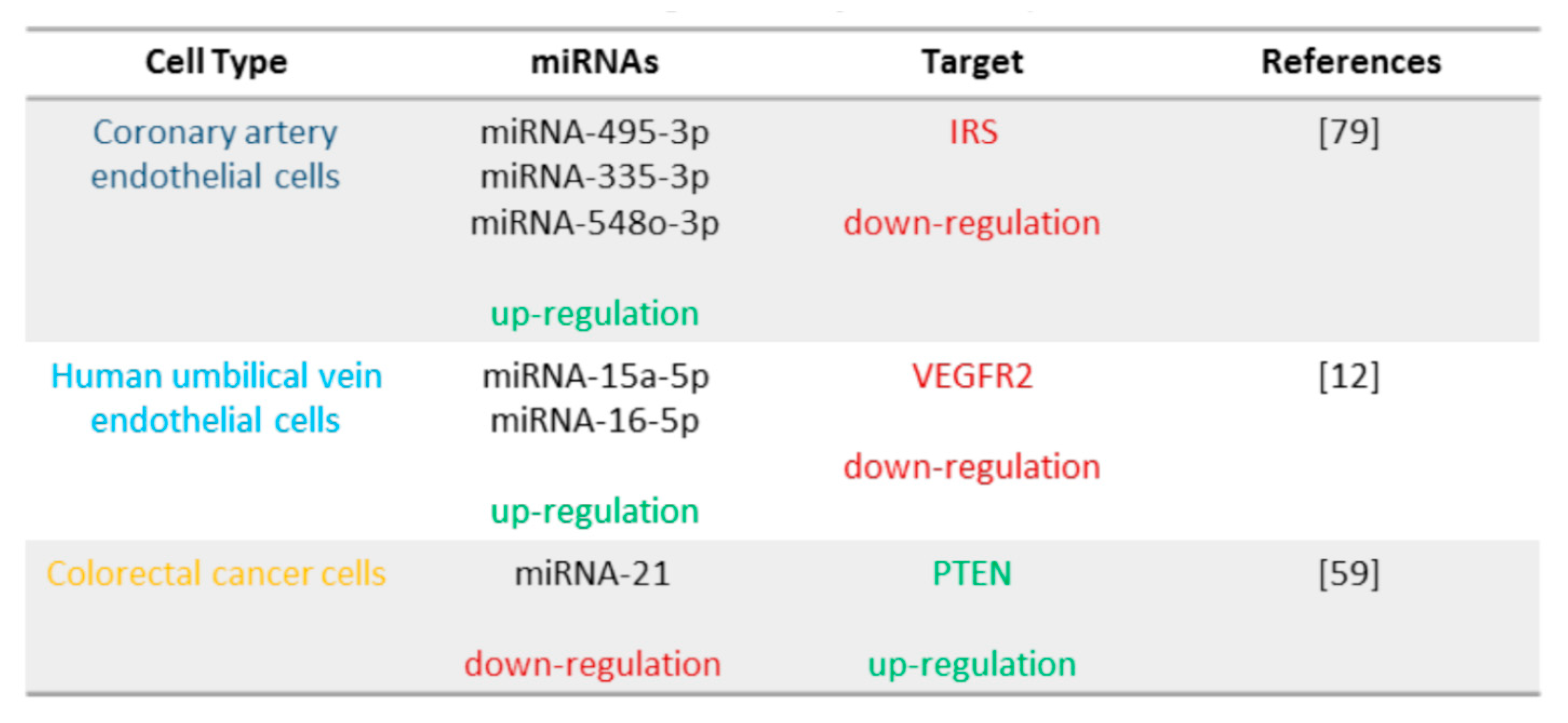
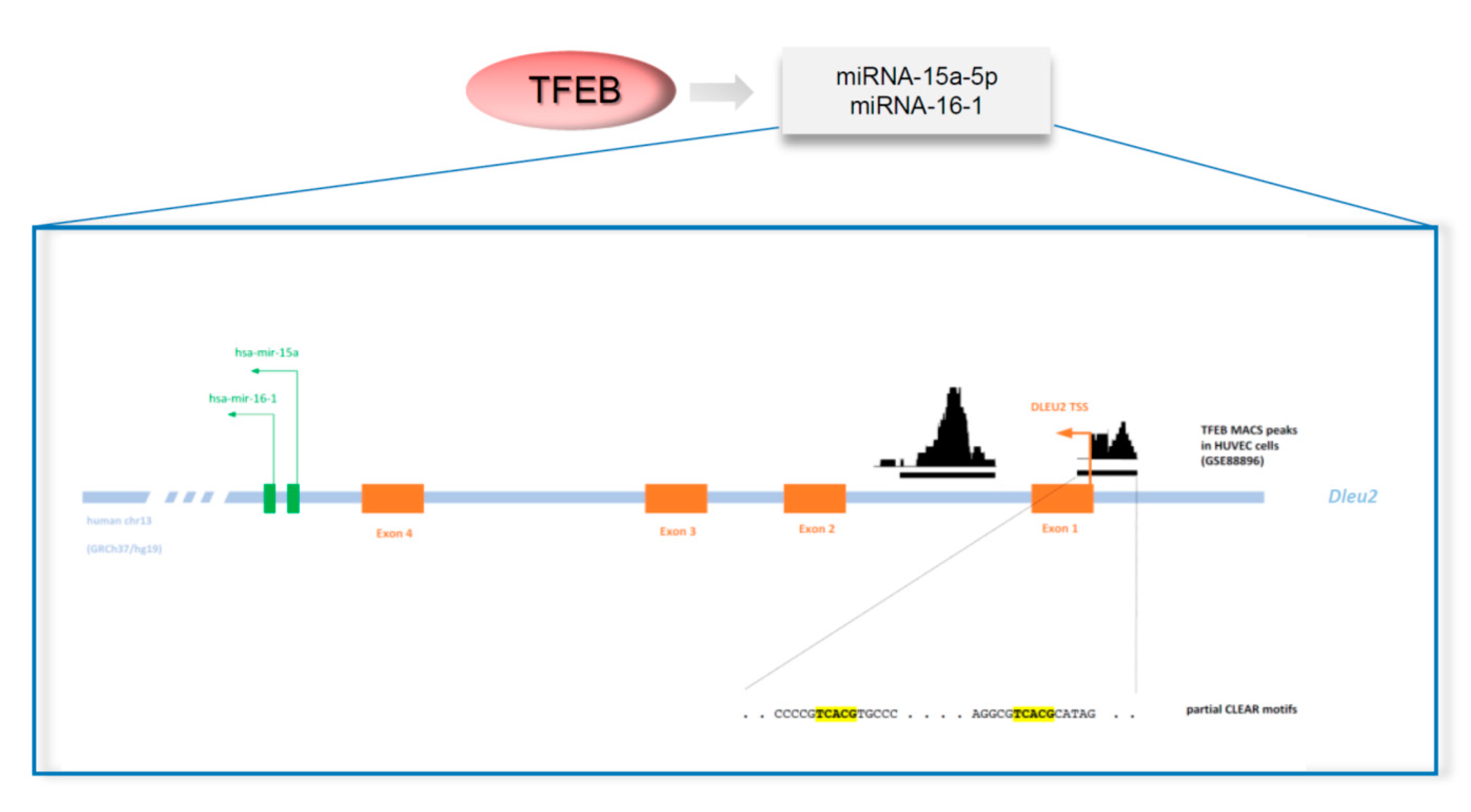
6.2. TFEB and VEGFR2: miRNA-15a-5p and miRNA-16-5p
6.3. TFEB and PTEN: miRNA-21
7. Conclusions and Perspectives
Author Contributions
Funding
Conflicts of Interest
References
- Glick, D.; Barth, S.; Macleod, K.F. Autophagy: Cellular and molecular mechanism. J. Pathol. 2010, 221, 3–12. [Google Scholar] [CrossRef] [PubMed]
- Saha, S.; Panigrahi, D.P.; Patil, S.; Bhutia, S.K. Autophagy in health and disease: A comprehensive review. Biomed. Pharmacother. 2018, 104, 485–495. [Google Scholar] [CrossRef] [PubMed]
- Slade, L.; Pulinilkunnil, T. The MiTF/TFE Family of Transcription Factors: Master Regulators of Organelle Signaling, Metabolism, and Stress Adaptation. Mol. Cancer Res. 2017, 15, 1637–1643. [Google Scholar] [CrossRef] [PubMed]
- Goding, C.R.; Arnheiter, H. MITF-the first 25 years. Genes Dev. 2019, 33, 983–1007. [Google Scholar] [CrossRef]
- Steingrímsson, E.; Tessarollo, L.; Reid, S.W.; Jenkins, N.A.; Copeland, N.G. The bHLH-Zip transcription factor Tfeb is essential for placental vascularization. Development 1998, 125, 4607–4616. [Google Scholar] [CrossRef]
- Hallsson, J.H.; Haflidadóttir, B.S.; Stivers, C.; Odenwald, W.; Arnheiter, H.; Pignoni, F.; Steingrímsson, E. The basic helix-loop-helix leucine zipper transcription factor Mitf is conserved in Drosophila and functions in eye development. Genetics 2004, 167, 233–241. [Google Scholar] [CrossRef] [PubMed]
- Lister, J.A.; Lane, B.M.; Nguyen, A.; Lunney, K. Embryonic expression of zebrafish MiT family genes tfe3b, tfeb, and tfec. Dev. Dyn. 2011, 240, 2529–2538. [Google Scholar] [CrossRef]
- Lapierre, L.R.; De Magalhaes Filho, C.D.; McQuary, P.R.; Chu, C.C.; Visvikis, O.; Chang, J.T.; Gelino, S.; Ong, B.; Davis, A.E.; Irazoqui, J.E.; et al. The TFEB orthologue HLH-30 regulates autophagy and modulates longevity in Caenorhabditis elegans. Nat. Commun. 2013, 4, 2267. [Google Scholar] [CrossRef]
- Kuiper, R.P.; Schepens, M.; Thijssen, J.; Schoenmakers, E.F.; van Kessel, A.G. Regulation of the MiTF/TFE bHLH-LZ transcription factors through restricted spatial expression and alternative splicing of functional domains. Nucleic Acids Res. 2004, 32, 2315–2322. [Google Scholar] [CrossRef]
- Fisher, D.E.; Carr, C.S.; Parent, L.A.; Sharp, P.A. TFEB has DNA-binding and oligomerization properties of a unique helix-loop-helix/leucine-zipper family. Genes Dev. 1991, 5, 2342–2352. [Google Scholar] [CrossRef]
- Palmieri, M.; Impey, S.; Kang, H.; di Ronza, A.; Pelz, C.; Sardiello, M.; Ballabio, A. Characterization of the CLEAR network reveals an integrated control of cellular clearance pathways. Hum. Mol. Genet. 2011, 20, 3852–3866. [Google Scholar] [CrossRef]
- Doronzo, G.; Astanina, E.; Corà, D.; Chiabotto, G.; Comunanza, V.; Noghero, A.; Neri, F.; Puliafito, A.; Primo, L.; Spampanato, C.; et al. TFEB controls vascular development by regulating the proliferation of endothelial cells. EMBO J. 2019, 38, e98250. [Google Scholar] [CrossRef] [PubMed]
- Sardiello, M.; Palmieri, M.; di Ronza, A.; Medina, D.L.; Valenza, M.; Gennarino, V.A.; Di Malta, C.; Donaudy, F.; Embrione, V.; Polishchuk, R.S.; et al. A gene network regulating lysosomal biogenesis and function. Science 2009, 325, 473–477. [Google Scholar] [CrossRef]
- Settembre, C.; Di Malta, C.; Polito, V.A.; Garcia Arencibia, M.; Vetrini, F.; Erdin, S.; Erdin, S.U.; Huynh, T.; Medina, D.; Colella, P.; et al. TFEB links autophagy to lysosomal biogenesis. Science 2011, 332, 1429–1433. [Google Scholar] [CrossRef] [PubMed]
- Astanina, E.; Bussolino, F.; Doronzo, G. Multifaceted activities of transcription factor eb in cancer onset and progression. Mol. Oncol. 2020, 15, 327–346. [Google Scholar] [CrossRef]
- Doronzo, G.; Astanina, E.; Bussolino, F. The Oncogene Transcription Factor EB Regulates Vascular Functions. Front. Physiol. 2021, 12, 640061. [Google Scholar] [CrossRef]
- Martina, J.A.; Diab, H.I.; Brady, O.A.; Puertollano, R. TFEB and TFE 3 are novel components of the integrated stress response. EMBO J. 2016, 35, 479–495. [Google Scholar] [CrossRef] [PubMed]
- Mansueto, G.; Armani, A.; Viscomi, C.; D’Orsi, L.; De Cegli, R.; Polishchuk, E.V.; Lamperti, C.; Di Meo, I.; Romanello, V.; Marchet, S.; et al. Transcription Factor EB Controls Metabolic Flexibility during Exercise. Cell Metab. 2017, 25, 182–196. [Google Scholar] [CrossRef] [PubMed]
- Napolitano, G.; Ballabio, A. TFEB at a glance. J. Cell Sci. 2016, 13, 2475–2481. [Google Scholar] [CrossRef]
- Yu, Z.; Wang, L.; Yang, D.L. The regulation of TFEB in lipid homeostasis of non-alcoholic fatty liver disease: Molecular mechanism and promising therapeutic targets. Life Sci. 2020, 246, 117418. [Google Scholar] [CrossRef]
- Nabar, N.R.; Kehrl, J.H. The Transcription Factor EB Links Cellular Stress to the Immune Response. Yale J. Biol. Med. 2017, 90, 301–315. [Google Scholar]
- Bartel, D.P. Metazoan MicroRNAs. Cell 2018, 173, 20–51. [Google Scholar] [CrossRef]
- Cora’, D.; Re, A.; Caselle, M.; Bussolino, F. MicroRNA-mediated regulatory circuits: Outlook and perspectives. Phys. Biol. 2017, 14, 045001. [Google Scholar] [CrossRef]
- Bracken, C.P.; Scott, H.S.; Goodall, G.J. A network-biology perspective of microRNA function and dysfunction in cancer. Nat. Rev. Genet. 2016, 17, 719–732. [Google Scholar] [CrossRef] [PubMed]
- Cosentino, G.; Plantamura, I.; Cataldo, A.; Iorio, M.V. MicroRNA and oxidative stress interplay in the context of breast cancer pathogenesis. Int. J. Mol. Sci. 2019, 20, 5143. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Li, X.; Xiao, Z.; Wang, Y.; Han, Y.; Li, J.; Zhu, W.; Leng, Q.; Wen, Y.; Wen, X. MicroRNA-488 inhibits proliferation and glycolysis in human prostate cancer cells by regulating PFKFB3. FEBS Open Bio 2019, 9, 1798–1807. [Google Scholar] [CrossRef] [PubMed]
- Cheleschi, S.; Tenti, S.; Mondanelli, N.; Corallo, C.; Barbarino, M.; Giannotti, S.; Gallo, I.; Giordano, A.; Fioravanti, A. MicroRNA-34a and MicroRNA-181a mediate visfatin-induced apoptosis and oxidative stress via NF-κB pathway in human osteoarthritic chondrocytes. Cells 2019, 8, 874. [Google Scholar] [CrossRef] [PubMed]
- Shirjang, S.; Mansoori, B.; Asghari, S.; Duijf, P.H.G.; Mohammadi, A.; Gjerstorff, M.; Baradaran, B. MicroRNAs in cancer cell death pathways: Apoptosis and necroptosis. Free Radic. Biol. Med. 2019, 139, 1–15. [Google Scholar]
- Wang, D.; Weng, Y.; Guo, S.; Qin, W.; Ni, J.; Yu, L.; Zhang, Y.; Zhao, Q.; Ben, J.; Ma, J. microRNA-1 regulates NCC migration and differentiation by targeting sec63. Int. J. Biol. Sci. 2019, 15, 2538–2547. [Google Scholar] [CrossRef]
- Zhao, Y.; Wang, Z.; Zhang, W.; Zhang, L. MicroRNAs play an essential role in autophagy regulation in various disease phenotypes. Biofactors 2019, 45, 844–856. [Google Scholar] [CrossRef]
- Akkoc, Y.; Gozuacik, D. MicroRNAs as major regulators of the autophagy pathway. Biochim. Biophys Acta Mol. Cell Res. 2020, 1867, 118662. [Google Scholar] [CrossRef]
- Sun, J.; Lu, H.; Liang, W.; Zhao, G.; Ren, L.; Hu, D.; Chang, Z.; Liu, Y.; Garcia-Barrio, M.T.; Zhang, J.; et al. Endothelial TFEB (Transcription Factor EB) Improves Glucose Tolerance via Upregulation of IRS (Insulin Receptor Substrate) 1 and IRS2. Arterioscler. Thromb. Vasc. Biol. 2020, 41, 783–795. [Google Scholar] [CrossRef]
- Zhang, Y.D.; Zhao, J.J. TFEB Participates in the A-Induced Pathogenesis of Alzheimer’s Disease by Regulating the Autophagy-Lysosome Pathway. DNA Cell Biol. 2015, 34, 661–668. [Google Scholar] [CrossRef] [PubMed]
- Shang, Q.; Shen, G.; Chen, G.; Zhang, Z.; Yu, X.; Zhao, W.; Zhang, P.; Chen, H.; Tang, K.; Yu, F.; et al. The emerging role of miR-128 in musculoskeletal diseases. J. Cell. Physiol. 2021, 236, 4231–4243. [Google Scholar] [CrossRef] [PubMed]
- Shi, Z.M.; Wang, J.; Yan, Z.; You, Y.P.; Li, C.Y.; Qian, X.; Yin, Y.; Zhao, P.; Wang, Y.Y.; Wang, X.F.; et al. MiR-128 inhibits tumor growth and angiogenesis by targeting p70S6K1. PLoS ONE 2012, 7, e32709. [Google Scholar] [CrossRef] [PubMed]
- Gennarino, V.A.; Sardiello, M.; Avellino, R.; Meola, N.; Maselli, V.; Anand, S.; Cutillo, L.; Ballabio, A.; Banfi, S. MicroRNA target prediction by expression analysis of host genes. Genome Res. 2009, 19, 481–490. [Google Scholar] [CrossRef]
- Tiribuzi, R.; Crispoltoni, L.; Porcellati, S.; Di Lullo, M.; Florenzano, F.; Pirro, M.; Bagaglia, F.; Kawarai, T.; Zampolini, M.; Orlacchio, A.; et al. miR128 up-regulation correlates with impaired amyloid β(1-42) degradation in monocytes from patients with sporadic Alzheimer’s disease. Neurobiol. Aging 2014, 35, 345–356. [Google Scholar] [CrossRef]
- Wang, W.X.; Huang, Q.; Hu, Y.; Stromberg, A.J.; Nelson, P.T. Patterns of microRNA expression in normal and early Alzheimer’s disease human temporal cortex: White matter versus gray matter. Acta Neuropathol. 2011, 121, 193–205. [Google Scholar] [CrossRef] [PubMed]
- Horita, M.; Farquharson, C.; Stephen, L.A. The role of miR-29 family in disease. J. Cell Biochem. 2021, 122, 696–715. [Google Scholar] [CrossRef] [PubMed]
- Kwon, J.J.; Willy, J.A.; Quirin, K.A.; Wek, R.C.; Korc, M.; Yin, X.M.; Kota, J. Novel role of miR-29a in pancreatic cancer autophagy and its therapeutic potential. Oncotarget 2016, 7, 71635–71650. [Google Scholar] [CrossRef]
- Cai, J.; Zhang, H.; Zhang, Y.F.; Zhou, Z.; Wu, S. MicroRNA-29 enhances autophagy and cleanses exogenous mutant alphaB-crystallin in retinal pigment epithelial cells. Exp. Cell Res. 2019, 374, 231–248. [Google Scholar] [CrossRef]
- Wu, K.; Zhao, Q.; Li, Z.; Li, N.; Xiao, Q.; Li, X.; Zhao, Q. Bioinformatic screening for key miRNAs and genes associated with myocardial infarction. FEBS Open Bio 2018, 8, 897–913. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Liu, H.; Murphy, J.T.; Foyil, S.R.; Godar, R.J.; Abuirqeba, H.; Weinheimer, C.J.; Barger, P.M.; Diwan, A. Regulation of the transcription factor EB-PGC1α axis by beclin-1 controls mitochondrial quality and cardiomyocyte death under stress. Mol. Cell Biol. 2015, 35, 956–976. [Google Scholar] [CrossRef]
- Liu, H.; Lei, C.; He, Q.; Pan, Z.; Xiao, D.; Tao, Y. Nuclear functions of mammalian MicroRNAs in gene regulation, immunity and cancer. Mol. Cancer 2018, 17, 64. [Google Scholar] [CrossRef] [PubMed]
- Mao, L.; Liu, S.; Hu, L.; Jia, L.; Wang, H.; Guo, M.; Chen, C.; Liu, Y.; Xu, L. miR-30 Family: A Promising Regulator in Development and Disease. BioMed Res. Int. 2018, 9623412. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.; Pu, M.; Tai, Y.; Chen, Y.; Lu, H.; Qiao, J.; Wang, G.; Chen, J.; Qi, X.; Huang, R.; et al. Nuclear miR-30b-5p suppresses TFEB-mediated lysosomal biogenesis and autophagy. Cell Death Differ. 2021, 28, 320–336. [Google Scholar] [CrossRef] [PubMed]
- Nardozzi, J.D.; Lott, K.; Cingolani, G. Phosphorylation meets nuclear import: A review. Cell Commun. Signal. 2010, 8, 32. [Google Scholar] [CrossRef]
- Puertollano, R.; Ferguson, S.M.; Brugarolas, J.; Ballabio, A. The complex relationship between TFEB transcription factor phosphorylation and subcellular localization. EMBO J. 2018, 37, e98804. [Google Scholar] [CrossRef]
- Visvikis, O.; Ihuegbu, N.; Labed, S.A.; Luhachack, L.G.; Alves, A.F.; Wollenberg, A.C.; Stuart, L.M.; Stormo, G.D.; Irazoqui, J.E. Innate host defense requires TFEB-mediated transcription of cytoprotective and antimicrobial genes. Immunity 2014, 40, 896–909. [Google Scholar] [CrossRef]
- Pastore, N.; Brady, O.A.; Diab, H.I.; Martina, J.A.; Sun, L.; Huynh, T.; Lim, J.A.; Zare, H.; Raben, N.; Ballabio, A.; et al. TFEB and TFE3 cooperate in the regulation of the innate immune response in activated macrophages. Autophagy 2016, 12, 1240–1258. [Google Scholar] [CrossRef]
- Gray, M.A.; Choy, C.H.; Dayam, R.M.; Ospina-Escobar, E.; Somerville, A.; Xiao, X.; Ferguson, S.M.; Botelho, R.J. Phagocytosis enhances lysosomal and bactericidal properties by activating the transcription factor TFEB. Curr. Biol. 2016, 26, 1955–1964. [Google Scholar] [CrossRef] [PubMed]
- Irazoqui, J.E. Key Roles of MiT Transcription Factors in Innate Immunity and Inflammation. Trends Immunol. 2020, 41, 157–171. [Google Scholar] [CrossRef] [PubMed]
- Nezich, C.L.; Wang, C.; Fogel, A.I.; Youle, R.J. MiT/TFE transcription factors are activated during mitophagy downstream of Parkin and Atg5. J. Cell Biol. 2015, 210, 435–450. [Google Scholar] [CrossRef] [PubMed]
- Laplante, M.; Sabatini, D.M. mTOR signaling in growth control and disease. Cell 2012, 149, 274–293. [Google Scholar] [CrossRef]
- Zhang, Y.; Huang, B.; Wang, H.Y.; Chang, A.; Zheng, X.F.S. Emerging Role of MicroRNAs in mTOR Signaling. Cell. Mol. Life Sci. 2017, 74, 2613–2625. [Google Scholar] [CrossRef]
- Mazar, J.; DeYoung, K.; Khaitan, D.; Meister, E.; Almodovar, A.; Goydos, J.; Ray, A.; Perera, R.J. The regulation of miRNA-211 expression and its role in melanoma cell invasiveness. PLoS ONE 2010, 5, e13779. [Google Scholar] [CrossRef]
- Ozturk, D.G.; Kocak, M.; Akcay, A.; Kinoglu, K.; Kara, E.; Buyuk, Y.; Kazan, H.; Gozuacik, D. MITF-MIR211 axis is a novel autophagy amplifier system during cellular stress. Autophagy 2019, 15, 375–390. [Google Scholar] [CrossRef]
- Ribas, J.; Ni, X.; Castanares, M.; Liu, M.M.; Esopi, D.; Yegnasubramanian, S.; Rodriguez, R.; Mendell, J.T.; Lupold, S.E. A novel source for miR-21 expression through the alternative polyadenylation of VMP1 gene transcripts. Nucleic. Acids Res. 2012, 40, 6821–6833. [Google Scholar] [CrossRef]
- Wang, C.; Peng, R.; Zeng, M.; Zhang, Z.; Liu, S.; Jiang, D.; Lu, Y.; Zou, F. An autoregulatory feedback loop of miR-21/VMP1 is responsible for the abnormal expression of miR-21 in colorectal cancer cells. Cell Death Dis. 2020, 11, 1067. [Google Scholar] [CrossRef]
- Hardie, D.G.; Schaffer, B.E.; Brunet, A. AMPK: An Energy-Sensing Pathway with Multiple Inputs and Outputs. Trends Cell Biol. 2016, 26, 190–201. [Google Scholar] [CrossRef]
- González, A.; Hall, M.N.; Lin, S.C.; Hardie, D.G. AMPK and TOR: The Yin and Yang of Cellular Nutrient Sensing and Growth Control. Cell Metab. 2020, 31, 472–492. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Chen, Y. AMPK and Autophagy. Adv. Exp. Med. Biol. 2019, 1206, 85–108. [Google Scholar]
- Collodet, C.; Foretz, M.; Deak, M.; Bultot, L.; Metairon, S.; Viollet, B.; Lefebvre, G.; Raymond, F.; Parisi, A.; Civiletto, G.; et al. AMPK promotes induction of the tumor suppressor FLCN through activation of TFEB independently of mTOR. FASEB J. 2019, 33, 12374–12391. [Google Scholar] [CrossRef]
- Paquette, M.; El-Houjeiri, L.C.; Zirden, L.; Puustinen, P.; Blanchette, P.; Jeong, H.; Dejgaard, K.; Siegel, P.M.; Pause, A. AMPK-dependent phosphorylation is required for transcriptional activation of TFEB and TFE3. Autophagy 2021, 18, 1–19. [Google Scholar] [CrossRef]
- Xu, Z.; Li, Z.; Wang, W.; Xia, Y.; He, Z.; Li, B.; Wang, S.; Huang, X.; Sun, G.; Xu, J.; et al. MIR-1265 regulates cellular proliferation and apoptosis by targeting calcium binding protein 39 in gastric cancer and, thereby, impairing oncogenic autophagy. Cancer Lett. 2019, 449, 226–236. [Google Scholar] [CrossRef]
- Bronisz, A.; Chiocca, E.A.; Godlewski, J. Response to energy depletion: miR-451/AMPK loop. Oncotarget 2015, 6, 17851–17852. [Google Scholar] [CrossRef]
- Li, S.; Zeng, X.; Ma, R.; Wang, L. MicroRNA-21 promotes the proliferation, migration and invasion of non-small cell lung cancer A549 cells by regulating autophagy activity via AMPK/ULK1 signaling pathway. Exp. Ther. Med. 2018, 16, 2038–2045. [Google Scholar] [CrossRef]
- Zhang, Y.J.; Pan, Q.; Yu, Y.; Zhong, X.P. microRNA-519d Induces Autophagy and Apoptosis of Human Hepatocellular Carcinoma Cells Through Activation of the AMPK Signaling Pathway via Rab10. Cancer Manag. Res. 2020, 12, 2589–2602. [Google Scholar] [CrossRef] [PubMed]
- Ouimet, M.; Koster, S.; Sakowski, E.; Ramkhelawon, B.; van Solingen, C.; Oldebeken, S.; Karunakaran, D.; Portal-Celhay, C.; Sheedy, F.J.; Ray, T.D.; et al. Mycobacterium tuberculosis induces the miR-33 locus to reprogram autophagy and host lipid metabolism. Nat. Immunol. 2016, 17, 677–686. [Google Scholar] [CrossRef]
- Dávalos, A.; Goedeke, L.; Smibert, P.; Ramírez, C.M.; Warrier, N.P.; Andreo, U.; Cirera-Salinas, D.; Rayner, K.; Suresh, U.; Pastor-Pareja, J.C.; et al. miR-33a/b contribute to the regulation of fatty acid metabolism and insulin signaling. Proc. Natl. Acad Sci. USA 2011, 108, 9232–9237. [Google Scholar] [CrossRef] [PubMed]
- Medina, D.L.; Ballabio, A. Lysosomal calcium regulates autophagy. Autophagy 2015, 11, 970–971. [Google Scholar] [CrossRef] [PubMed]
- Naso, F.; Intartaglia, D.; Falanga, D.; Soldati, C.; Polishchuk, E.; Giamundo, G.; Tiberi, P.; Marrocco, E.; Scudieri, P.; Di Malta, C.; et al. Light-responsive microRNA miR-211 targets Ezrin to modulate lysosomal biogenesis and retinal cell clearance. EMBO J. 2020, 39, e102468. [Google Scholar] [CrossRef]
- Mizushima, N. Autophagy: Process and function. Genes Dev. 2007, 21, 2861–2873. [Google Scholar] [CrossRef]
- Ghafouri-Fard, S.; Shoorei, H.; Mohaqiq, M.; Majidpoor, J.; Moosavi, M.A.; Taheri, M. Exploring the role of non-coding RNAs in autophagy. Autophagy 2021, 18, 1–22. [Google Scholar] [CrossRef] [PubMed]
- Shan, C.; Chen, X.; Cai, H.; Hao, X.; Li, J.; Zhang, Y.; Gao, J.; Zhou, Z.; Li, X.; Liu, C.; et al. The Emerging Roles of Autophagy-Related MicroRNAs in Cancer. Int. J. Biol. Sci. 2021, 17, 134–150. [Google Scholar] [CrossRef]
- Lu, H.; Fan, Y.; Qiao, C.; Liang, W.; Hu, W.; Zhu, T.; Zhang, J.; Chen, Y.E. TFEB inhibits endothelial cell inflammation and reduces atherosclerosis. Sci. Signal. 2017, 10, eaah4214. [Google Scholar] [CrossRef]
- Fan, Y.; Lu, H.; Liang, W.; Garcia-Barrio, M.T.; Guo, Y.; Zhang, J.; Zhu, T.; Hao, Y.; Zhang, J.; Chen, Y.E. Endothelial TFEB (Transcription Factor EB) positively regulates postischemic angiogenesis. Circ. Res. 2018, 122, 945–957. [Google Scholar] [CrossRef]
- Wang, X.; Xiao, H.; Wu, D.; Zhang, D.; Zhang, Z. miR-335-5p Regulates Cell Cycle and Metastasis in Lung Adenocarcinoma by Targeting CCNB2. OncoTargets Ther. 2020, 13, 6255–6263. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Wu, J.; Jiao, K.; Wu, Q.; Ma, J.; Chen, D.; Kang, J.; Zhao, G.; Shi, Y.; Fan, D.; et al. MicroRNA-495-3p inhibits multidrug resistance by modulating autophagy through GRP78/mTOR axis in gastric cancer. Cell Death Dis. 2018, 9, 1070. [Google Scholar] [CrossRef]
- Chamorro-Jorganes, A.; Araldi, E.; Suárez, Y. MicroRNAs as pharmacological targets in endothelial cell function and dysfunction. Pharmacol. Res. 2013, 75, 15–27. [Google Scholar] [CrossRef]
- Dang, L.T.; Lawson, N.D.; Fish, J.E. MicroRNA control of vascular endothelial growth factor signaling output during vascular development. Arterioscler. Thromb. Vasc. Biol. 2013, 33, 193–200. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lerner, M.; Harada, M.; Lovén, J.; Castro, J.; Davis, Z.; Oscier, D.; Henriksson, M.; Sangfelt, O.; Grandér, D.; Corcoran, M.M. DLEU2, frequently deleted in malignancy, functions as a critical host gene of the cell cycle inhibitory microRNAs miR-15a and miR-16–1. Exp. Cell Res. 2009, 315, 2941–2952. [Google Scholar] [CrossRef]
- Zhou, B.; Chen, H.; Wei, D.; Kuang, Y.; Zhao, X.; Li, G.; Xie, J.; Chen, P. A novel miR-219-SMC4-JAK2/Stat3 regulatory pathway in human hepatocellular carcinoma. J. Exp. Clin. Cancer Res. 2014, 33, 55. [Google Scholar] [CrossRef] [PubMed]
- Yue, J.; Tigyi, G. Conservation of miR-15a/16-1 and miR-15b/16-2 clusters. Mamm. Genome 2010, 21, 88–94. [Google Scholar] [CrossRef]
- Di Malta, C.; Cinque, L.; Settembre, C. Transcriptional Regulation of Autophagy: Mechanisms and Diseases. Front. Cell Dev. Biol. 2019, 7, 114. [Google Scholar] [CrossRef] [PubMed]
- Ferracin, M.; Negrini, M. Micromarkers 2.0: An update on the role of microRNAs in cancer diagnosis and prognosis. Expert Rev. Mol. Diagn. 2015, 15, 1369–1381. [Google Scholar] [CrossRef] [PubMed]
- Bhatti, G.K.; Khullar, N.; Sidhu, I.S.; Navik, U.S.; Reddy, A.P.; Reddy, P.H.; Bhatti, J.S. Emerging role of non-coding RNA in health and disease. Metab. Brain Dis. 2021, 21, 1–16. [Google Scholar]
- Chen, S.; Xue, Y.; Wu, X.; Le, C.; Bhutkar, A.; Bell, E.L.; Zhang, F.; Langer, R.; Sharp, P.A. Global microRNA depletion suppresses tumor angiogenesis. Genes Dev. 2014, 28, 1054–1067. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Corà, D.; Bussolino, F.; Doronzo, G. TFEB Signalling-Related MicroRNAs and Autophagy. Biomolecules 2021, 11, 985. https://doi.org/10.3390/biom11070985
Corà D, Bussolino F, Doronzo G. TFEB Signalling-Related MicroRNAs and Autophagy. Biomolecules. 2021; 11(7):985. https://doi.org/10.3390/biom11070985
Chicago/Turabian StyleCorà, Davide, Federico Bussolino, and Gabriella Doronzo. 2021. "TFEB Signalling-Related MicroRNAs and Autophagy" Biomolecules 11, no. 7: 985. https://doi.org/10.3390/biom11070985
APA StyleCorà, D., Bussolino, F., & Doronzo, G. (2021). TFEB Signalling-Related MicroRNAs and Autophagy. Biomolecules, 11(7), 985. https://doi.org/10.3390/biom11070985