Can Glycosylation Mask the Detection of MHC Expressing p53 Peptides by T Cell Receptors?
Abstract
:1. Introduction
2. Methods
2.1. Model Building
2.1.1. Model of p53 and Its Glycosylation
2.1.2. Model of p161 Complexed to A24
- A dimeric complex of A24 and an antigenic peptide (RFPLTFGWCF) from HIV and a trimeric complex of A24, peptide andTCR. Both these complexes were constructed from a crystal structure of the trimeric complex of A24 with the HIV peptide and TCR (PDB ID: 3VXU, resolution 2.7 Å [32]). The A24-HIV peptide dimer and the A24-HIV peptide-TCR trimer were simulated to examine the ability of our force field to generate stable complexes of these systems during the MD simulations;
- A dimeric complex of A24 and p161 and a trimeric complex of A24, p161 and TCR were constructed to examine the structural dynamics of p161 in complex with A24 and TCR. The dimer was constructed using the crystal structure of the complex between A24 and a SARS peptide (PDB ID: 3I6L, resolution 2.4 Å [31]), while the trimer was built using the crystal structure of the trimeric complex of A24-HIV-TCR (PDB ID: 3VXU, resolution 2.7 Å). The reason we did not build the dimer from the same structure as the trimer is because the peptide in the trimer is 10 amino acids long, while our p161 peptide is only 9 amino acids long;
- A dimeric complex of A24 and Galp161 (a monosaccharide GalNAc attached to the Oγ atom of the residue S166 in p161) and a trimeric complex of A24, Galp161 and TCR were constructed to examine the structural dynamics of the effects of glycosylation of p161 with GalNAc in complex with A24 and TCR;
- A dimeric complex of A24 and Glcp161 (a monosaccharide GlcNAc attached to the Oγ atom of the residue S166 in p161) and a trimeric complex of A24, Glcp161 and TCR were constructed to examine the structural dynamics of the effects of glycosylation of p161 with GlcNAc in complex with A24 and TCR.
2.2. Molecular Dynamics (MD) Simulations
3. Results
3.1. Benchmarks with the Complex of A24 and the HIV Peptide
3.2. Structural Dynamics of A24 Complexed to the p161 Peptides
3.2.1. Hydrogen Bond (Hbond) Interactions
3.2.2. Energetics of Interactions of Peptides and A24
3.3. The Interactions of TCR with the A24-Peptide Complexes
3.3.1. RMSD and RMSF
3.3.2. Binding Energy
3.3.3. Hbond Interactions between A24 and Peptide
3.3.4. Hbond Interactions between Peptide and TCR
3.3.5. Hbond Interactions between A24 and TCR
3.4. The Proximity of TCR to the A24-Peptide Complexes
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dutz, J.P.; Tsomides, T.J.; Kageyama, S.; Rasmussen, M.H.; Eisen, H.N. A cytotoxic T lymphocyte clone can recognize the same naturally occurring self peptide in association with a self and nonself class I MHC protein. Mol. Immunol. 1994, 31, 967–975. [Google Scholar] [CrossRef]
- Rock, K.L.; York, I.A.; Goldberg, A.L. Post-proteasomal antigen processing for major histocompatibility complex class I presentation. Nat. Immunol. 2004, 5, 670–677. [Google Scholar] [CrossRef] [PubMed]
- Kubo, R.T.; Sette, A.; Grey, H.M.; Appella, E.; Sakaguchi, K.; Zhu, N.Z.; Arnott, D.; Sherman, N.; Shabanowitz, J.; Michel, H.; et al. Definition of specific peptide motifs for four major HLA-A alleles. J. Immunol. 1994, 152, 3913–3924. [Google Scholar] [PubMed]
- Lane, D.P. Cancer. p53, guardian of the genome. Nature 1992, 358, 15–16. [Google Scholar] [CrossRef]
- Vousden, K.H.; Lane, D.P. p53 in health and disease. Nat. Rev. Mol. Cell Biol. 2007, 8, 275–283. [Google Scholar] [CrossRef]
- Mantovani, F.; Collavin, L.; Del Sal, G. Mutant p53 as a guardian of the cancer cell. Cell Death Differ. 2019, 26, 199–212. [Google Scholar] [CrossRef] [PubMed]
- De Leo, A.B. p53-based immunotherapy of cancer. Approaches ro reversing unresponsiveness to T lymphocytes and preventing tumor escape. Adv. Otorhinolaryngol. 2005, 62, 134–150. [Google Scholar] [PubMed]
- Hsiue, E.H.; Wright, K.M.; Douglass, J.; Hwang, M.S.; Mog, B.J.; Pearlman, A.H.; Paul, S.; DiNapoli, S.R.; Konig, M.F.; Wang, Q.; et al. Targeting a neoantigen derived from a common TP53 mutation. Science 2021, 371, 6533. [Google Scholar] [CrossRef]
- Mizukoshi, E.; Nakamoto, Y.; Arai, K.; Yamashita, T.; Sakai, A.; Sakai, Y.; Kagaya, T.; Yamashita, T.; Honda, M.; Kaneko, S. Comparative analysis of various tumor-associated antigen-specific t-cell responses in patients with hepatocellular carcinoma. Hepatology 2011, 53, 1206–1216. [Google Scholar] [CrossRef] [PubMed]
- Terashima, T.; Mizukoshi, E.; Arai, K.; Yamashita, T.; Yoshida, M.; Ota, H.; Onishi, I.; Kayahara, M.; Ohtsubo, K.; Kagaya, T.; et al. P53, hTERT, WT-1, and VEGFR2 are the most suitable targets for cancer vaccine therapy in HLA-A24 positive pancreatic adenocarcinoma. Cancer Immunol. Immunother. 2014, 63, 479–489. [Google Scholar] [CrossRef] [PubMed]
- Hakomori, S. Glycosylation defining cancer malignancy: New wine in an old bottle. Proc. Natl. Acad. Sci. USA 2002, 99, 10231–10233. [Google Scholar] [CrossRef] [Green Version]
- Wolfert, M.A.; Boons, G.J. Adaptive immune activation: Glycosylation does matter. Nat. Chem. Biol. 2013, 9, 776–784. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lisowska, E. The role of glycosylation in protein antigenic properties. Cell. Mol. Life Sci. 2002, 59, 445–455. [Google Scholar] [CrossRef]
- Haurum, J.S.; Hoier, I.B.; Arsequell, G.; Neisig, A.; Valencia, G.; Zeuthen, J.; Neefjes, J.; Elliott, T. Presentation of cytosolic glycosylated peptides by human class I major histocompatibility complex molecules in vivo. J. Exp. Med. 1999, 190, 145–150. [Google Scholar] [CrossRef]
- Jensen, T.; Galli-Stampino, L.; Mouritsen, S.; Frische, K.; Peters, S.; Meldal, M.; Werdelin, O. T cell recognition of Tn-glycosylated peptide antigens. Eur. J. Immunol. 1996, 26, 1342–1349. [Google Scholar] [CrossRef]
- Haurum, J.S.; Tan, L.; Arsequell, G.; Frodsham, P.; Lellouch, A.C.; Moss, P.A.; Dwek, R.A.; McMichael, A.J.; Elliott, T. Peptide anchor residue glycosylation: Effect on class I major histocompatibility complex binding and cytotoxic T lymphocyte recognition. Eur. J. Immunol. 1995, 25, 3270–3276. [Google Scholar] [CrossRef]
- Olvera, A.; Cedeño, S.; Llano, A.; Mothe, B.; Sanchez, J.; Arsequell, G.; Brander, C. Does Antigen Glycosylation Impact the HIV-Specific T Cell Immunity? Front. Immunol. 2021, 11, 3561–3568. [Google Scholar] [CrossRef] [PubMed]
- Apostolopoulos, V.; Yuriev, E.; Ramsland, P.A.; Halton, J.; Osinski, C.; Li, W.; Plebanski, M.; Paulsen, H.; McKenzie, I.F.C. A glycopeptide in complex with MHC class I uses the GalNAc residue as an anchor. Proc. Natl. Acad. Sci. USA 2003, 100, 15029–15034. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, W.H.; Kim, J.E.; Nam, H.W.; Ju, J.W.; Kim, H.S.; Kim, Y.S.; Cho, J.W. Modification of p53 with O-linked N-acetylglucosamine regulates p53 activity and stability. Nat. Cell Biol. 2006, 8, 1074–1083. [Google Scholar] [CrossRef] [PubMed]
- Shtraizent, N.; De Rossi, C.; Nayar, S.; Sachidanandam, R.; Katz, L.S.; Prince, A.; Koh, A.P.; Vincek, A.; Hadas, Y.; Hoshida, Y.; et al. MPI depletion enhances O-GlcNAcylation of p53 and suppresses the Warburg effect. Elife 2017, 6, e22477. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Li, H.; Lu, L.; Jiang, J. Structures of human O-GlcNAcase and its complexes reveal a new substrate recognition mode. Nat. Struct. Mol. Biol. 2017, 24, 362–369. [Google Scholar] [CrossRef] [PubMed]
- Schimpl, M.; Borodkin, V.S.; Gray, L.J.; van Aalten, D.M. Synergy of peptide and sugar in O-GlcNAcase substrate recognition. Chem. Biol. 2012, 19, 173–178. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cejas, R.B.; Lorenz, V.; Garay, Y.C.; Irazoqui, F.J. Biosynthesis of O-N-acetylgalactosamine glycans in the human cell nucleus. J. Biol. Chem. 2019, 294, 2997–3011. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fults, D.; Brockmeyer, D.; Tullous, M.W.; Pedone, C.A.; Cawthon, R.M. p53 mutation and loss of heterozygosity on chromosomes 17 and 10 during human astrocytoma progression. Cancer Res. 1992, 52, 674–679. [Google Scholar]
- Mullany, L.K.; Wong, K.K.; Marciano, D.C.; Katsonis, P.; King-Crane, E.R.; Ren, Y.A.; Lichtarge, O.; Richards, J.S. Specific TP53 Mutants Overrepresented in Ovarian Cancer Impact CNV, TP53 Activity, Responses to Nutlin-3a, and Cell Survival. Neoplasia 2015, 17, 789–803. [Google Scholar] [CrossRef] [Green Version]
- Kishimoto, Y.; Murakami, Y.; Shiraishi, M.; Hayashi, K.; Sekiya, T. Aberrations of the p53 tumor suppressor gene in human non-small cell carcinomas of the lung. Cancer Res. 1992, 52, 4799–4804. [Google Scholar]
- GLYCAM-Web Server. 2019. Available online: http://glycam.org/ (accessed on 28 June 2021).
- Pancera, M.; Zhou, T.; Druz, A.; Georgiev, I.S.; Soto, C.; Gorman, J.; Huang, J.; Acharya, P.; Chuang, G.Y.; Ofek, G.; et al. Structure and immune recognition of trimeric pre-fusion HIV-1 Env. Nature 2014, 514, 455–461. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kitayner, M.; Rozenberg, H.; Kessler, N.; Rabinovich, D.; Shaulov, L.; Haran, T.E.; Shakked, Z. Structural basis of DNA recognition by p53 tetramers. Mol. Cell 2006, 22, 741–753. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Ku, X.; Tomioka, A.; Xie, W.; Liang, T.; Zou, X.; Cui, Y.; Sato, T.; Kaji, H.; Narimatsu, H.; et al. O-linked N-acetylgalactosamine modification is present on the tumor suppressor p53. Biochim. Biophys. Acta Gen. Subj. 2020, 1864, 129635. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Wu, P.; Gao, F.; Qi, J.; Kawana-Tachikawa, A.; Xie, J.; Vavricka, C.J.; Iwamoto, A.; Li, T.; Gao, G.F. Novel immunodominant peptide presentation strategy: A featured HLA-A*2402-restricted cytotoxic T-lymphocyte epitope stabilized by intrachain hydrogen bonds from severe acute respiratory syndrome coronavirus nucleocapsid protein. J. Virol. 2010, 84, 11849–11857. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shimizu, A.; Kawana-Tachikawa, A.; Yamagata, A.; Han, C.; Zhu, D.; Sato, Y.; Nakamura, H.; Koibuchi, T.; Carlson, J.; Martin, E.; et al. Structure of TCR and antigen complexes at an immunodominant CTL epitope in HIV-1 infection. Sci. Rep. 2013, 3, 3097. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mazza, C.; Auphan-Anezin, N.; Gregoire, C.; Guimezanes, A.; Kellenberger, C.; Roussel, A.; Kearney, A.; van der Merwe, P.A.; Schmitt-Verhulst, A.M.; Malissen, B. How much can a T-cell antigen receptor adapt to structurally distinct antigenic peptides? EMBO J. 2007, 26, 1972–1983. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eswar, N.; Webb, B.; Marti-Renom, M.A.; Madhusudhan, M.S.; Eramian, D.; Shen, M.Y.; Pieper, U.; Sali, A. Comparative protein structure modeling using MODELLER. In Current Protocols in Protein Science; John Wiley & Sons: Hoboken, NJ, USA, 2007; Chapter 2. [Google Scholar]
- Shen, M.Y.; Sali, A. Statistical potential for assessment and prediction of protein structures. Protein Sci. 2006, 15, 2507–2524. [Google Scholar] [CrossRef] [Green Version]
- Word, J.M.; Lovell, S.C.; Richardson, J.S.; Richardson, D.C. Asparagine and glutamine: Using hydrogen atom contacts in the choice of side-chain amide orientation. J. Mol. Biol. 1999, 285, 1735–1747. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, V.B.; Arendall, W.B., 3rd; Headd, J.J.; Keedy, D.A.; Immormino, R.M.; Kapral, G.J.; Murray, L.W.; Richardson, J.S.; Richardson, D.C. MolProbity: All-atom structure validation for macromolecular crystallography. Acta Cryst. D Biol. Crystallogr. 2010, 66 Pt 1, 12–21. [Google Scholar] [CrossRef] [Green Version]
- Kirschner, K.N.; Yongye, A.B.; Tschampel, S.M.; Gonzalez-Outeirino, J.; Daniels, C.R.; Foley, B.L.; Woods, R.J. GLYCAM06: A generalizable biomolecular force field. Carbohydrates. J. Comput. Chem. 2008, 29, 622–655. [Google Scholar] [CrossRef] [Green Version]
- Case, D.A.; Betz, R.M.; Cerutti, D.S.; Cheatham, T.E., III; Darden, T.A.; Duke, R.E.; Giese, T.J.; Gohlke, H.; Goetz, A.W.; Homeyer, N.; et al. AMBER 2016; University of California: San Francisco, CA, USA, 2016. [Google Scholar]
- Maier, J.A.; Martinez, C.; Kasavajhala, K.; Wickstrom, L.; Hauser, K.E.; Simmerling, C. ff14SB: Improving the Accuracy of Protein Side Chain and Backbone Parameters from ff99SB. J. Chem. Theory Comput. 2015, 11, 3696–3713. [Google Scholar] [CrossRef] [Green Version]
- Jorgensen, W.L.; Chandrasekhar, J.; Madura, J.D.; Impey, R.W.; Klein, M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983, 79, 926–935. [Google Scholar] [CrossRef]
- Izaguirre, J.A.; Catarello, D.P.; Wozniak, J.M.; Skeel, R.D. Langevin stabilization of molecular dynamics. J. Chem. Phys. 2001, 114, 2090–2098. [Google Scholar] [CrossRef]
- Berendsen, H.J.C.; Postma, J.P.M.; Gunsteren, W.F.V.; DiNola, A.; Haak, J.R. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 1984, 81, 3684–3690. [Google Scholar] [CrossRef] [Green Version]
- Darden, T.; York, D.; Pedersen, L. Particle mesh Ewald: An N⋅log(N) method for Ewald sums in large systems. J. Chem. Phys. 1993, 98, 10089–10092. [Google Scholar] [CrossRef] [Green Version]
- Miyamoto, S.; Kollman, P.A. Settle: An analytical version of the SHAKE and RATTLE algorithm for rigid water models. J. Comput. Chem. 1992, 13, 952–962. [Google Scholar] [CrossRef]
- Tsui, V.; Case, D.A. Molecular Dynamics Simulations of Nucleic Acids with a Generalized Born Solvation Model. J. Am. Chem. Soc. 2000, 122, 2489–2498. [Google Scholar] [CrossRef]
- Bashford, D.; Case, D.A. Generalized born models of macromolecular solvation effects. Annu. Rev. Phys. Chem. 2000, 51, 129–152. [Google Scholar] [CrossRef] [PubMed]
- Wang, E.; Sun, H.; Wang, J.; Wang, Z.; Liu, H.; Zhang, J.Z.H.; Hou, T. End-Point Binding Free Energy Calculation with MM/PBSA and MM/GBSA: Strategies and Applications in Drug Design. Chem. Rev. 2019, 119, 9478–9508. [Google Scholar] [CrossRef] [PubMed]
- Genheden, S.; Ryde, U. The MM/PBSA and MM/GBSA methods to estimate ligand-binding affinities. Expert Opin. Drug Discov. 2015, 10, 449–461. [Google Scholar] [CrossRef] [PubMed]
- Garstka, M.A.; Fish, A.; Celie, P.H.; Joosten, R.P.; Janssen, G.M.; Berlin, I.; Hoppes, R.; Stadnik, M.; Janssen, L.; Ovaa, H.; et al. The first step of peptide selection in antigen presentation by MHC class I molecules. Proc. Natl. Acad. Sci. USA 2015, 112, 1505–1510. [Google Scholar] [CrossRef] [Green Version]
- Knapp, B.; Deane, C.M. T-Cell Receptor Binding Affects the Dynamics of the Peptide/MHC-I Complex. J. Chem. Inf. Model 2016, 56, 46–53. [Google Scholar] [CrossRef]
- Dominguez, J.L.; Knapp, B. How peptide/MHC presence affects the dynamics of the LC13 T-cell receptor. Sci. Rep. 2019, 9, 2638. [Google Scholar] [CrossRef]
- von Boehmer, H.; Kisielow, P. Self-nonself discrimination by T cells. Science 1990, 248, 1369–1373. [Google Scholar] [CrossRef]
- Douglass, J.; Hsiue, E.H.; Mog, B.J.; Hwang, M.S.; DiNapoli, S.R.; Pearlman, A.H.; Miller, M.S.; Wright, K.M.; Azurmendi, P.A.; Wang, Q.; et al. Bispecific antibodies targeting mutant RAS neoantigens. Sci. Immunol. 2021, 6, eabd5515. [Google Scholar] [CrossRef] [PubMed]
- Harndahl, M.; Rasmussen, M.; Roder, G.; Dalgaard Pedersen, I.; Sorensen, M.; Nielsen, M.; Buus, S. Peptide-MHC class I stability is a better predictor than peptide affinity of CTL immunogenicity. Eur. J. Immunol. 2012, 42, 1405–1416. [Google Scholar] [CrossRef]
- Hafstrand, I.; Badia-Martinez, D.; Josey, B.J.; Norstrom, M.; Buratto, J.; Pellegrino, S.; Duru, A.D.; Sandalova, T.; Achour, A. Crystal structures of H-2Db in complex with the LCMV-derived peptides GP92 and GP392 explain pleiotropic effects of glycosylation on antigen presentation and immunogenicity. PLoS ONE 2017, 12, e0189584. [Google Scholar] [CrossRef] [Green Version]
- Zhu, K.; Wang, J.; Zhu, J.; Jiang, J.; Shou, J.; Chen, X. p53 induces TAP1 and enhances the transport of MHC class I peptides. Oncogene 1999, 18, 7740–7747. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blagih, J.; Buck, M.D.; Vousden, K.H. p53, cancer and the immune response. J. Cell Sci. 2020, 133, jcs237453. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dhatchinamoorthy, K.; Colbert, J.D.; Rock, K.L. Cancer Immune Evasion Through Loss of MHC Class I Antigen Presentation. Front. Immunol. 2021, 12, 636568. [Google Scholar] [CrossRef]
- Speetjens, F.M.; Kuppen, P.J.K.; Welters, M.J.P.; Essahsah, F.; Voet van den Brink, A.M.E.G.; Lantrua, M.G.K.; Valentijn, A.R.P.M.; Oostendorp, J.; Fathers, L.M.; Nijman, H.W.; et al. Induction of p53-Specific Immunity by a p53 Synthetic Long Peptide Vaccine in Patients Treated for Metastatic Colorectal Cancer. Clin. Cancer Res. 2009, 15, 1086–1095. [Google Scholar] [CrossRef] [Green Version]
- Fedoseyeva, E.V.; Boisgérault, F.; Anosova, N.G.; Wollish, W.S.; Arlotta, P.; Jensen, P.E.; Ono, S.J.; Benichou, G. CD4+ T Cell Responses to Self- and Mutated p53 Determinants During Tumorigenesis in Mice. J. Immunol. 2000, 164, 5641–5651. [Google Scholar] [CrossRef] [Green Version]
- Chaumet, A.; Wright, G.; Seet, S.H.; Tham, K.M.; Gounko, N.V.; Bard, F. Nuclear envelope-associated endosomes deliver surface proteins to the nucleus. Nat. Commun. 2015, 6, 8218. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kisielow, P. How does the immune system learn to distinguish between good and evil? The first definitive studies of T cell central tolerance and positive selection. Immunogenetics 2019, 71, 513–518. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peixoto, A.; Relvas-Santos, M.; Azevedo, R.; Santos, L.L.; Ferreira, J.A. Protein Glycosylation and Tumor Microenvironment Alterations Driving Cancer Hallmarks. Front. Oncol. 2019, 9, 380. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Theobald, M.; Offringa, R. Anti-p53-directed immunotherapy of malignant disease. Expert Rev. Mol. Med. 2003, 5, 1–13. [Google Scholar] [CrossRef] [PubMed]
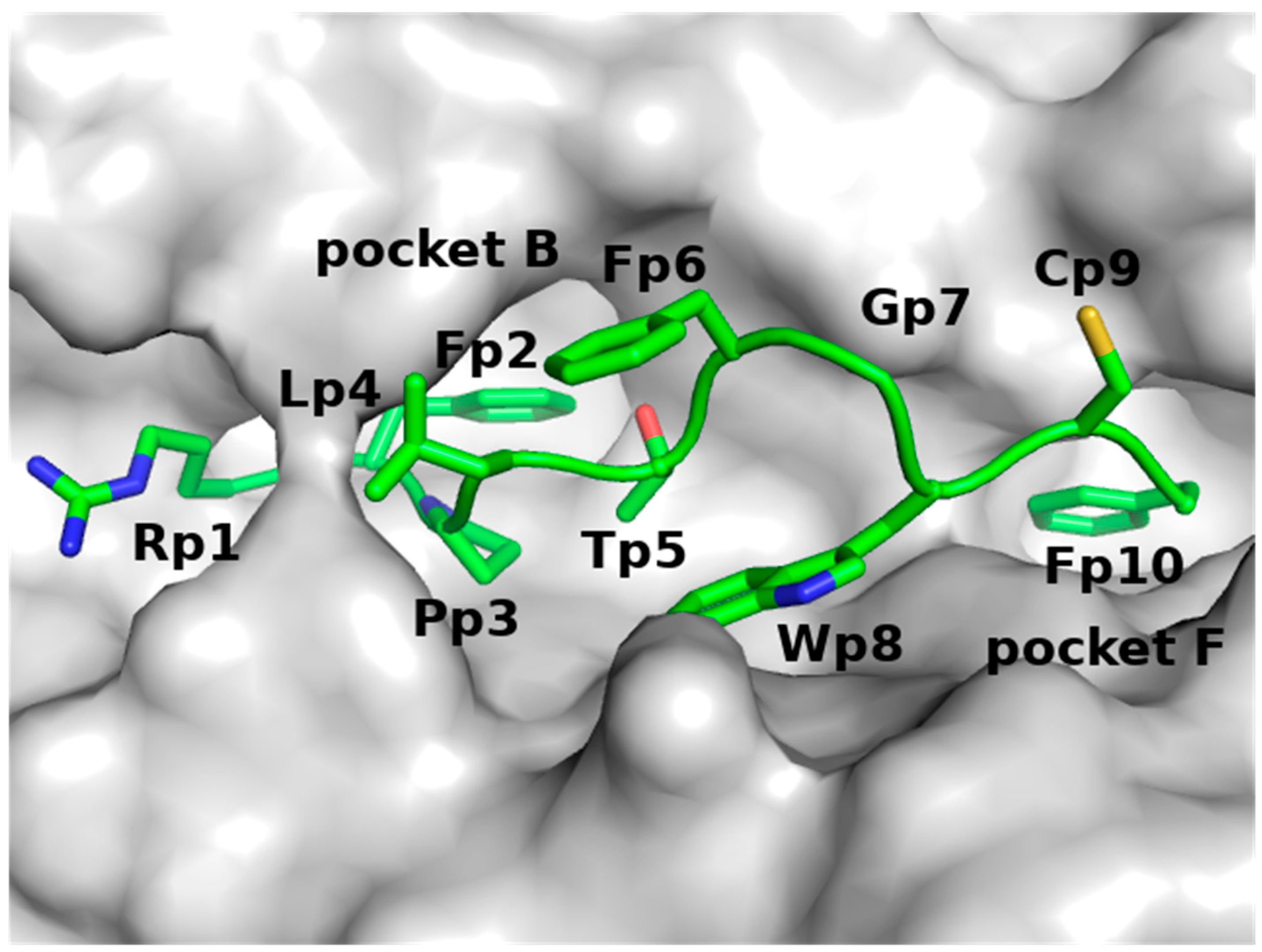
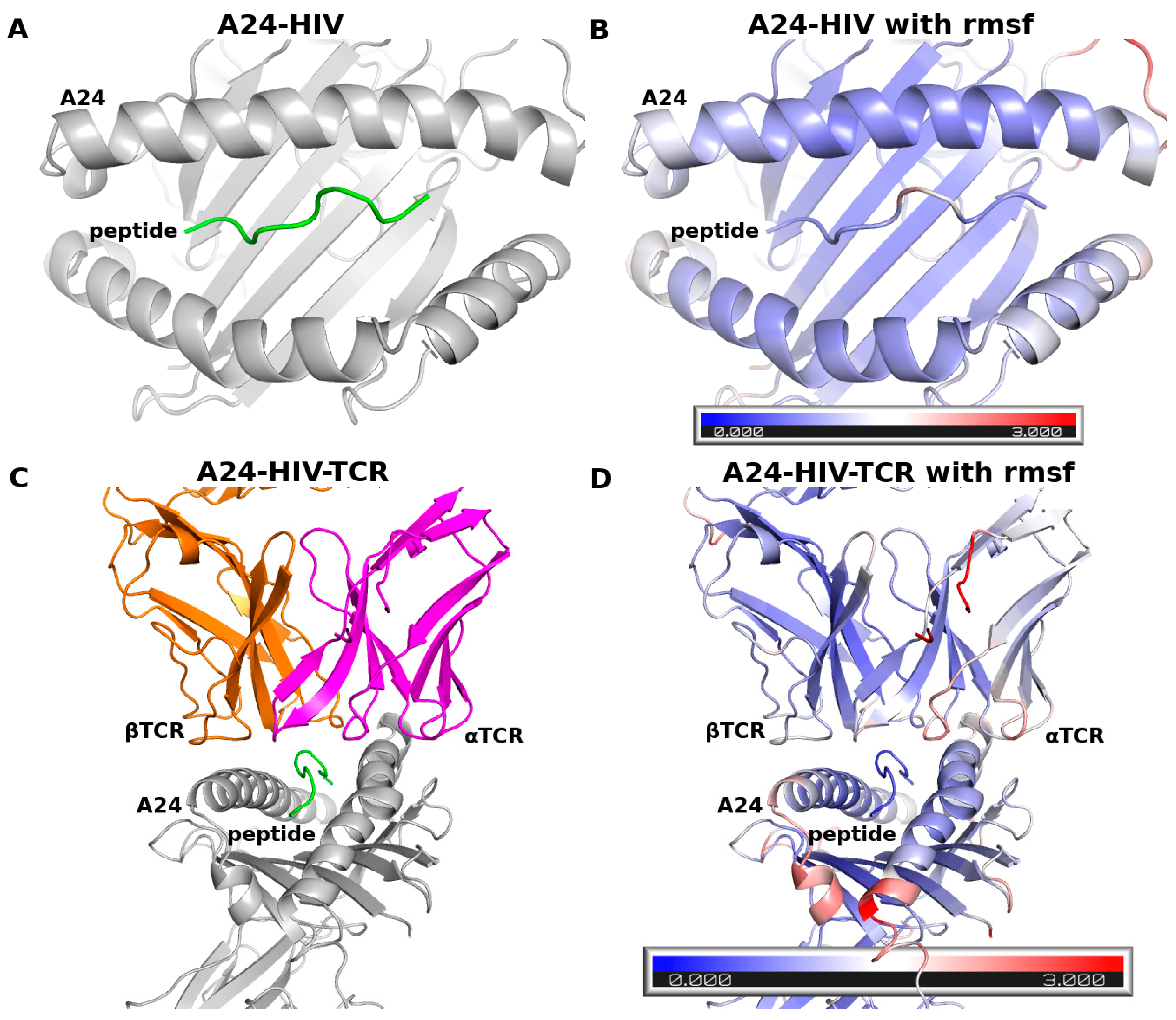

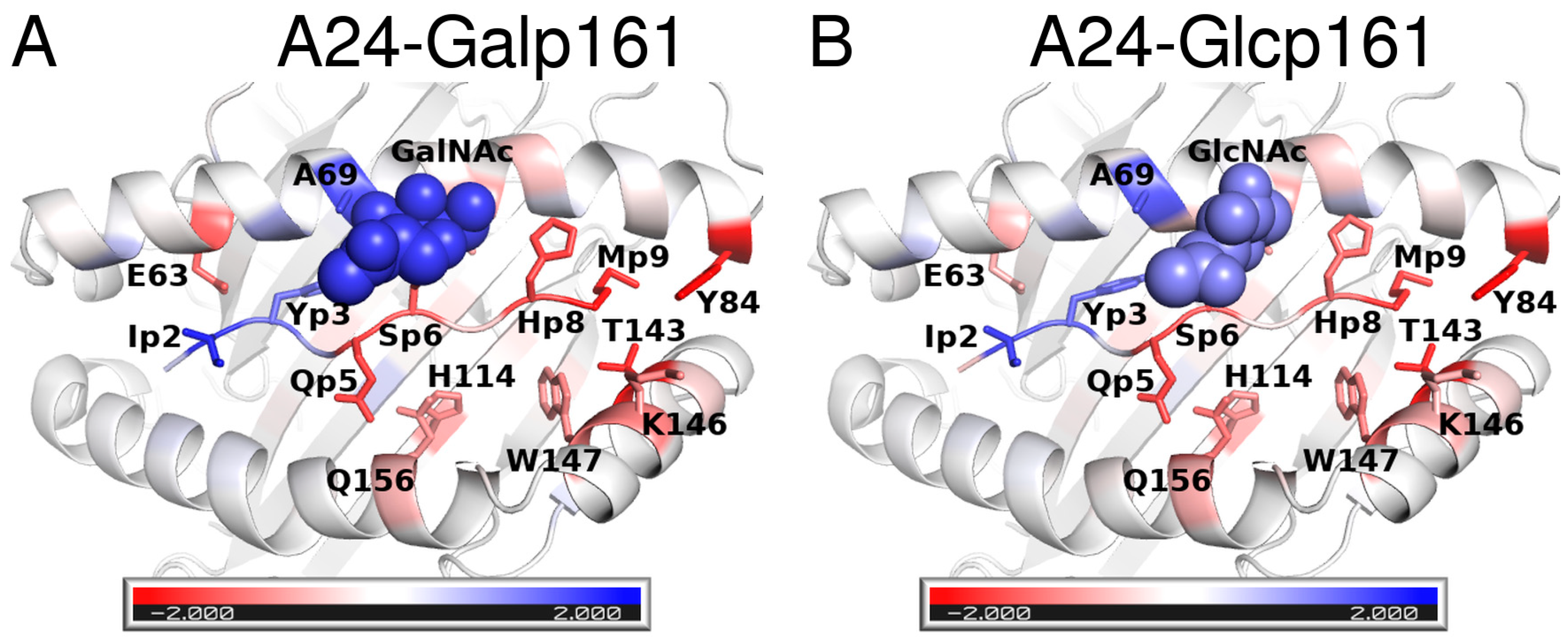
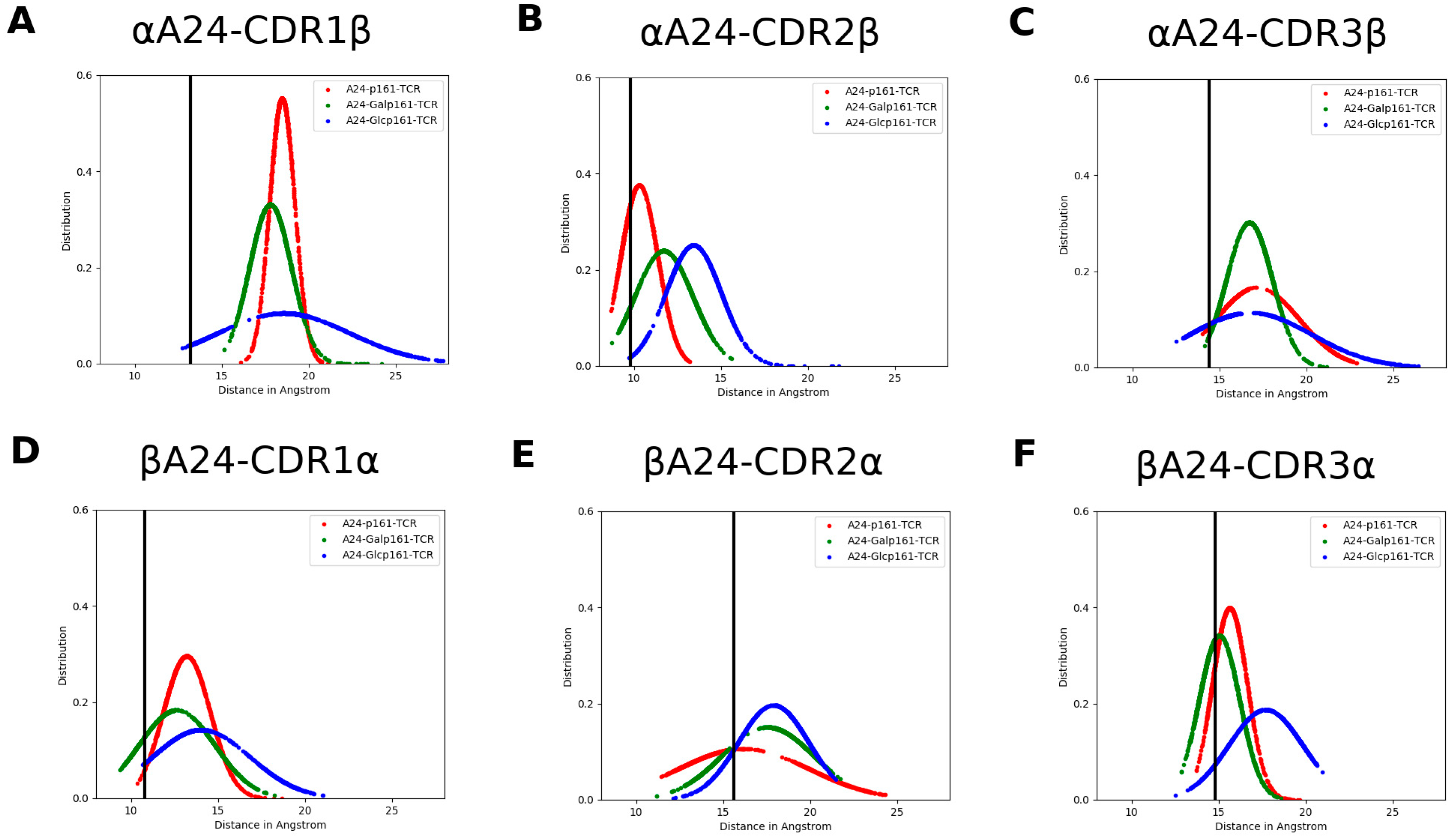
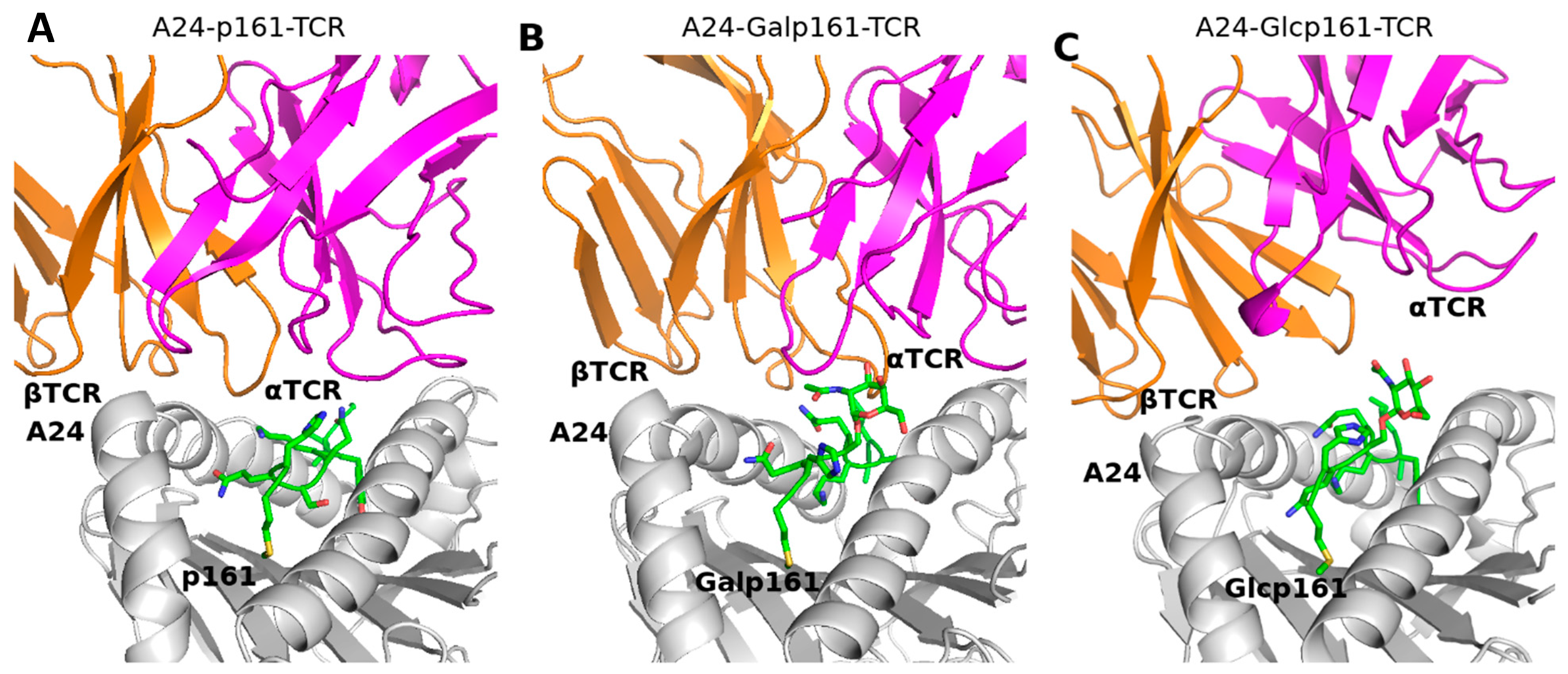
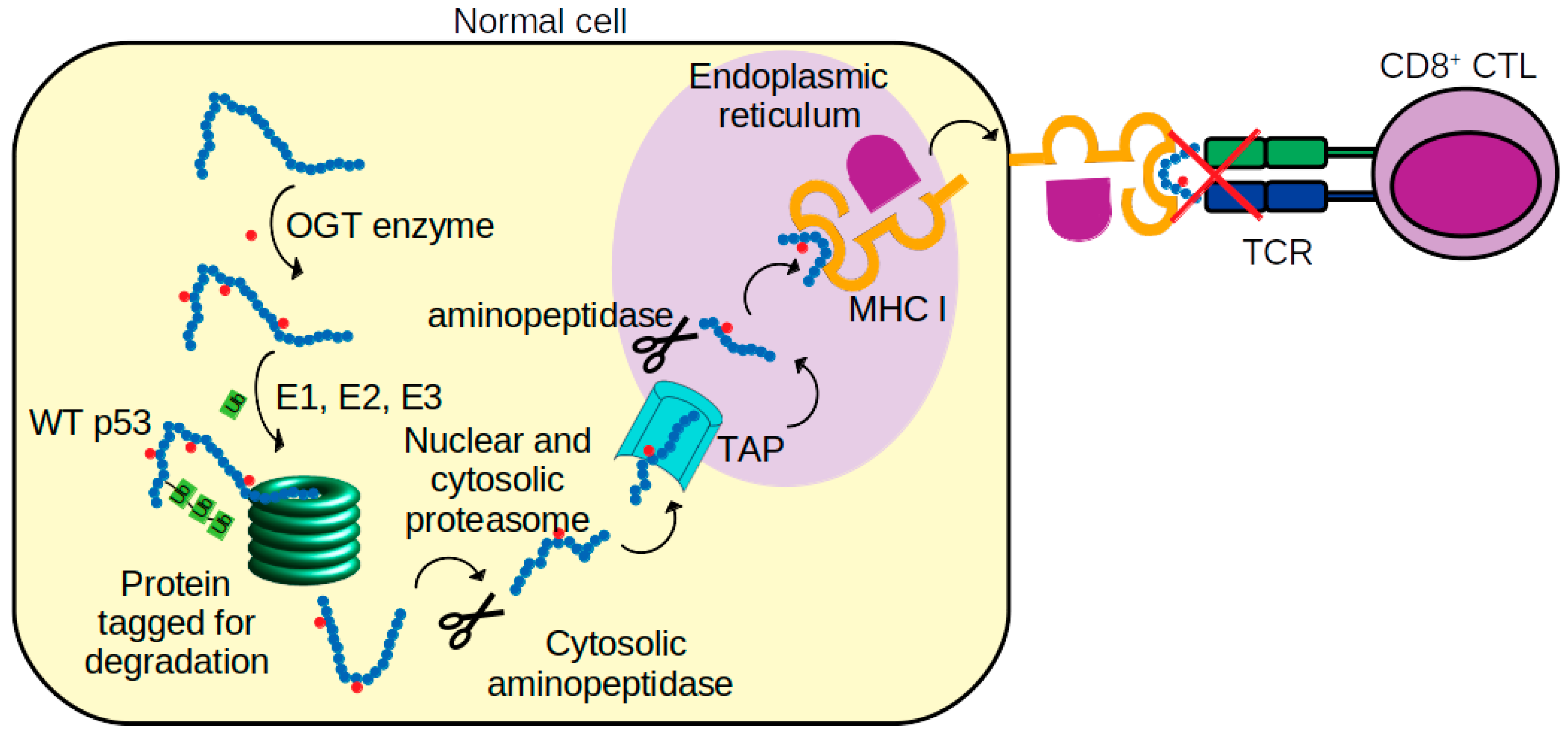
| Residue Name | Chain | Residue Number | GlyCAM Score for Wild Type Monomeric DBD | Glycosylation Status | Difference Compared to Wild Type Monomeric DBD | |
|---|---|---|---|---|---|---|
| Y163C | Q167K | |||||
| S*VP | A | 96 | 99.8 | + | 0 | 0 |
| VPS*QK | A | 99 | 57.7 | + | 0 | 0 |
| QKT*YQ | A | 102 | 51.9 | + | 0 | 0 |
| QGS*YG | A | 106 | 89.2 | + | 0 | 0 |
| LHS*GT | A | 116 | 31.5 | 0 | 0 | 0 |
| SGT*AK | A | 118 | 19.4 | − | 0 | 0 |
| AKS*VT | A | 121 | 56.3 | + | 0 | 0 |
| SVT*CT | A | 123 | 56.8 | + | 0 | 0 |
| TCT*YS | A | 125 | 1.3 | − | 0 | 0 |
| TYS*PA | A | 127 | 0 | − | 0 | 0 |
| AKT*CP | A | 140 | 41.6 | + | 0 | 0 |
| VDS*TP | A | 149 | 69.3 | + | 0 | 0 |
| DST*PP | A | 150 | 63.2 | + | 0 | 0 |
| PGT*RV | A | 155 | 1.3 | − | 0 | 0 |
| KQS*QH | A | 166 | 91.3 | + | 0 | 6.9 |
| HMT*EV | A | 170 | 83.9 | + | 0 | 0 |
| RCS*DS | A | 183 | 111.7 | + | 0 | 0 |
| SDS*DG | A | 185 | 68.3 | + | 0 | 0 |
| RNT*FR | A | 211 | 37.2 | 0 | 0 | 0 |
| RHS*VV | A | 215 | 5.2 | − | 0 | 0 |
| VGS*DC | A | 227 | 41 | + | 0 | 0 |
| DCT*TI | A | 230 | 17.5 | − | 0 | 0 |
| CTT*IH | A | 231 | 35.5 | 0 | 0 | 0 |
| CNS*SC | A | 240 | 13.4 | − | 0 | 0 |
| NSS*CM | A | 241 | 62.2 | + | 0 | 0 |
| ILT*II | A | 253 | 0.7 | − | 0 | 0 |
| IIT*LE | A | 256 | 14.5 | − | 0 | 0 |
| EDS*SG | A | 260 | 70.4 | + | 0 | 0 |
| DSS*GN | A | 261 | 81.2 | + | 0 | 0 |
| RNS*FE | A | 269 | 14.7 | − | 0 | 0 |
| RRT*EE | A | 284 | 70.8 | + | 0 | 0 |
| Protein | Peptide | TCR | Number of Waters | Total Number of Atoms | Simulation Time (μs) |
|---|---|---|---|---|---|
| A24 | HIV | − | 14,130 | 48,538 | 0.5 * 3 = 1.5 |
| A24 | p161 | − | 14,746 | 50,364 | 0.5 * 3 = 1.5 |
| A24 | Galp161 | − | 14,742 | 50,379 | 0.5 * 3 = 1.5 |
| A24 | Glcp161 | − | 14,736 | 50,361 | 0.5 * 3 = 1.5 |
| A24 | HIV | TCR | 64,105 | 205,288 | 1.0 * 3 = 3.0 |
| A24 | p161 | TCR | 64,673 | 206,970 | 1.0 * 3 = 3.0 |
| A24 | Galp161 | TCR | 64,705 | 207,093 | 1.0 * 3 = 3.0 |
| A24 | Glcp161 | TCR | 64,675 | 207,003 | 1.0 * 3 = 3.0 |
| Energy | A24-p161 | A24-Galp161 | A24-Glcp161 |
|---|---|---|---|
| ELE | −448.3 (11.0) | −400.3 (18.6) | −397.0 (6.8) |
| VDW | −97.5 (3.0) | −102.3 (0.8) | −99.5 (1.1) |
| GBSOL | 459.8 (11.5) | 427.6 (17.6) | 421.8 (4.4) |
| ΔH | −86.0 (4.0) | −75.0 (0.7) | −74.6 (1.7) |
| Interaction | Energy | A24-p161-TCR | A24-Galp161-TCR | A24-Glcp161-TCR |
|---|---|---|---|---|
| A24 vs. pep | ELE | −414.0 (10.5) | −427.9 (27.4) | −410.4 (3.9) |
| VDW | −96.5 (0.4) | −91.6 (7.6) | −101.4 (3.3) | |
| GBSOL | 430.3 (5.6) | 443.4 (14.1) | 436.1 (4.8) | |
| ΔH | −80.2 (6.1) | −76.1 (8.6) | −75.7 (1.9) | |
| pep vs. TCR | ELE | −74.1 (12.2) | −79.5 (3.1) | −63.9 (5.2) |
| VDW | −18.1 (3.7) | −26.9 (2.4) | −27.5 (10.2) | |
| GBSOL | 87.5 (12.2) | 96.7 (3.4) | 80.1 (1.5) | |
| ΔH | −4.7 (2.4) | −9.7 (1.6) | −11.3 (6.1) | |
| A24-pep vs. TCR | ELE | 209.8 (44.9) | 171.4 (43.9) | 224.1 (60.4) |
| VDW | −91.0 (5.8) | −99.4 (4.6) | −84.2 (17.9) | |
| GBSOL | −145.4 (41.4) | −111.5 (40.0) | −163.8 (59.6) | |
| ΔH | −26.5 (3.3) | −39.5 (8.6) | −23.9 (15.5) | |
| A24 vs. TCR | ELE | 284.0 (42.9) | 250.9 (44.2) | 288.0 (65.6) |
| VDW | −72.9 (2.6) | −72.5 (4.5) | −56.8 (7.7) | |
| GBSOL | −228.6 (40.4) | −203.2 (40.7) | −239.6 (60.5) | |
| ΔH | −17.6 (2.3) | −24.8 (8.4) | −8.4 (10.9) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nguyen, T.B.; Lane, D.P.; Verma, C.S. Can Glycosylation Mask the Detection of MHC Expressing p53 Peptides by T Cell Receptors? Biomolecules 2021, 11, 1056. https://doi.org/10.3390/biom11071056
Nguyen TB, Lane DP, Verma CS. Can Glycosylation Mask the Detection of MHC Expressing p53 Peptides by T Cell Receptors? Biomolecules. 2021; 11(7):1056. https://doi.org/10.3390/biom11071056
Chicago/Turabian StyleNguyen, Thanh Binh, David P. Lane, and Chandra S. Verma. 2021. "Can Glycosylation Mask the Detection of MHC Expressing p53 Peptides by T Cell Receptors?" Biomolecules 11, no. 7: 1056. https://doi.org/10.3390/biom11071056
APA StyleNguyen, T. B., Lane, D. P., & Verma, C. S. (2021). Can Glycosylation Mask the Detection of MHC Expressing p53 Peptides by T Cell Receptors? Biomolecules, 11(7), 1056. https://doi.org/10.3390/biom11071056






