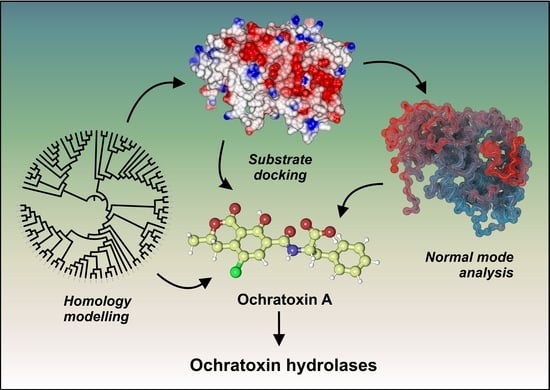
Systematic Structure-Based Search for Ochratoxin-Degrading Enzymes in Proteomes from Filamentous Fungi
Abstract
1. Introduction
2. Materials and Methods
2.1. Fungal Proteome Interrogation
2.2. Homology Modeling, Model Refinement and Substrate Docking
2.3. Molecular Dynamics Simulations
2.4. Calculation of Substrate-Binding Free Energy from Molecular Dynamics Simulations
2.5. Structure Comparison
2.6. Structure Analysis
3. Results
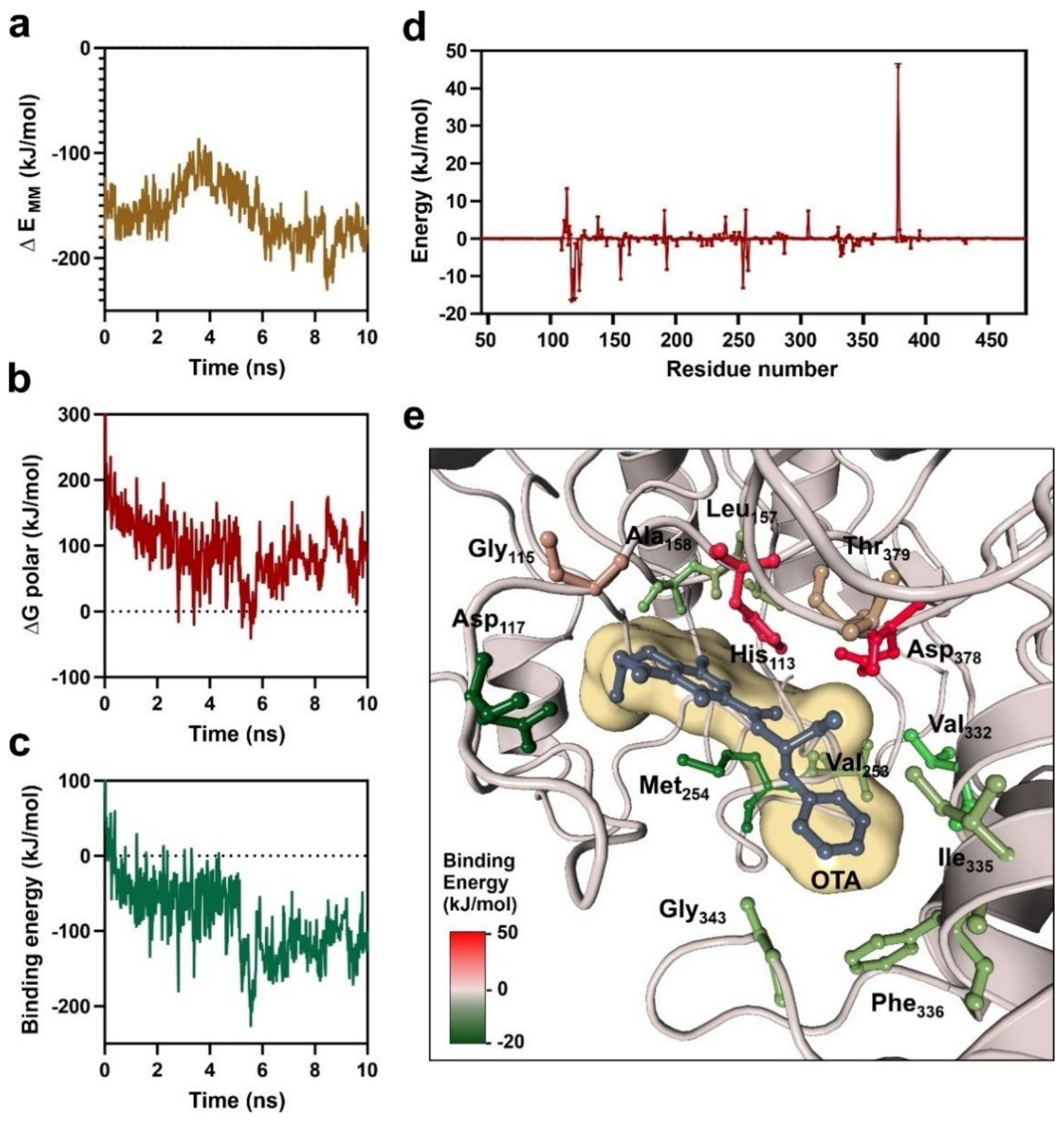
3.1. Catalytic Center of Ochratoxinase from A. niger
3.2. Characterization of OTA-Binding Pocket by Molecular Dynamics
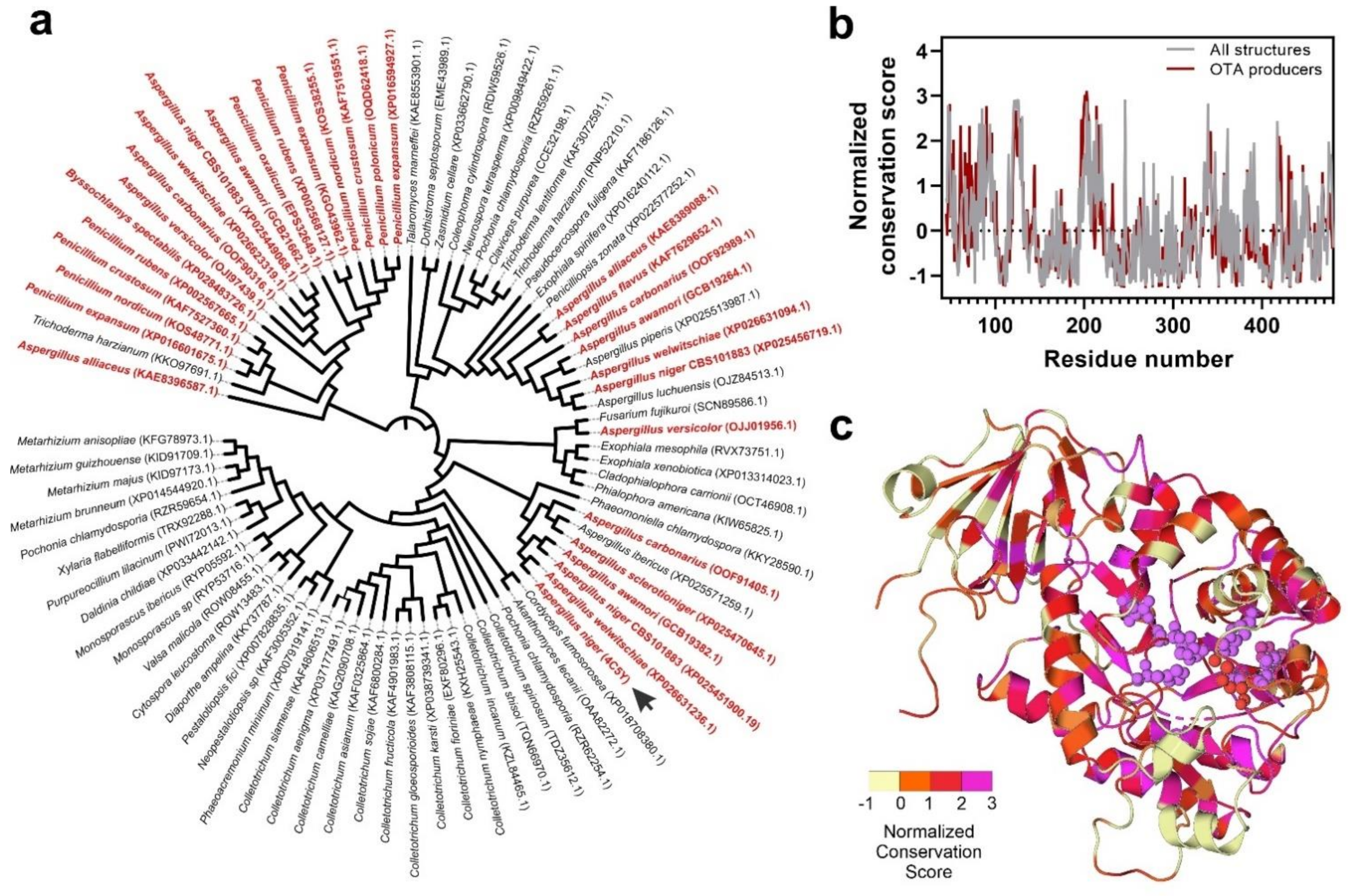
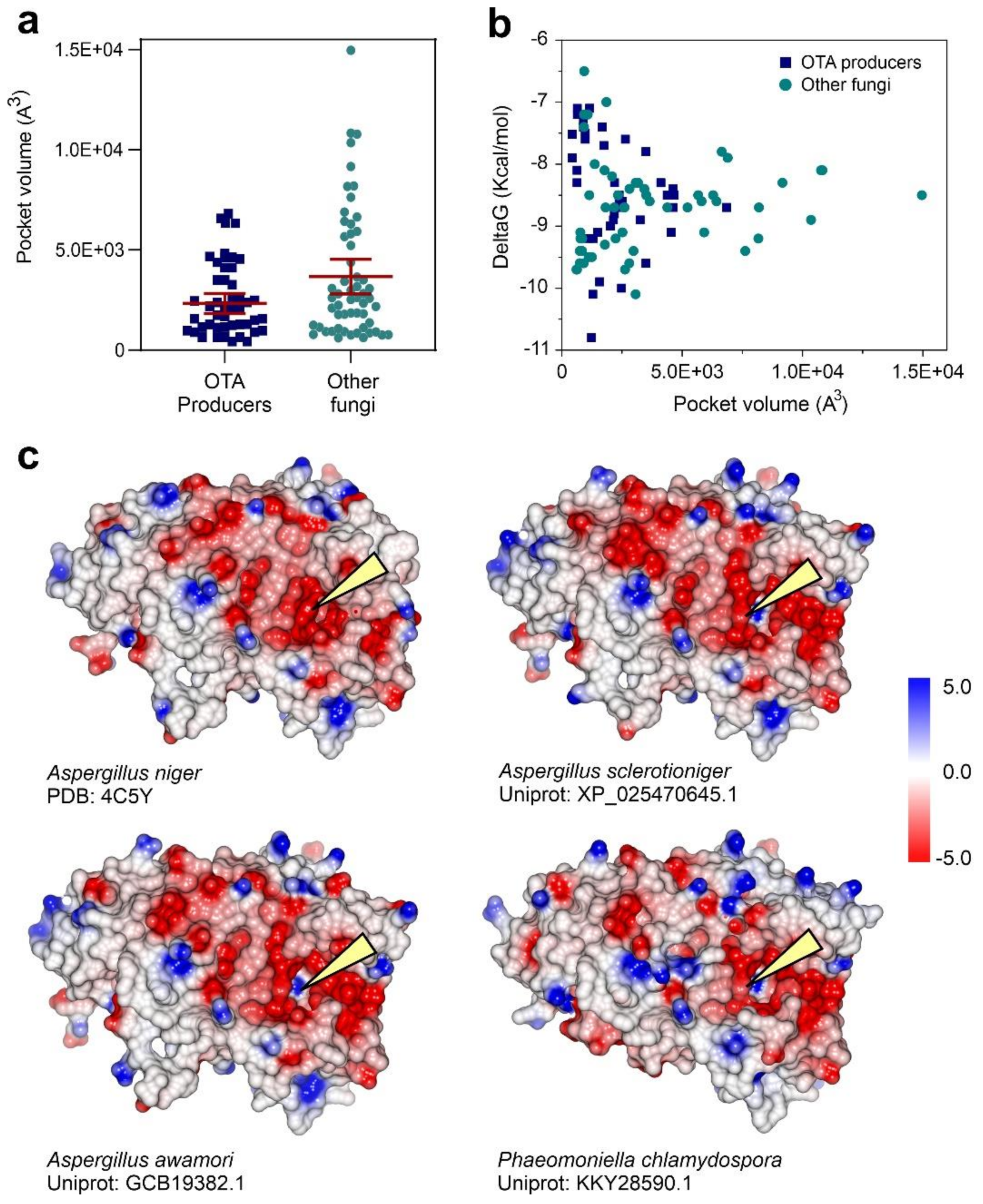
3.3. Fungal Proteome Mining for the Presence of Putative Ochratoxinases
3.4. Substrate Docking and Characterization of Substrate-Binding Cavities
3.5. Characterization of Low-Frequency Domain Motions
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- van der Merwe, K.J.; Steyn, P.S.; Fourie, L.; Scott, D.B.; Theron, J.J. Ochratoxin A, a toxic metabolite produced by Aspergillus ochraceus Wilh. Nature 1965, 205, 1112–1113. [Google Scholar] [CrossRef] [PubMed]
- Casas-Junco, P.P.; Ragazzo-Sanchez, J.A.; Ascencio-Valle, F.J.; Calderon-Santoyo, M. Determination of potentially mycotoxigenic fungi in coffee (Coffea arabica L.) from Nayarit. Food Sci. Biotechnol. 2018, 27, 891–898. [Google Scholar] [CrossRef] [PubMed]
- Ferrara, M.; Gallo, A.; Perrone, G.; Magista, D.; Baker, S.E. Comparative Genomic Analysis of Ochratoxin A Biosynthetic Cluster in Producing Fungi: New Evidence of a Cyclase Gene Involvement. Front. Microbiol. 2020, 11, 581309. [Google Scholar] [CrossRef] [PubMed]
- Konrad, I.; Roschenthaler, R. Inhibition of phenylalanine tRNA synthetase from Bacillus subtilis by ochratoxin A. FEBS Lett. 1977, 83, 341–347. [Google Scholar] [CrossRef]
- Liu, J.; Wang, Y.; Cui, J.; Xing, L.; Shen, H.; Wu, S.; Lian, H.; Wang, J.; Yan, X.; Zhang, X. Ochratoxin A induces oxidative DNA damage and G1 phase arrest in human peripheral blood mononuclear cells in vitro. Toxicol. Lett. 2012, 211, 164–171. [Google Scholar] [CrossRef]
- Li, H.; Mao, X.; Liu, K.; Sun, J.; Li, B.; Malyar, R.M.; Liu, D.; Pan, C.; Gan, F.; Liu, Y.; et al. Ochratoxin A induces nephrotoxicity in vitro and in vivo via pyroptosis. Arch. Toxicol. 2021, 95, 1489–1502. [Google Scholar] [CrossRef]
- Park, S.; Lim, W.; You, S.; Song, G. Ochratoxin A exerts neurotoxicity in human astrocytes through mitochondria-dependent apoptosis and intracellular calcium overload. Toxicol. Lett. 2019, 313, 42–49. [Google Scholar] [CrossRef]
- Rutigliano, L.; Valentini, L.; Martino, N.A.; Pizzi, F.; Zanghi, A.; Dell’Aquila, M.E.; Minervini, F. Ochratoxin A at low concentrations inhibits in vitro growth of canine umbilical cord matrix mesenchymal stem cells through oxidative chromatin and DNA damage. Reprod. Toxicol. 2015, 57, 121–129. [Google Scholar] [CrossRef]
- Zeljezic, D.; Domijan, A.M.; Peraica, M. DNA damage by ochratoxin A in rat kidney assessed by the alkaline comet assay. Braz. J. Med. Biol. Res. 2006, 39, 1563–1568. [Google Scholar] [CrossRef]
- Duarte, S.C.; Lino, C.M.; Pena, A. Mycotoxin food and feed regulation and the specific case of ochratoxin A: A review of the worldwide status. Food Addit. Contam. Part A Chem. Anal. Control Expo. Risk Assess. 2010, 27, 1440–1450. [Google Scholar] [CrossRef]
- Abrunhosa, L.; Paterson, R.; Venâncio, A. Biodegradation of Ochratoxin A for Food and Feed Decontamination. Toxins 2010, 2, 1078–1099. [Google Scholar] [CrossRef]
- Dahal, S.; Lee, H.J.; Gu, K.; Ryu, D. Heat Stability of Ochratoxin A in an Aqueous Buffered Model System. J. Food Prot. 2016, 79, 1748–1752. [Google Scholar] [CrossRef]
- Vidal, A.; Sanchis, V.; Ramos, A.J.; Marin, S. Thermal stability and kinetics of degradation of deoxynivalenol, deoxynivalenol conjugates and ochratoxin A during baking of wheat bakery products. Food Chem. 2015, 178, 276–286. [Google Scholar] [CrossRef]
- Varga, J.; Kocsube, S.; Peteri, Z.; Vagvolgyi, C.; Toth, B. Chemical, physical and biological approaches to prevent ochratoxin induced toxicoses in humans and animals. Toxins 2010, 2, 1718–1750. [Google Scholar] [CrossRef]
- Maatouk, I.; Mehrez, A.; Amara, A.B.; Chayma, R.; Abid, S.; Jerbi, T.; Landoulsi, A. Effects of Gamma Irradiation on Ochratoxin A Stability and Cytotoxicity in Methanolic Solutions and Potential Application in Tunisian Millet Samples. J. Food Prot. 2019, 82, 1433–1439. [Google Scholar] [CrossRef]
- Solfrizzo, M.; Avantaggiato, G.; Panzarini, G.; Visconti, A. Removal of ochratoxin A from contaminated red wines by repassage over grape pomaces. J. Agric. Food Chem. 2010, 58, 317–323. [Google Scholar] [CrossRef]
- Raj, J.; Vasiljevic, M.; Tassis, P.; Farkas, H.; Bosnjak-Neumuller, J.; Manner, K. Effects of a modified clinoptilolite zeolite on growth performance, health status and detoxification of aflatoxin B1 and ochratoxin A in male broiler chickens. Br. Poult. Sci. 2021. [Google Scholar] [CrossRef]
- Jalili, M.; Jinap, S.; Son, R. The effect of chemical treatment on reduction of aflatoxins and ochratoxin A in black and white pepper during washing. Food Addit. Contam Part A Chem. Anal. Control Expo. Risk Assess. 2011, 28, 485–493. [Google Scholar] [CrossRef]
- Amezqueta, S.; Gonzalez-Penas, E.; Lizarraga, T.; Murillo-Arbizu, M.; Lopez de Cerain, A. A simple chemical method reduces ochratoxin A in contaminated cocoa shells. J. Food Prot. 2008, 71, 1422–1426. [Google Scholar] [CrossRef]
- Upadhaya, S.D.; Song, J.Y.; Park, M.A.; Seo, J.K.; Yang, L.; Lee, C.H.; Cho, K.J.; Ha, J.K. Isolation, screening and identification of Swine gut microbiota with ochratoxin a biodegradation ability. Asian Australas. J. Anim. Sci. 2012, 25, 114–121. [Google Scholar] [CrossRef]
- Kiessling, K.H.; Pettersson, H.; Sandholm, K.; Olsen, M. Metabolism of aflatoxin, ochratoxin, zearalenone, and three trichothecenes by intact rumen fluid, rumen protozoa, and rumen bacteria. Appl. Environ. Microbiol. 1984, 47, 1070–1073. [Google Scholar] [CrossRef] [PubMed]
- Hohler, D.; Sudekum, K.H.; Wolffram, S.; Frohlich, A.A.; Marquardt, R.R. Metabolism and excretion of ochratoxin A fed to sheep. J. Anim. Sci. 1999, 77, 1217–1223. [Google Scholar] [CrossRef] [PubMed]
- Cho, S.M.; Jeong, S.E.; Lee, K.R.; Sudhani, H.P.; Kim, M.; Hong, S.Y.; Chung, S.H. Biodegradation of Ochratoxin A by Aspergillus tubingensis Isolated from Meju. J. Microbiol. Biotechnol. 2016, 26, 1687–1695. [Google Scholar] [CrossRef] [PubMed]
- De Bellis, P.; Tristezza, M.; Haidukowski, M.; Fanelli, F.; Sisto, A.; Mule, G.; Grieco, F. Biodegradation of Ochratoxin A by Bacterial Strains Isolated from Vineyard Soils. Toxins 2015, 7, 5079–5093. [Google Scholar] [CrossRef] [PubMed]
- Abrunhosa, L.; Ines, A.; Rodrigues, A.I.; Guimaraes, A.; Pereira, V.L.; Parpot, P.; Mendes-Faia, A.; Venancio, A. Biodegradation of ochratoxin A by Pediococcus parvulus isolated from Douro wines. Int. J. Food Microbiol. 2014, 188, 45–52. [Google Scholar] [CrossRef] [PubMed]
- Lyagin, I.; Efremenko, E. Enzymes for Detoxification of Various Mycotoxins: Origins and Mechanisms of Catalytic Action. Molecules 2019, 24, 2362. [Google Scholar] [CrossRef] [PubMed]
- Pitout, M.J. The hydrolysis of ochratoxin a by some proteolytic enzymes. Biochem. Pharmacol. 1969, 18, 485–491. [Google Scholar] [CrossRef]
- Abrunhosa, L.; Santos, L.; Venancio, A. Degradation of Ochratoxin A by Proteases and by a Crude Enzyme of Aspergillus niger. Food Biotechnol. 2006, 20, 231–242. [Google Scholar] [CrossRef]
- Abrunhosa, L.; Venancio, A. Isolation and purification of an enzyme hydrolyzing ochratoxin A from Aspergillus niger. Biotechnol. Lett. 2007, 29, 1909–1914. [Google Scholar] [CrossRef]
- Dobritzsch, D.; Wang, H.; Schneider, G.; Yu, S. Structural and functional characterization of ochratoxinase, a novel mycotoxin-degrading enzyme. Biochem. J. 2014, 462, 441–452. [Google Scholar] [CrossRef]
- Jenuth, J.P. The NCBI. Publicly available tools and resources on the Web. Methods Mol. Biol. 2000, 132, 301–312. [Google Scholar] [CrossRef]
- Kelley, L.A.; Mezulis, S.; Yates, C.M.; Wass, M.N.; Sternberg, M.J. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015, 10, 845–858. [Google Scholar] [CrossRef]
- Xu, D.; Zhang, Y. Improving the physical realism and structural accuracy of protein models by a two-step atomic-level energy minimization. Biophys. J. 2011, 101, 2525–2534. [Google Scholar] [CrossRef]
- Emsley, P.; Lohkamp, B.; Scott, W.G.; Cowtan, K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010, 66, 486–501. [Google Scholar] [CrossRef]
- Liu, Y.; Grimm, M.; Dai, W.T.; Hou, M.C.; Xiao, Z.X.; Cao, Y. CB-Dock: A web server for cavity detection-guided protein-ligand blind docking. Acta Pharmacol. Sin. 2020, 41, 138–144. [Google Scholar] [CrossRef]
- Rakhshani, H.; Dehghanian, E.; Rahati, A. Enhanced GROMACS: Toward a better numerical simulation framework. J. Mol. Model. 2019, 25, 355. [Google Scholar] [CrossRef]
- Kim, S.; Lee, J.; Jo, S.; Brooks, C.L., 3rd; Lee, H.S.; Im, W. CHARMM-GUI ligand reader and modeler for CHARMM force field generation of small molecules. J. Comput. Chem. 2017, 38, 1879–1886. [Google Scholar] [CrossRef]
- Brown, D.K.; Penkler, D.L.; Amamuddy, O.S.; Ross, C.; Atilgan, A.R.; Atilgan, C.; Bishop, Ö.T. MD-TASK: A software suite for analyzing molecular dynamics trajectories. Bioinformatics 2017, 33, 2768–2771. [Google Scholar] [CrossRef]
- Wang, C.; Greene, D.; Xiao, L.; Qi, R.; Luo, R. Recent Developments and Applications of the MMPBSA Method. Front. Mol. Biosci. 2017, 4, 87. [Google Scholar] [CrossRef]
- Wang, C.; Nguyen, P.H.; Pham, K.; Huynh, D.; Le, T.B.; Wang, H.; Ren, P.; Luo, R. Calculating protein-ligand binding affinities with MMPBSA: Method and error analysis. J. Comput. Chem. 2016, 37, 2436–2446. [Google Scholar] [CrossRef]
- Kumari, R.; Kumar, R.; Lynn, A. g_mmpbsa—A GROMACS Tool for High-Throughput MM-PBSA Calculations. J. Chem. Inf. Model. 2014, 54, 1951–1962. [Google Scholar] [CrossRef] [PubMed]
- Akdel, M.; Durairaj, J.; de Ridder, D.; van Dijk, A.D.J. Caretta—A multiple protein structure alignment and feature extraction suite. Comput. Struct. Biotechnol. J. 2020, 18, 981–992. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021. [Google Scholar] [CrossRef] [PubMed]
- Ashkenazy, H.; Abadi, S.; Martz, E.; Chay, O.; Mayrose, I.; Pupko, T.; Ben-Tal, N. ConSurf 2016: An improved methodology to estimate and visualize evolutionary conservation in macromolecules. Nucleic Acids Res. 2016, 44, W344–W350. [Google Scholar] [CrossRef]
- Jurrus, E.; Engel, D.; Star, K.; Monson, K.; Brandi, J.; Felberg, L.E.; Brookes, D.H.; Wilson, L.; Chen, J.; Liles, K.; et al. Improvements to the APBS biomolecular solvation software suite. Protein Sci. 2018, 27, 112–128. [Google Scholar] [CrossRef]
- Yuan, S.; Chan, H.C.S.; Filipek, S.; Vogel, H. PyMOL and Inkscape Bridge the Data and the Data Visualization. Structure 2016, 24, 2041–2042. [Google Scholar] [CrossRef]
- Potterton, L.; McNicholas, S.; Krissinel, E.; Gruber, J.; Cowtan, K.; Emsley, P.; Murshudov, G.N.; Cohen, S.; Perrakis, A.; Noble, M. Developments in the CCP4 molecular-graphics project. Acta Crystallogr. D Biol. Crystallogr. 2004, 60, 2288–2294. [Google Scholar] [CrossRef]
- Hinsen, K. Analysis of domain motions by approximate normal mode calculations. Proteins 1998, 33, 417–429. [Google Scholar] [CrossRef]
- Tiwari, S.P.; Fuglebakk, E.; Hollup, S.M.; Skjaerven, L.; Cragnolini, T.; Grindhaug, S.H.; Tekle, K.M.; Reuter, N. WEBnm@ v2.0: Web server and services for comparing protein flexibility. BMC Bioinform. 2014, 15, 427. [Google Scholar] [CrossRef]
- Tomasello, G.; Armenia, I.; Molla, G. The Protein Imager: A full-featured online molecular viewer interface with server-side HQ-rendering capabilities. Bioinformatics 2020, 36, 2909–2911. [Google Scholar] [CrossRef]
- Liu, A.; Huo, L. Amidohydrolase Superfamily. In ELS; John Wiley & Sons, Ltd: Chichester, UK, 2014. [Google Scholar] [CrossRef]
- Laskowski, R.A.; Swindells, M.B. LigPlot+: Multiple ligand-protein interaction diagrams for drug discovery. J. Chem. Inf. Model. 2011, 51, 2778–2786. [Google Scholar] [CrossRef]
- Westermaier, Y.; Ruiz-Carmona, S.; Theret, I.; Perron-Sierra, F.; Poissonnet, G.; Dacquet, C.; Boutin, J.A.; Ducrot, P.; Barril, X. Binding mode prediction and MD/MMPBSA-based free energy ranking for agonists of REV-ERBα/NCoR. J. Comput. Aided Mol. Des. 2017, 31, 755–775. [Google Scholar] [CrossRef]
- Genheden, S.; Ryde, U. The MM/PBSA and MM/GBSA methods to estimate ligand-binding affinities. Expert Opin. Drug Discov. 2015, 10, 449–461. [Google Scholar] [CrossRef]
- Balakrishnan, R.; Christie, K.R.; Costanzo, M.C.; Dolinski, K.; Dwight, S.S.; Engel, S.R.; Fisk, D.G.; Hirschman, J.E.; Hong, E.L.; Nash, R.; et al. Fungal BLAST and Model Organism BLASTP Best Hits: New comparison resources at the Saccharomyces Genome Database (SGD). Nucleic Acids Res. 2005, 33, D374–D377. [Google Scholar] [CrossRef]
- Jimenez-Roldan, J.E.; Freedman, R.B.; Romer, R.A.; Wells, S.A. Rapid simulation of protein motion: Merging flexibility, rigidity and normal mode analyses. Phys. Biol. 2012, 9, 016008. [Google Scholar] [CrossRef]
- Stander, M.A.; Bornscheuer, U.T.; Henke, E.; Steyn, P.S. Screening of commercial hydrolases for the degradation of ochratoxin A. J. Agric. Food Chem. 2000, 48, 5736–5739. [Google Scholar] [CrossRef]
- Bejaoui, H.; Mathieu, F.; Taillandier, P.; Lebrihi, A. Biodegradation of ochratoxin A by Aspergillus section Nigri species isolated from French grapes: A potential means of ochratoxin A decontamination in grape juices and musts. FEMS Microbiol. Lett. 2006, 255, 203–208. [Google Scholar] [CrossRef]
- Dellafiora, L.; Gonaus, C.; Streit, B.; Galaverna, G.; Moll, W.D.; Vogtentanz, G.; Schatzmayr, G.; Dall’Asta, C.; Prasad, S. An In Silico Target Fishing Approach to Identify Novel Ochratoxin A Hydrolyzing Enzyme. Toxins 2020, 12, 258. [Google Scholar] [CrossRef]
- Xiang, D.F.; Patskovsky, Y.; Xu, C.; Fedorov, A.A.; Fedorov, E.V.; Sisco, A.A.; Sauder, J.M.; Burley, S.K.; Almo, S.C.; Raushel, F.M. Functional identification and structure determination of two novel prolidases from cog1228 in the amidohydrolase superfamily. Biochemistry 2010, 49, 6791–6803. [Google Scholar] [CrossRef][Green Version]
- Kumar, K.; Woo, S.M.; Siu, T.; Cortopassi, W.A.; Duarte, F.; Paton, R.S. Cation-pi interactions in protein-ligand binding: Theory and data-mining reveal different roles for lysine and arginine. Chem. Sci. 2018, 9, 2655–2665. [Google Scholar] [CrossRef]
- Lavanya, P.; Ramaiah, S.; Anbarasu, A. Cation-pi interactions in beta-lactamases: The role in structural stability. Cell Biochem. Biophys. 2013, 66, 147–155. [Google Scholar] [CrossRef]
- Kamps, J.J.; Khan, A.; Choi, H.; Lesniak, R.K.; Brem, J.; Rydzik, A.M.; McDonough, M.A.; Schofield, C.J.; Claridge, T.D.; Mecinovic, J. Cation-pi Interactions Contribute to Substrate Recognition in gamma-Butyrobetaine Hydroxylase Catalysis. Chemistry 2016, 22, 1270–1276. [Google Scholar] [CrossRef]
- Sommer, T.; Bjerregaard-Andersen, K.; Uribe, L.; Etzerodt, M.; Diezemann, G.; Gauss, J.; Cascella, M.; Morth, J.P. A fundamental catalytic difference between zinc and manganese dependent enzymes revealed in a bacterial isatin hydrolase. Sci. Rep. 2018, 8. [Google Scholar] [CrossRef]
- Gil-Serna, J.; Garcia-Diaz, M.; Gonzalez-Jaen, M.T.; Vazquez, C.; Patino, B. Description of an orthologous cluster of ochratoxin A biosynthetic genes in Aspergillus and Penicillium species. A comparative analysis. Int. J. Food Microbiol. 2018, 268, 35–43. [Google Scholar] [CrossRef] [PubMed]
- Lohkamp, B.; Andersen, B.; Piskur, J.; Dobritzsch, D. The crystal structures of dihydropyrimidinases reaffirm the close relationship between cyclic amidohydrolases and explain their substrate specificity. J. Biol. Chem. 2006, 281, 13762–13776. [Google Scholar] [CrossRef] [PubMed]
- Arima, J.; Shimone, K.; Miyatani, K.; Tsunehara, Y.; Isoda, Y.; Hino, T.; Nagano, S. Crystal structure of D-stereospecific amidohydrolase from Streptomyces sp. 82F2-insight into the structural factors for substrate specificity. FEBS J. 2016, 283, 337–349. [Google Scholar] [CrossRef] [PubMed]
- Shinohara, Y.; Miyanaga, A.; Kudo, F.; Eguchi, T. The crystal structure of the amidohydrolase VinJ shows a unique hydrophobic tunnel for its interaction with polyketide substrates. FEBS Lett. 2014, 588, 995–1000. [Google Scholar] [CrossRef] [PubMed]
- Gatti-Lafranconi, P.; Hollfelder, F. Flexibility and reactivity in promiscuous enzymes. ChemBioChem 2013, 14, 285–292. [Google Scholar] [CrossRef]
- Hernick, M.; Fierke, C.A. Zinc hydrolases: The mechanisms of zinc-dependent deacetylases. Arch. Biochem. Biophys. 2005, 433, 71–84. [Google Scholar] [CrossRef]
- Mitic, N.; Smith, S.J.; Neves, A.; Guddat, L.W.; Gahan, L.R.; Schenk, G. Catalytic mechanism of binuclear metallohydrolases. Chem. Rev. 2006, 106. [Google Scholar] [CrossRef]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Leitão, A.L.; Enguita, F.J. Systematic Structure-Based Search for Ochratoxin-Degrading Enzymes in Proteomes from Filamentous Fungi. Biomolecules 2021, 11, 1040. https://doi.org/10.3390/biom11071040
Leitão AL, Enguita FJ. Systematic Structure-Based Search for Ochratoxin-Degrading Enzymes in Proteomes from Filamentous Fungi. Biomolecules. 2021; 11(7):1040. https://doi.org/10.3390/biom11071040
Chicago/Turabian StyleLeitão, Ana Lúcia, and Francisco J. Enguita. 2021. "Systematic Structure-Based Search for Ochratoxin-Degrading Enzymes in Proteomes from Filamentous Fungi" Biomolecules 11, no. 7: 1040. https://doi.org/10.3390/biom11071040
APA StyleLeitão, A. L., & Enguita, F. J. (2021). Systematic Structure-Based Search for Ochratoxin-Degrading Enzymes in Proteomes from Filamentous Fungi. Biomolecules, 11(7), 1040. https://doi.org/10.3390/biom11071040