Properties of the Extracellular Polymeric Substance Layer from Minimally Grown Planktonic Cells of Listeria monocytogenes
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacteria Strains and Media
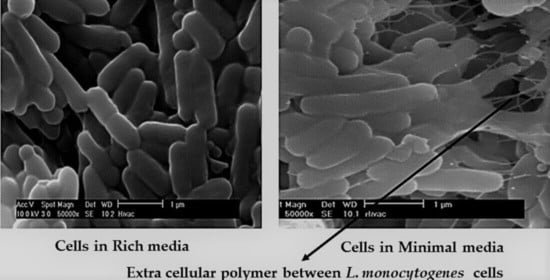
2.2. SEM of Cells Grown in Different Media
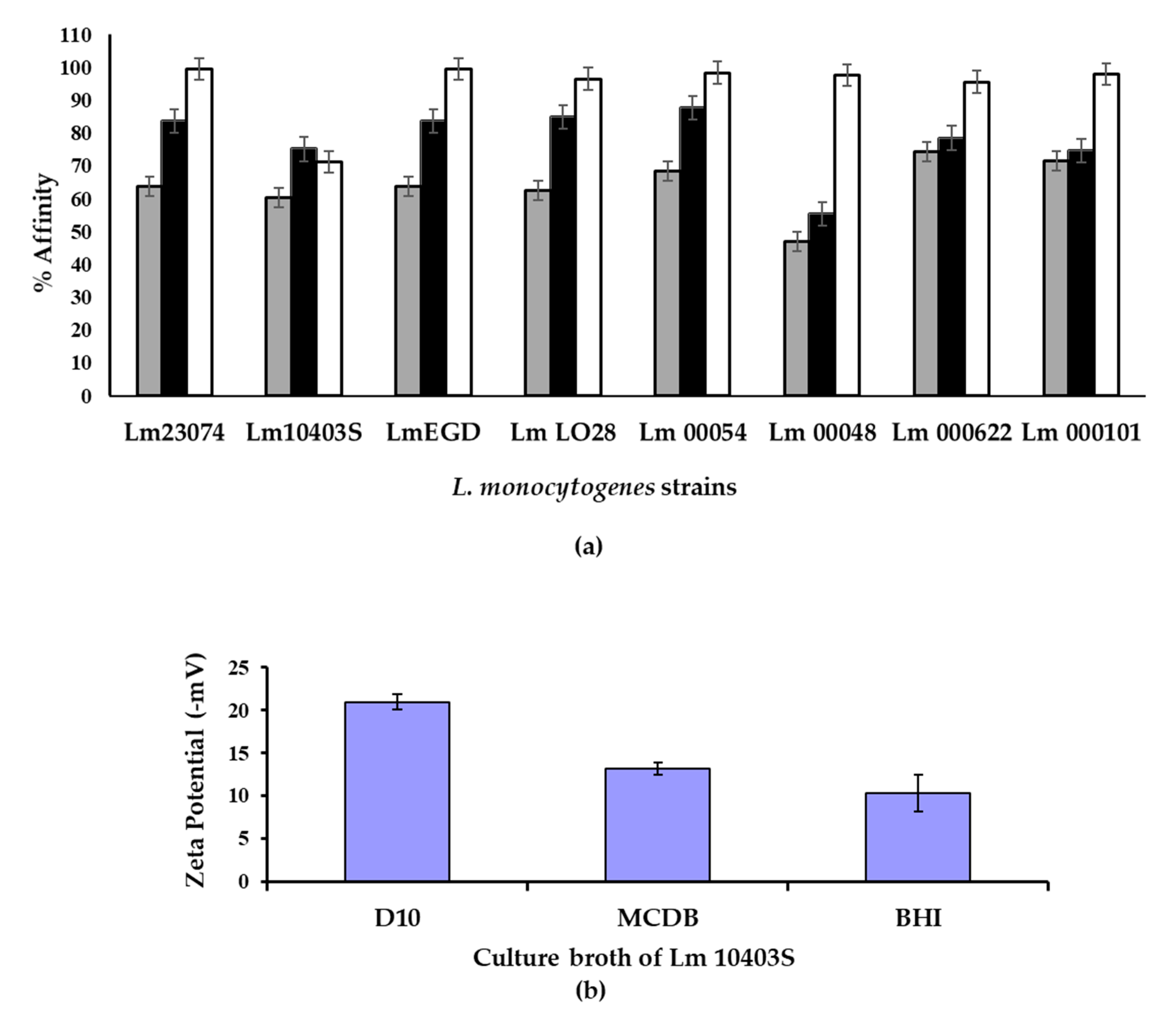
2.3. Hydrophobicity, Zeta Potential and Attachment Assays
2.4. Extraction and Purification of Capsular Polymer from L. monocytogenes
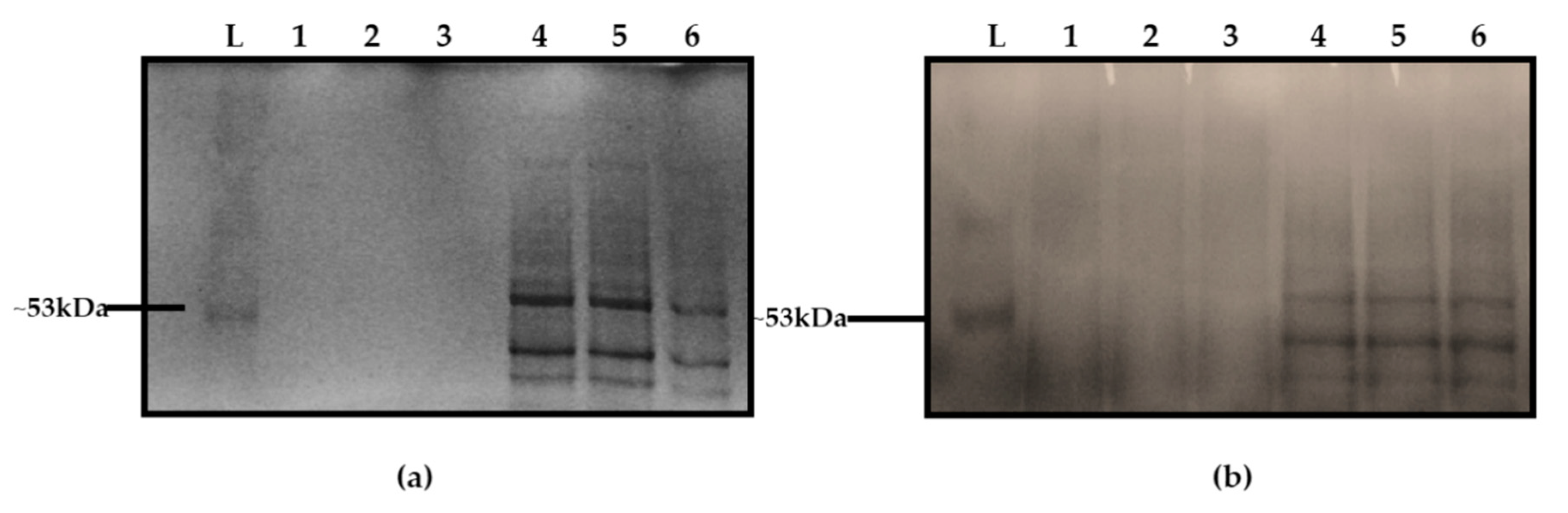
2.5. Sodium Dodecyl Sulphate Polyacrylamide Gel Electrophoresis Analysis of Listeria Crude Extract (Purified)
2.6. Amino Acids and Sugar Analysis of Crude/Purified EPS Extract
2.7. High Resolution Liquid State Nuclear Magnetic Resonance (NMR)
2.8. X-ray Diffraction Evaluation of the EPS of L. monocytogenes
2.9. Attenuated Total Reflectance-Fourier Transform Infrared (ATR-FTIR) Spectra of the Purified EPS of L. monocytogenes
3. Results
3.1. Initial Detection of the EPS in Planktonic Cells
3.2. Cell Charge, Hydrophobicity and Attachment of Cells
3.3. Initial Comparison of the EPS of L. monocytogenes with PGA
3.4. Amino Acid and Sugar Analysis
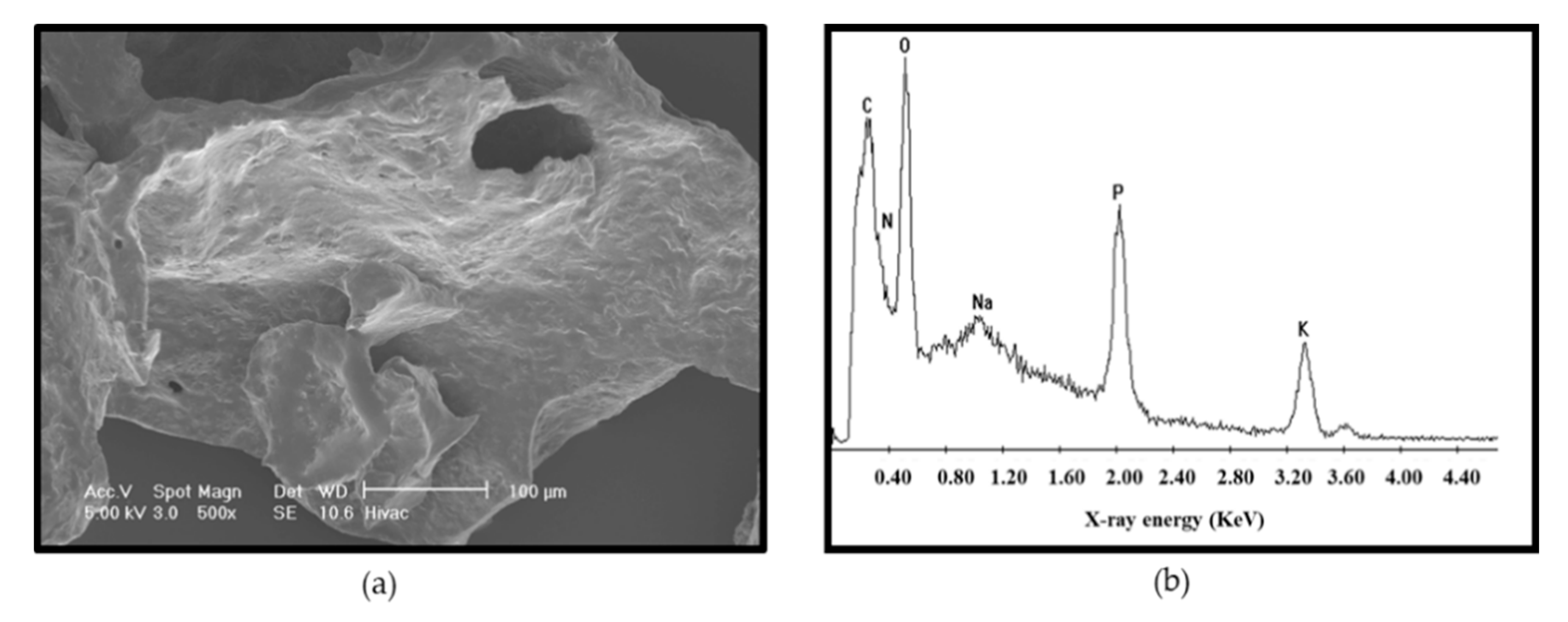
3.5. Microstructure of the Purified EPS
3.6. X-ray Diffraction Analysis of the EPS of L. monocytogenes
3.7. NMR Analysis of the EPS of L. monocytogenes
3.8. ATR-FTIR Analysis of Purified L. monocytogenes
4. Discussion
4.1. Cell Hydrophobicity and Surface Charge
4.2. Constituents of the EPS of L. monocytogenes
4.3. Role of the EPS in the Food Processing Environment
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Leclercq, A.; Moura, A.; Vales, G.; Tessaud-Rita, N.; Aguilhon, C.; Marc Lecuit, M. Listeria thailandensis sp. nov. Int. J. Syst. Evol. Microbiol. 2019, 69, 74–81. [Google Scholar] [CrossRef] [PubMed]
- Nwaiwu, O. What are the recognized species of the genus Listeria? Access Microbiol. 2020, 2. [Google Scholar] [CrossRef]
- Friesema, I.H.; Kuiling, S.; van der Ende, A.; Heck, M.E.; Spanjaard, L.; van Pelt, W. Risk factors for sporadic Listeriosis in the Netherlands, 2008 to 2013. Eurosurveillance 2015, 20. [Google Scholar] [CrossRef][Green Version]
- Allam, M.; Tau, N.; Smouse, S.L.; Mtshali, P.S.; Mnyameni, F.; Khumalo, Z.T.H.; Ismail, A.; Govender, N.; Thomas, J.; Smith, A.M. Whole-Genome sequences of Listeria monocytogenes sequence type 6 isolates associated with a large foodborne outbreak in South Africa, 2017 to 2018. Genome Announc. 2018, 6, e00538-18. [Google Scholar] [CrossRef]
- Thomas, M.K.; Vriezen, R.; Farber, J.M.; Currie, A.; Schlech, W.; Fazil, A. Economic cost of a Listeria monocytogenes outbreak in Canada, 2008. Foodborne Pathog. Dis. 2015, 12, 966–971. [Google Scholar] [CrossRef] [PubMed]
- Martín, B.; Bover-Cid, S.; Aymerich, T. MLVA Subtyping of Listeria Monocytogenes isolates from meat products and meat processing plants. Food Res. Int. 2018, 106, 225–232. [Google Scholar] [CrossRef] [PubMed]
- Fagerlund, A.; Langsrud, S.; Moen, B.; Heir, E.; Møretrø, T. Complete genome sequences of six Listeria monocytogenes sequence type 9 isolates from meat processing plants in Norway. Genome Announc. 2018, 6. [Google Scholar] [CrossRef]
- Chen, B.-Y.; Wang, C.-Y.; Wang, C.-L.; Fan, Y.-C.; Weng, I.-T.; Chou, C.-H. Prevalence and persistence of Listeria monocytogenes in ready-to-eat Tilapia Sashimi Processing Plants. J. Food Prot. 2016, 79, 1898–1903. [Google Scholar] [CrossRef]
- Almeida, G.; Magalhães, R.; Carneiro, L.; Santos, I.; Silva, J.; Ferreira, V.; Hogg, T.; Teixeira, P. Foci of contamination of Listeria monocytogenes in different cheese processing plants. Int. J. Food Microbiol. 2013, 167, 303–309. [Google Scholar] [CrossRef]
- Taguchi, M.; Kanki, M.; Yamaguchi, Y.; Inamura, H.; Koganei, Y.; Sano, T.; Nakamura, H.; Asakura, H. Prevalence of Listeria monocytogenes in retail lightly pickled vegetables and its successful control at processing plants. J. Food Prot. 2017, 80, 467–475. [Google Scholar] [CrossRef] [PubMed]
- Fox, E.M.; Wall, P.G.; Fanning, S. Control of Listeria species food safety at a poultry food production facility. Food Microbiol. 2015, 51, 81–86. [Google Scholar] [CrossRef]
- Noll, M.; Kleta, S.; Al Dahouk, S. Antibiotic susceptibility of 259 Listeria monocytogenes strains isolated from food, food-processing plants and human samples in Germany. J. Infect. Public Health 2018, 11, 572–577. [Google Scholar] [CrossRef]
- Carpentier, B.; Cerf, O. Review-Persistence of Listeria monocytogenes in food industry equipment and premises. Int. J. Food Microbiol. 2011, 145, 1–8. [Google Scholar] [CrossRef]
- Overney, A.; Jacques-André-Coquin, J.; Ng, P.; Carpentier, B.; Guillier, L.; Firmesse, O. Impact of environmental factors on the culturability and viability of Listeria monocytogenes under conditions encountered in food processing plants. Int. J. Food Microbiol. 2017, 244, 74–81. [Google Scholar] [CrossRef]
- Ferreira, V.; Wiedmann, M.; Teixeira, P.; Stasiewicz, M.J. Listeria monocytogenes persistence in food-associated environments: Epidemiology, strain characteristics, and implications for public health. J. Food Prot. 2014, 77, 150–170. [Google Scholar] [CrossRef]
- Decho, A.W.; Gutierrez, T. Microbial extracellular polymeric substances (EPSs) in ocean systems. Front. Microbiol. 2017, 8, 922. [Google Scholar] [CrossRef] [PubMed]
- Koo, H.; Yamada, K.M. Dynamic cell–matrix interactions modulate microbial biofilm and tissue 3d microenvironments. Curr. Opin. Cell Biol. 2016, 42, 102–112. [Google Scholar] [CrossRef]
- Pal, A.; Paul, A.K. Microbial extracellular polymeric substances: Central elements in heavy metal bioremediation. Indian J. Microbiol. 2008, 48, 49. [Google Scholar] [CrossRef] [PubMed]
- Yin, X.; Weitzel, F.; Jiménez-López, C.; Griesshaber, E.; Fernández-Díaz, L.; Rodríguez-Navarro, A.; Ziegler, A.; Schmahl, W.W. Directing effect of bacterial extracellular polymeric substances (EPS) on calcite organization and eps–carbonate composite aggregate formation. Cryst. Growth Des. 2020, 20, 1467–1484. [Google Scholar] [CrossRef]
- Wingender, J.; Neu, T.R.; Flemming, H.-C. (Eds.) What are bacterial extracellular polymeric substances? In Microbial Extracellular Polymeric Substances; Springer: Berlin, Germany, 1999; pp. 1–19. [Google Scholar] [CrossRef]
- Bhaskar, P.V.; Bhosle, N.B. Microbial extracellular polymeric substances in marine biogeochemical processes. Curr. Sci. 2005, 88, 45–53. [Google Scholar]
- Colagiorgi, A.; Di Ciccio, P.; Zanardi, E.; Ghidini, S.; Ianieri, A. A Look inside the listeria monocytogenes biofilms extracellular matrix. Microorganisms 2016, 4, 22. [Google Scholar] [CrossRef]
- Grigore-Gurgu, L.; Bucur, F.; Borda, D.; Alexa, E.-A.; Neagu, C.; Nicolau, A.I. Biofilms Formed by Pathogens in Food and Food Processing Environments, Bacterial Biofilms; Dincer, S., Özdenefe, M., Arkut, A., Eds.; Intech Open: London, UK, 2019; pp. 1–32. [Google Scholar]
- Di Martino, P. Extracellular polymeric substances, a key element in understanding biofilm phenotype. AIMS Microbiol. 2018, 4, 274–288. [Google Scholar] [CrossRef]
- Trivett, T.; Meyer, E. Citrate cycle and related metabolism of Listeria monocytogenes. J. Bacteriol. 1971, 107, 770–779. [Google Scholar] [CrossRef]
- Chavant, P.; Martinie, B.; Meylheuc, T.; Bellon-Fontaine, M.-N.; Hebraud, M. Listeria monocytogenes LO28: Surface physicochemical properties and ability to form biofilms at different temperatures and growth phases. Appl. Environ. Microbiol. 2002, 68, 728–737. [Google Scholar] [CrossRef]
- Doumith, M.; Buchrieser, C.; Glaser, P.; Jacquet, C.; Martin, P. Differentiation of the major Listeria monocytogenes serovars by multiplex PCR. J. Clin. Microbiol. 2004, 42, 3819–3822. [Google Scholar] [CrossRef]
- Pitcher, D.G.; Saunders, N.A.; Owen, R.J. Rapid extraction of bacterial genomic DNA with guanidium thiocyanate. Lett. Appl. Microbiol. 1989, 8, 151–156. [Google Scholar] [CrossRef]
- Briandet, R.; Meylheuc, T.; Maher, C.; Bellon-Fontaine, M.N. Listeria monocytogenes Scott A: Cell surface charge, hydrophobicity, and electron donor and acceptor characteristics under different environmental growth conditions. Appl. Environ. Microbiol. 1999, 65, 5328–5333. [Google Scholar] [CrossRef]
- Djordjevic, D.; Wiedmann, M.; McLandsborough, L.A. Microtiter Plate assay for assessment of Listeria monocytogenes biofilm formation. Appl. Environ. Microbiol. 2002, 68, 2950–2958. [Google Scholar] [CrossRef]
- Goto, A.; Kunioka, M. Biosynthesis and hydrolysis of poly(γ-glutamic acid) from Bacillus subtilis IF03335. Biosci. Biotechnol. Biochem. 1992, 56, 1031–1035. [Google Scholar] [CrossRef]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef]
- Englyst, H.; Wiggins, H.S.; Cummings, J.H. determination of the non-starch polysaccharides in plant foods by gas-liquid chromatography of constituent sugars as alditol acetates. Analyst 1982, 107, 307–318. [Google Scholar] [CrossRef]
- Nwaiwu, O.; Lad, M.; Davis, A.; Foster, T.; Rees, C. Preliminary analysis of structure and chemical composition of extracellular polymeric substance produced by listeria monocytogenes. In International Symposium on Problems of Listeriosis; Universidade Católica Portuguesa: Porto, Portugal, 2010; p. 144. [Google Scholar]
- Ukuku, D.O.; Fett, W.F. Relationship of cell surface charge and hydrophobicity to strength of attachment of bacteria to cantaloupe rind. J. Food Prot. 2002, 65, 1093–1099. [Google Scholar] [CrossRef]
- Forfang, K.; Zimmermann, B.; Kosa, G.; Kohler, A.; Shapaval, V. FTIR Spectroscopy for evaluation and monitoring of lipid extraction efficiency for oleaginous fungi. PLoS ONE 2017, 12, e0170611. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.Y.; Frank, J.F. Effect of growth nutrients on attachment of Listeria monocytogenes to stainless steel. J. Food Prot. 1994, 57, 720–724. [Google Scholar] [CrossRef]
- Combrouse, T.; Sadovskaya, I.; Faille, C.; Kol, O.; Guérardel, Y.; Midelet-Bourdin, G. Quantification of the extracellular matrix of the Listeria monocytogenes biofilms of different phylogenic lineages with optimization of culture conditions. J. Appl. Microbiol. 2013, 114, 1120–1131. [Google Scholar] [CrossRef]
- Doijad, S.P.; Barbuddhe, S.B.; Garg, S.; Poharkar, K.V.; Kalorey, D.R.; Kurkure, N.V.; Rawool, D.B.; Chakraborty, T. Biofilm-forming abilities of Listeria monocytogenes serotypes isolated from different sources. PLoS ONE 2015, 10, e0137046. [Google Scholar] [CrossRef]
- Huang, Y.; Chakraborty, S.; Liang, H. Methods to probe the formation of biofilms: Applications in foods and related surfaces. Anal. Methods 2020, 12, 416–432. [Google Scholar] [CrossRef]
- Zeraik, A.; Nitschke, M. Influence of growth media and temperature on bacterial adhesion to polystyrene surfaces. Braz. Arch. Biol. Technol. 2012, 55, 569–576. [Google Scholar] [CrossRef]
- Frank, J.F.; Koffi, R.A. Surface-adherent growth of Listeria monocytogenes is associated with increased resistance to surfactant sanitizers and heat. J. Food Prot. 1990, 53, 550–554. [Google Scholar] [CrossRef] [PubMed]
- Lee, B.-H.; Hébraud, M.; Bernardi, T. Increased adhesion of Listeria monocytogenes strains to abiotic surfaces under cold stress. Front. Microbiol. 2017, 8, 2221. [Google Scholar] [CrossRef]
- Public Health England. Identification of Listeria Species, and Other Non-Sporing Gram Positive Rods (Except Corynebacterium). Available online: https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/534131/ID_3i3.2.pdf (accessed on 10 December 2020).
- Smith, C.; Metzger, J. Demonstration of a capsular structure on Listeria monocytogenes. Pathol. Microbiol. 1962, 25, 499–506. [Google Scholar] [CrossRef]
- Fadaee-Shohada, M.; Hirst, R.; Rutman, A.; Roberts, I.; O’Callaghan, C.; Andrew, P. Scanning Electron Microscope Image of Listeria and Extracellular Material. Available online: https://doi.org/10.1371/journal.pone.0010450.g002 (accessed on 28 December 2020).
- Sallen, B.; Rajoharison, A.; Desvarenne, S.; Quinn, F.; Mabilat, C. Comparative analysis of 16S and 23S RRNA sequences of Listeria species. Int. J. Syst. Bacteriol. 1996, 46 3, 669–674. [Google Scholar] [CrossRef]
- Begley, M.; Gahan, C.G.M.; Hill, C. Bile stress response in Listeria monocytogenes LO28: Adaptation, cross-protection, and identification of genetic loci involved in bile resistance. Appl. Environ. Microbiol. 2002, 68, 6005–6012. [Google Scholar] [CrossRef]
- Makino, S.; Uchida, I.; Terakado, N.; Sasakawa, C.; Yoshikawa, M. Molecular characterization and protein analysis of the cap region, which is essential for encapsulation in Bacillus anthracis. J. Bacteriol. 1989, 171, 722–730. [Google Scholar] [CrossRef]
- Chae, M.S.; Schraft, H.; Truelstrup Hansen, L.; Mackereth, R. Effects of physicochemical surface characteristics of Listeria monocytogenes strains on attachment to glass. Food Microbiol. 2006, 23, 250–259. [Google Scholar] [CrossRef]
- Harmsen, M.; Lappann, M.; Knøchel, S.; Molin, S. Role of extracellular DNA during biofilm formation by Listeria monocytogenes. Appl. Environ. Microbiol. 2010, 76, 2271–2279. [Google Scholar] [CrossRef] [PubMed]
- Borucki, M.K.; Peppin, J.D.; White, D.; Loge, F.; Call, D.R. Variation in biofilm formation among strains of Listeria monocytogenes. Appl. Environ. Microbiol. 2003, 69, 7336–7342. [Google Scholar] [CrossRef]
- Cachat, E.; Barker, M.; Read, T.D.; Priest, F.G. A Bacillus thuringiensis strain producing a polyglutamate capsule resembling that of Bacillus anthracis. FEMS Microbiol. Lett. 2008, 285, 220–226. [Google Scholar] [CrossRef]
- Vazquez-Armenta, F.J.; Bernal-Mercado, A.T.; Tapia-Rodriguez, M.R.; Gonzalez-Aguilar, G.A.; Lopez-Zavala, A.A.; Martinez-Tellez, M.A.; Hernandez-Oñate, M.A.; Ayala-Zavala, J.F. Quercetin reduces adhesion and inhibits biofilm development by listeria monocytogenes by reducing the amount of extracellular proteins. Food Control 2018, 90, 266–273. [Google Scholar] [CrossRef]
- Hether, N.W.; Jackson, L.L. Lipoteichoic acid from Listeria monocytogenes. J. Bacteriol. 1983, 156, 809–817. [Google Scholar] [CrossRef]
- Köseoğlu, V.K.; Heiss, C.; Azadi, P.; Topchiy, E.; Güvener, Z.T.; Lehmann, T.E.; Miller, K.W.; Gomelsky, M. Listeria monocytogenes exopolysaccharide: Origin, structure, biosynthetic machinery and c-Di-GMP-dependent regulation. Mol. Microbiol. 2015, 96, 728–743. [Google Scholar] [CrossRef] [PubMed]
- Joseph, B.; Mertins, S.; Stoll, R.; Schär, J.; Umesha, K.R.; Luo, Q.; Müller-Altrock, S.; Goebel, W. Glycerol metabolism and PrfA activity in Listeria monocytogenes. J. Bacteriol. 2008, 190, 5412–5430. [Google Scholar] [CrossRef]
- Webb, A.J.; Karatsa-Dodgson, M.; Gründling, A. Two-enzyme systems for glycolipid and polyglycerolphosphate lipoteichoic acid synthesis in Listeria monocytogenes. Mol. Microbiol. 2009, 74, 299–314. [Google Scholar] [CrossRef]
- Susi, H.; Byler, D.M.B.T.-M. Resolution-enhanced Fourier transform infrared spectroscopy of enzymes. In Enzyme Structure Part K; Academic Press: Cambridge, MA, USA, 1986; Volume 130, pp. 290–311. [Google Scholar]
- Krimm, S.; Bandekar, J. Vibrational spectroscopy and conformation of peptides, polypeptides, and proteins. In Advances in Protein Chemistry; Anfinsen, C.B., Edsall, J.T., Richards, F.M., Eds.; Academic Press: Cambridge, MA, USA, 1986; Volume 38, pp. 181–364. [Google Scholar]
- Kong, J.; Yu, S. Fourier transform infrared spectroscopic analysis of protein secondary structures. Acta Biochim. Biophys. Sin. 2007, 39, 549–559. [Google Scholar] [CrossRef]
- Olszewska, M.A.; Zhao, T.; Doyle, M.P. Inactivation and induction of sublethal injury of Listeria monocytogenes in biofilm treated with various sanitizers. Food Control 2016, 70, 371–379. [Google Scholar] [CrossRef]
- Khan, F.; Jeong, M.-C.; Park, S.-K.; Kim, S.-K.; Kim, Y.-M. Contribution of Chito-oligosaccharides to biofilm formation, antibiotics resistance and disinfectants tolerance of Listeria monocytogenes. Microb. Pathog. 2019, 136, 103673. [Google Scholar] [CrossRef]
- Colagiorgi, A.; Bruini, I.; Di Ciccio, P.A.; Zanardi, E.; Ghidini, S.; Ianieri, A. Listeria monocytogenes biofilms in the wonderland of food industry. Pathogens 2017, 6, 41. [Google Scholar] [CrossRef]
- Heir, E.; Møretrø, T.; Simensen, A.; Langsrud, S. Listeria monocytogenes strains show large variations in competitive growth in mixed culture biofilms and suspensions with bacteria from food processing environments. Int. J. Food Microbiol. 2018, 275, 46–55. [Google Scholar] [CrossRef]
- Andrade, J.C.; João, A.L.; de Alonso, C.S.; Barreto, A.S.; Henriques, A.R. genetic subtyping, biofilm-forming ability and biocide susceptibility of Listeria monocytogenes strains isolated from a ready-to-eat food industry. Antibiotics 2020, 9, 416. [Google Scholar] [CrossRef]
- Kassinger, S.J.; van Hoek, M.L. Biofilm architecture: An emerging synthetic biology target. Synth. Syst. Biotechnol. 2020, 5, 1–10. [Google Scholar] [CrossRef]
- Dos Reis-Teixeira, F.B.; Conceição, N.; da Silva, L.P.; Alves, V.F.; De Martinis, E.C.P. Potential of oxygen and nitrogen reactive intermediates to disperse Listeria monocytogenes from biofilms. Braz. J. Microbiol. 2019, 50, 501–506. [Google Scholar] [CrossRef] [PubMed]
- Bonifácio, D.; Martins, C.; David, B.; Lemos, C.; Neves, M.G.P.M.S.; Almeida, A.; Pinto, D.C.G.A.; Faustino, M.A.F.; Cunha, Â. Photodynamic inactivation of Listeria innocua biofilms with food-grade photosensitizers: A curcumin-rich extract of curcuma longa vs commercial curcumin. J. Appl. Microbiol. 2018, 125, 282–294. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-López, P.; Carballo-Justo, A.; Draper, L.A.; Cabo, M.L. Removal of Listeria monocytogenes dual-species biofilms using combined enzyme-benzalkonium chloride treatments. Biofouling 2017, 33, 45–58. [Google Scholar] [CrossRef] [PubMed]
- Carniello, V.; Peterson, B.W.; van der Mei, H.C.; Busscher, H.J. Physico-Chemistry from Initial Bacterial Adhesion to Surface-Programmed Biofilm Growth. Adv. Colloid Interface Sci. 2018, 261, 1–14. [Google Scholar] [CrossRef]
- Camargo, A.C.; Todorov, S.D.; Chihib, N.E.; Drider, D.; Nero, L.A. Lactic acid bacteria (LAB) and their bacteriocins as alternative biotechnological tools to control listeria monocytogenes biofilms in food processing facilities. Mol. Biotechnol. 2018, 60, 712–726. [Google Scholar] [CrossRef]
- Rocha, K.R.; Perini, H.F.; de Souza, C.M.; Schueler, J.; Tosoni, N.F.; Furlaneto, M.C.; Furlaneto-Maia, L. Inhibitory effect of bacteriocins from Enterococci on developing and preformed biofilms of Listeria monocytogenes, Listeria ivanovii and Listeria innocua. World J. Microbiol. Biotechnol. 2019, 35, 96. [Google Scholar] [CrossRef]
- Bolocan, A.S.; Pennone, V.; O’Connor, P.M.; Coffey, A.; Nicolau, A.I.; McAuliffe, O.; Jordan, K. Inhibition of Listeria monocytogenes biofilms by bacteriocin-producing bacteria isolated from mushroom substrate. J. Appl. Microbiol. 2017, 122, 279–293. [Google Scholar] [CrossRef]
- Oloketuyi, S.F.; Khan, F. Inhibition strategies of Listeria monocytogenes biofilms—Current knowledge and future outlooks. J. Basic Microbiol. 2017, 57, 728–743. [Google Scholar] [CrossRef]

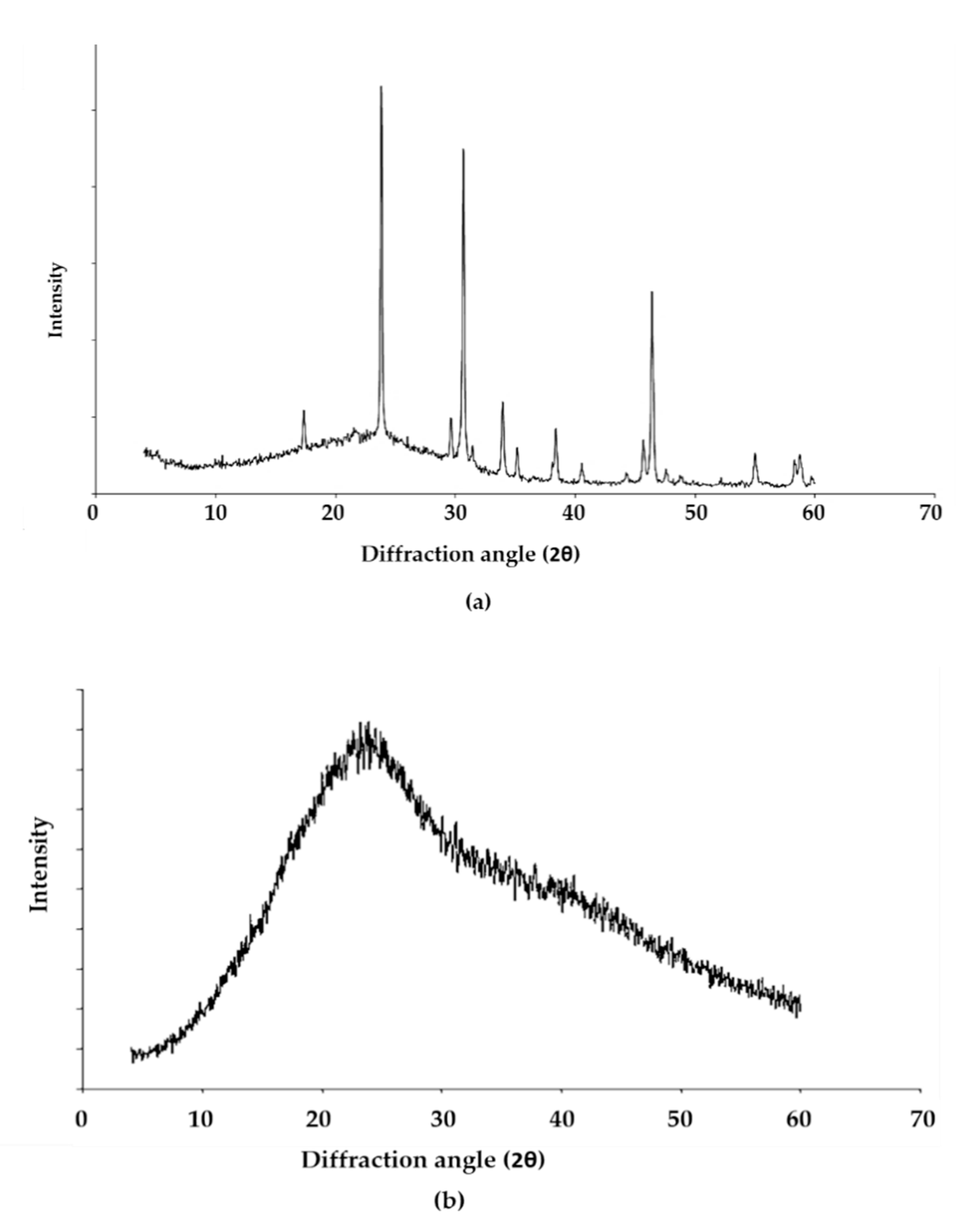
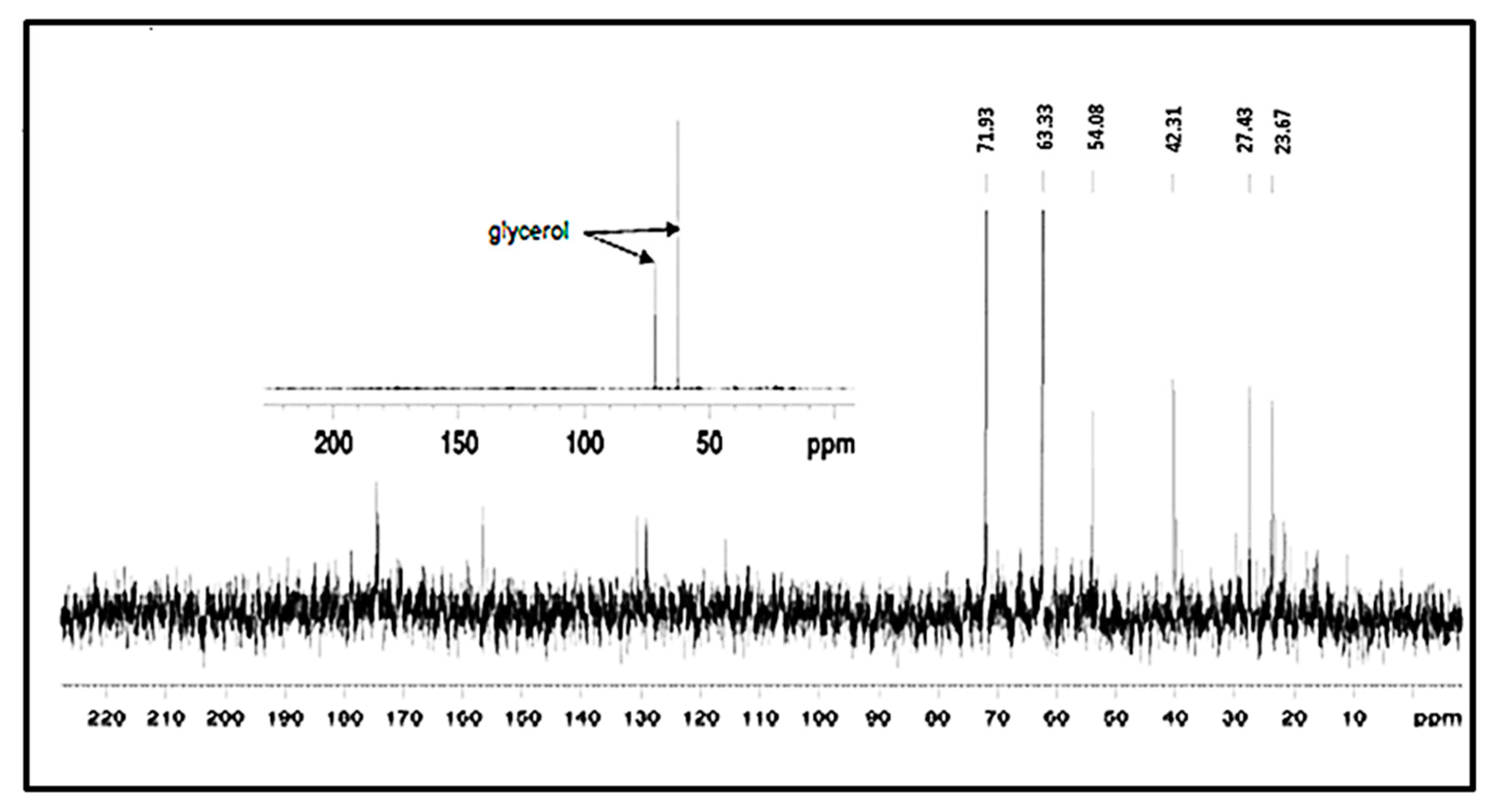

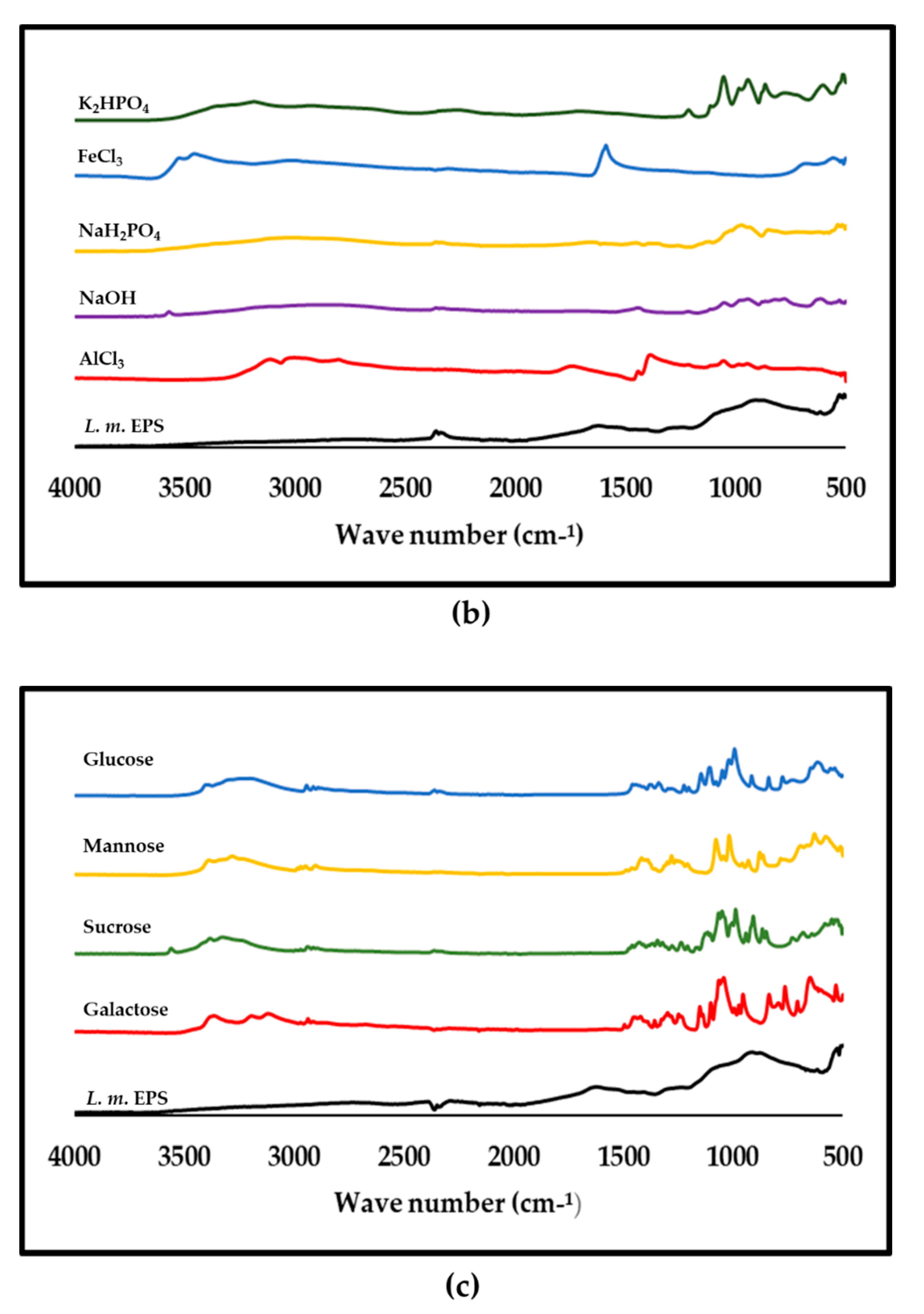
| (a) | ||
| Amino Acid | Listeria EPS | PGA |
| Cystine | 0.88 | 0.09 |
| Aspartic acid | 1.84 | 0.34 |
| Methionine | 0.38 | 0.00 |
| Threonine | 1.10 | 0.00 |
| Serine | 1.10 | 0.00 |
| Glutamic acid | 3.10 | 193.22 |
| Glycine | 1.72 | 0.07 |
| Alanine | 1.77 | 0.00 |
| Valine | 1.07 | 0.00 |
| Isoleucine | 1.09 | 0.10 |
| Leucine | 1.15 | 0.00 |
| Tyrosine | 1.26 | 0.00 |
| Phenylalanine | 1.24 | 0.04 |
| Lysine | 2.06 | 0.05 |
| Histidine | 0.31 | 0.00 |
| Arginine | 3.96 | 0.00 |
| (b) | ||
| Sugar Content (% w/w) of Listeria EPS and PGA | ||
| Sugar | Listeria EPS | PGA |
| Sucrose | 0.05 | 1.11 |
| Glucose | 0.44 | 0.09 |
| Fructose | 0.06 | 0.13 |
| Complex sugars | nd | nd |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nwaiwu, O.; Wong, L.; Lad, M.; Foster, T.; MacNaughtan, W.; Rees, C. Properties of the Extracellular Polymeric Substance Layer from Minimally Grown Planktonic Cells of Listeria monocytogenes. Biomolecules 2021, 11, 331. https://doi.org/10.3390/biom11020331
Nwaiwu O, Wong L, Lad M, Foster T, MacNaughtan W, Rees C. Properties of the Extracellular Polymeric Substance Layer from Minimally Grown Planktonic Cells of Listeria monocytogenes. Biomolecules. 2021; 11(2):331. https://doi.org/10.3390/biom11020331
Chicago/Turabian StyleNwaiwu, Ogueri, Lawrence Wong, Mita Lad, Timothy Foster, William MacNaughtan, and Catherine Rees. 2021. "Properties of the Extracellular Polymeric Substance Layer from Minimally Grown Planktonic Cells of Listeria monocytogenes" Biomolecules 11, no. 2: 331. https://doi.org/10.3390/biom11020331
APA StyleNwaiwu, O., Wong, L., Lad, M., Foster, T., MacNaughtan, W., & Rees, C. (2021). Properties of the Extracellular Polymeric Substance Layer from Minimally Grown Planktonic Cells of Listeria monocytogenes. Biomolecules, 11(2), 331. https://doi.org/10.3390/biom11020331