Probiotics and Trained Immunity
Abstract
1. Introduction
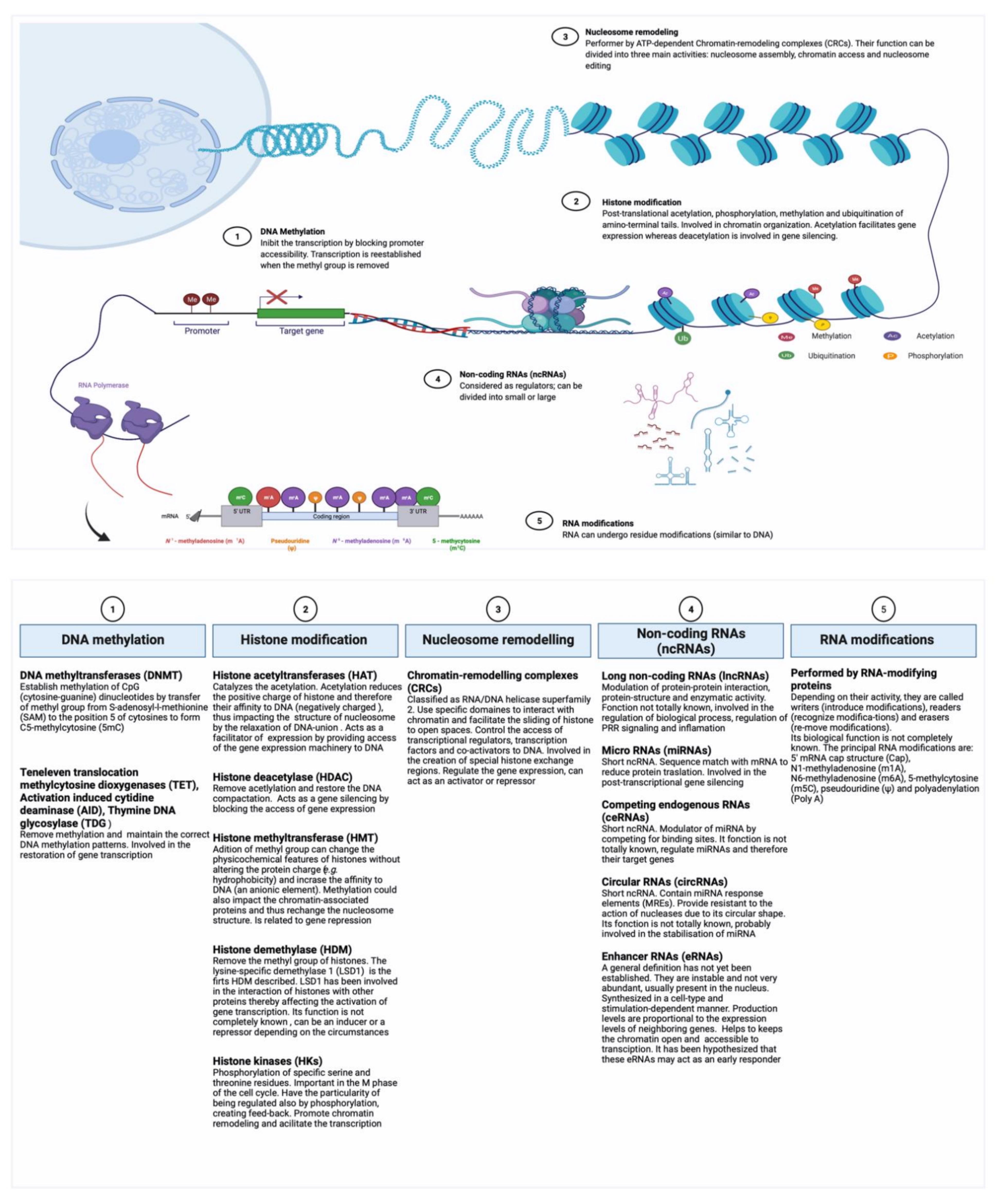
2. Epigenetic Mechanisms Potentially Involved in Trained Immunity
2.1. Epigenetic Regulation
2.2. Probiotics as Potential Epigenetic Regulators
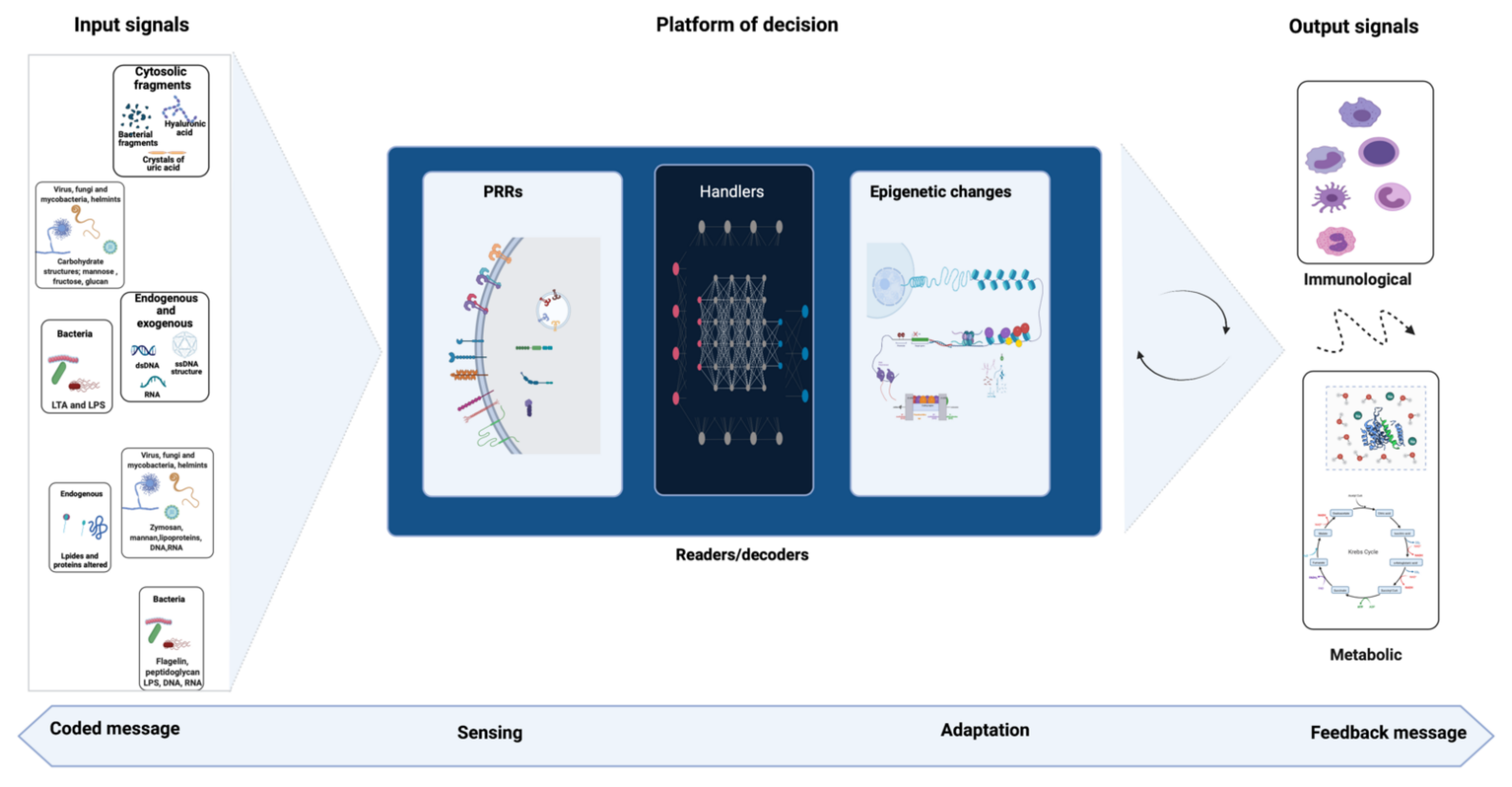
3. Signal Detection and Immune Response
4. Immune Cells Subjected to Trained Immunity
5. Hypothetical Role of Probiotics in Trained Immunity
6. Conclusions and Perspectives
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gasteiger, G.; D’Osualdo, A.; Schubert, D.A.; Weber, A.; Bruscia, E.M.; Hartl, D. Cellular Innate Immunity: An Old Game with New Players. J. Innate Immun. 2017, 9, 111–125. [Google Scholar] [CrossRef]
- Netea, M.G.; Quintin, J.; van der Meer, J.W. Trained immunity: A memory for innate host defense. Cell Host Microbe 2011, 9, 355–361. [Google Scholar] [CrossRef]
- Horvath, P.; Barrangou, R. CRISPR/Cas, the immune system of bacteria and archaea. Science 2010, 327, 167–170. [Google Scholar] [CrossRef]
- Sorek, R.; Lawrence, C.M.; Wiedenheft, B. CRISPR-mediated adaptive immune systems in bacteria and archaea. Annu. Rev. Biochem. 2013, 82, 237–266. [Google Scholar] [CrossRef]
- Koonin, E.V.; Makarova, K.S.; Zhang, F. Diversity, classification and evolution of CRISPR-Cas systems. Curr. Opin. Microbiol. 2017, 37, 67–78. [Google Scholar] [CrossRef]
- Reimer-Michalski, E.M.; Conrath, U. Innate immune memory in plants. Semin. Immunol. 2016, 28, 319–327. [Google Scholar] [CrossRef]
- Weinhold, B. Epigenetics: The science of change. Environ. Health Perspect. 2006, 114, A160–A167. [Google Scholar] [CrossRef]
- Handy, D.E.; Castro, R.; Loscalzo, J. Epigenetic modifications: Basic mechanisms and role in cardiovascular disease. Circulation 2011, 123, 2145–2156. [Google Scholar] [CrossRef]
- Netea, M.G.; Joosten, L.A.B.; van der Meer, J.W.M. Hypothesis: Stimulation of trained immunity as adjunctive immunotherapy in cancer. J. Leukoc. Biol. 2017, 102, 1323–1332. [Google Scholar] [CrossRef]
- Netea, M.G.; Dominguez-Andres, J.; Barreiro, L.B.; Chavakis, T.; Divangahi, M.; Fuchs, E.; Joosten, L.A.B.; van der Meer, J.W.M.; Mhlanga, M.M.; Mulder, W.J.M.; et al. Defining trained immunity and its role in health and disease. Nat. Rev. Immunol. 2020, 20, 375–388. [Google Scholar] [CrossRef]
- Netea, M.G.; van der Meer, J.W. Trained Immunity: An Ancient Way of Remembering. Cell Host Microbe 2017, 21, 297–300. [Google Scholar] [CrossRef]
- Quintin, J.; Saeed, S.; Martens, J.H.A.; Giamarellos-Bourboulis, E.J.; Ifrim, D.C.; Logie, C.; Jacobs, L.; Jansen, T.; Kullberg, B.J.; Wijmenga, C.; et al. Candida albicans infection affords protection against reinfection via functional reprogramming of monocytes. Cell Host Microbe 2012, 12, 223–232. [Google Scholar] [CrossRef]
- Zhang, Q.; Cao, X. Epigenetic regulation of the innate immune response to infection. Nat. Rev. Immunol. 2019, 19, 417–432. [Google Scholar] [CrossRef]
- Jia, G.; Fu, Y.; Zhao, X.; Dai, Q.; Zheng, G.; Yang, Y.; Yi, C.; Lindahl, T.; Pan, T.; Yang, Y.G.; et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 2011, 7, 885–887. [Google Scholar] [CrossRef]
- Liu, L.; Song, B.; Ma, J.; Song, Y.; Zhang, S.Y.; Tang, Y.; Wu, X.; Wei, Z.; Chen, K.; Su, J.; et al. Bioinformatics approaches for deciphering the epitranscriptome: Recent progress and emerging topics. Comput. Struct. Biotechnol. J. 2020, 18, 1587–1604. [Google Scholar] [CrossRef]
- Burgess, A.; David, R.; Searle, I.R. Deciphering the epitranscriptome: A green perspective. J. Integr. Plant Biol. 2016, 58, 822–835. [Google Scholar] [CrossRef]
- Kleinnijenhuis, J.; Quintin, J.; Preijers, F.; Joosten, L.A.; Ifrim, D.C.; Saeed, S.; Jacobs, C.; van Loenhout, J.; de Jong, D.; Stunnenberg, H.G.; et al. Bacille Calmette-Guerin induces NOD2-dependent nonspecific protection from reinfection via epigenetic reprogramming of monocytes. Proc. Natl. Acad. Sci. USA 2012, 109, 17537–17542. [Google Scholar] [CrossRef] [PubMed]
- DiNardo, A.R.; Netea, M.G.; Musher, D.M. Postinfectious Epigenetic Immune Modifications - A Double-Edged Sword. N. Engl. J. Med. 2021, 384, 261–270. [Google Scholar] [CrossRef]
- Khader, S.A.; Divangahi, M.; Hanekom, W.; Hill, P.C.; Maeurer, M.; Makar, K.W.; Mayer-Barber, K.D.; Mhlanga, M.M.; Nemes, E.; Schlesinger, L.S.; et al. Targeting innate immunity for tuberculosis vaccination. J. Clin. Investig. 2019, 129, 3482–3491. [Google Scholar] [CrossRef] [PubMed]
- Koeken, V.; van Crevel, R.; Netea, M.G.; Li, Y. Resolving trained immunity with systems biology. Eur. J. Immunol. 2021, 51, 773–784. [Google Scholar] [CrossRef]
- Lyko, F. The DNA methyltransferase family: A versatile toolkit for epigenetic regulation. Nat. Rev. Genet. 2018, 19, 81–92. [Google Scholar] [CrossRef]
- Hauser, A.T.; Robaa, D.; Jung, M. Epigenetic small molecule modulators of histone and DNA methylation. Curr. Opin. Chem. Biol. 2018, 45, 73–85. [Google Scholar] [CrossRef]
- Rice, J.C.; Allis, C.D. Histone methylation versus histone acetylation: New insights into epigenetic regulation. Curr. Opin. Cell Biol. 2001, 13, 263–273. [Google Scholar] [CrossRef]
- Grunstein, M. Histone acetylation in chromatin structure and transcription. Nature 1997, 389, 349–352. [Google Scholar] [CrossRef] [PubMed]
- Biel, M.; Wascholowski, V.; Giannis, A. Epigenetics—An epicenter of gene regulation: Histones and histone-modifying enzymes. Angew. Chem. Int. Ed. Engl. 2005, 44, 3186–3216. [Google Scholar] [CrossRef] [PubMed]
- Perillo, B.; Tramontano, A.; Pezone, A.; Migliaccio, A. LSD1: More than demethylation of histone lysine residues. Exp. Mol. Med. 2020, 52, 1936–1947. [Google Scholar] [CrossRef]
- Combes, G.; Alharbi, I.; Braga, L.G.; Elowe, S. Playing polo during mitosis: PLK1 takes the lead. Oncogene 2017, 36, 4819–4827. [Google Scholar] [CrossRef] [PubMed]
- Davie, J.R.; Spencer, V.A. Control of histone modifications. J. Cell Biochem. 1999, 141–148. [Google Scholar] [CrossRef]
- Clapier, C.R.; Iwasa, J.; Cairns, B.R.; Peterson, C.L. Mechanisms of action and regulation of ATP-dependent chromatin-remodelling complexes. Nat. Rev. Mol. Cell Biol. 2017, 18, 407–422. [Google Scholar] [CrossRef]
- Becker, P.B.; Horz, W. ATP-dependent nucleosome remodeling. Annu. Rev. Biochem. 2002, 71, 247–273. [Google Scholar] [CrossRef]
- Esteller, M. Non-coding RNAs in human disease. Nat. Rev. Genet. 2011, 12, 861–874. [Google Scholar] [CrossRef] [PubMed]
- Sen, R.; Ghosal, S.; Das, S.; Balti, S.; Chakrabarti, J. Competing endogenous RNA: The key to posttranscriptional regulation. Sci. World J. 2014, 2014, 896206. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Yang, T.; Wang, W.; Xi, W.; Zhang, T.; Li, Q.; Yang, A.; Wang, T. Circular RNAs in immune responses and immune diseases. Theranostics 2019, 9, 588–607. [Google Scholar] [CrossRef] [PubMed]
- Sartorelli, V.; Lauberth, S.M. Enhancer RNAs are an important regulatory layer of the epigenome. Nat. Struct. Mol. Biol. 2020, 27, 521–528. [Google Scholar] [CrossRef]
- Meng, H.; Bartholomew, B. Emerging roles of transcriptional enhancers in chromatin looping and promoter-proximal pausing of RNA polymerase II. J. Biol. Chem. 2018, 293, 13786–13794. [Google Scholar] [CrossRef]
- Esteve-Puig, R.; Bueno-Costa, A.; Esteller, M. Writers, readers and erasers of RNA modifications in cancer. Cancer Lett. 2020, 474, 127–137. [Google Scholar] [CrossRef]
- Nie, F.; Feng, P.; Song, X.; Wu, M.; Tang, Q.; Chen, W. RNAWRE: A resource of writers, readers and erasers of RNA modifications. Database 2020, 2020. [Google Scholar] [CrossRef]
- Shenderov, B.A. Gut indigenous microbiota and epigenetics. Microb. Ecol. Health Dis. 2012, 23, 618. [Google Scholar] [CrossRef]
- Carbonero, F. Human epigenetics and microbiome: The potential for a revolution in both research areas by integrative studies. Future Sci. OA 2017, 3, Fso207. [Google Scholar] [CrossRef]
- Nackerdien, Z.E. Perspectives on microbes as oncogenic infectious agents and implications for breast cancer. Med. Hypotheses 2008, 71, 302–306. [Google Scholar] [CrossRef]
- Licciardi, P.V.; Wong, S.S.; Tang, M.L.; Karagiannis, T.C. Epigenome targeting by probiotic metabolites. Gut Pathog. 2010, 2, 24. [Google Scholar] [CrossRef][Green Version]
- Canani, R.B.; Costanzo, M.D.; Leone, L.; Bedogni, G.; Brambilla, P.; Cianfarani, S.; Nobili, V.; Pietrobelli, A.; Agostoni, C. Epigenetic mechanisms elicited by nutrition in early life. Nutr. Res. Rev. 2011, 24, 198–205. [Google Scholar] [CrossRef]
- Worthley, D.L.; Le Leu, R.K.; Whitehall, V.L.; Conlon, M.; Christophersen, C.; Belobrajdic, D.; Mallitt, K.A.; Hu, Y.; Irahara, N.; Ogino, S.; et al. A human, double-blind, placebo-controlled, crossover trial of prebiotic, probiotic, and synbiotic supplementation: Effects on luminal, inflammatory, epigenetic, and epithelial biomarkers of colorectal cancer. Am. J. Clin. Nutr. 2009, 90, 578–586. [Google Scholar] [CrossRef] [PubMed]
- Rosenberg, E.; Zilber-Rosenberg, I. Symbiosis and development: The hologenome concept. Birth Defects Res. C Embryo Today 2011, 93, 56–66. [Google Scholar] [CrossRef] [PubMed]
- Azad, M.B.; Kozyrskyj, A.L. Perinatal programming of asthma: The role of gut microbiota. Clin. Dev. Immunol. 2012, 2012, 932072. [Google Scholar] [CrossRef]
- West, C.E.; D’Vaz, N.; Prescott, S.L. Dietary immunomodulatory factors in the development of immune tolerance. Curr. Allergy Asthma Rep. 2011, 11, 325–333. [Google Scholar] [CrossRef] [PubMed]
- Bebek, G.; Bennett, K.L.; Funchain, P.; Campbell, R.; Seth, R.; Scharpf, J.; Burkey, B.; Eng, C. Microbiomic subprofiles and MDR1 promoter methylation in head and neck squamous cell carcinoma. Hum. Mol. Genet. 2012, 21, 1557–1565. [Google Scholar] [CrossRef]
- Hamon, M.A.; Cossart, P. Histone modifications and chromatin remodeling during bacterial infections. Cell Host Microbe 2008, 4, 100–109. [Google Scholar] [CrossRef] [PubMed]
- Candelli, M.; Franza, L.; Pignataro, G.; Ojetti, V.; Covino, M.; Piccioni, A.; Gasbarrini, A.; Franceschi, F. Interaction between Lipopolysaccharide and Gut Microbiota in Inflammatory Bowel Diseases. Int. J. Mol. Sci. 2021, 22, 6242. [Google Scholar] [CrossRef]
- Ghadimi, D.; Helwig, U.; Schrezenmeir, J.; Heller, K.J.; de Vrese, M. Epigenetic imprinting by commensal probiotics inhibits the IL-23/IL-17 axis in an in vitro model of the intestinal mucosal immune system. J. Leukoc. Biol. 2012, 92, 895–911. [Google Scholar] [CrossRef] [PubMed]
- Kumar, M.; Nagpal, R.; Verma, V.; Kumar, A.; Kaur, N.; Hemalatha, R.; Gautam, S.K.; Singh, B. Probiotic metabolites as epigenetic targets in the prevention of colon cancer. Nutr. Rev. 2013, 71, 23–34. [Google Scholar] [CrossRef]
- Panwar, H.; Rashmi, H.M.; Batish, V.K.; Grover, S. Probiotics as potential biotherapeutics in the management of type 2 diabetes - prospects and perspectives. Diabetes Metab. Res. Rev. 2013, 29, 103–112. [Google Scholar] [CrossRef] [PubMed]
- Sestito, S.; D’Auria, E.; Baldassarre, M.E.; Salvatore, S.; Tallarico, V.; Stefanelli, E.; Tarsitano, F.; Concolino, D.; Pensabene, L. The Role of Prebiotics and Probiotics in Prevention of Allergic Diseases in Infants. Front. Pediatr. 2020, 8, 583946. [Google Scholar] [CrossRef] [PubMed]
- Bhat, M.I.; Kapila, R. Dietary metabolites derived from gut microbiota: Critical modulators of epigenetic changes in mammals. Nutr. Rev. 2017, 75, 374–389. [Google Scholar] [CrossRef] [PubMed]
- Lenoir, M.; Martín, R.; Torres-Maravilla, E.; Chadi, S.; González-Dávila, P.; Sokol, H.; Langella, P.; Chain, F.; Bermúdez-Humarán, L.G. Butyrate mediates anti-inflammatory effects of Faecalibacterium prausnitzii in intestinal epithelial cells through Dact3. Gut Microbes 2020, 12, 1–16. [Google Scholar] [CrossRef]
- Amarante-Mendes, G.P.; Adjemian, S.; Branco, L.M.; Zanetti, L.C.; Weinlich, R.; Bortoluci, K.R. Pattern Recognition Receptors and the Host Cell Death Molecular Machinery. Front. Immunol. 2018, 9, 2379. [Google Scholar] [CrossRef]
- Medzhitov, R.; Janeway, C.A., Jr. Decoding the patterns of self and nonself by the innate immune system. Science 2002, 296, 298–300. [Google Scholar] [CrossRef]
- Koenderman, L.; Buurman, W.; Daha, M.R. The innate immune response. Immunol. Lett. 2014, 162, 95–102. [Google Scholar] [CrossRef]
- Canton, J.; Neculai, D.; Grinstein, S. Scavenger receptors in homeostasis and immunity. Nat. Rev. Immunol. 2013, 13, 621–634. [Google Scholar] [CrossRef]
- Aderem, A.; Ulevitch, R.J. Toll-like receptors in the induction of the innate immune response. Nature 2000, 406, 782–787. [Google Scholar] [CrossRef]
- Matzinger, P. The danger model: A renewed sense of self. Science 2002, 296, 301–305. [Google Scholar] [CrossRef]
- Zindel, J.; Kubes, P. DAMPs, PAMPs, and LAMPs in Immunity and Sterile Inflammation. Annu. Rev. Pathol. 2020, 15, 493–518. [Google Scholar] [CrossRef]
- PrabhuDas, M.R.; Baldwin, C.L.; Bollyky, P.L.; Bowdish, D.M.E.; Drickamer, K.; Febbraio, M.; Herz, J.; Kobzik, L.; Krieger, M.; Loike, J.; et al. A Consensus Definitive Classification of Scavenger Receptors and Their Roles in Health and Disease. J. Immunol. 2017, 198, 3775–3789. [Google Scholar] [CrossRef] [PubMed]
- Del Fresno, C.; Cueto, F.J.; Sancho, D. A Proposal for Nomenclature in Myeloid C-Type Lectin Receptors. Front. Immunol. 2019, 10, 2098. [Google Scholar] [CrossRef] [PubMed]
- Figdor, C.G.; van Kooyk, Y.; Adema, G.J. C-type lectin receptors on dendritic cells and Langerhans cells. Nat. Rev. Immunol. 2002, 2, 77–84. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Lu, Y.; Thulasi Raman, S.N.; Xu, F.; Wu, Q.; Li, Z.; Brownlie, R.; Liu, Q.; Zhou, Y. Nuclear-resident RIG-I senses viral replication inducing antiviral immunity. Nat. Commun. 2018, 9, 3199. [Google Scholar] [CrossRef] [PubMed]
- Uhlén, M.; Fagerberg, L.; Hallström, B.M.; Lindskog, C.; Oksvold, P.; Mardinoglu, A.; Sivertsson, Å.; Kampf, C.; Sjöstedt, E.; Asplund, A.; et al. Proteomics. Tissue-based map of the human proteome. Science 2015, 347, 1260419. [Google Scholar] [CrossRef] [PubMed]
- Pombinho, R.; Sousa, S.; Cabanes, D. Scavenger Receptors: Promiscuous Players during Microbial Pathogenesis. Crit. Rev. Microbiol. 2018, 44, 685–700. [Google Scholar] [CrossRef] [PubMed]
- Hoving, J.C.; Wilson, G.J.; Brown, G.D. Signalling C-type lectin receptors, microbial recognition and immunity. Cell. Microbiol. 2014, 16, 185–194. [Google Scholar] [CrossRef] [PubMed]
- Geijtenbeek, T.B.; Gringhuis, S.I. Signalling through C-type lectin receptors: Shaping immune responses. Nat. Rev. Immunol. 2009, 9, 465–479. [Google Scholar] [CrossRef]
- Ganguly, K.; Kishore, U.; Madan, T. Interplay between C-type lectin receptors and microRNAs in cellular homeostasis and immune response. FEBS J. 2021, 288, 4210–4229. [Google Scholar] [CrossRef]
- Brown, G.D.; Willment, J.A.; Whitehead, L. C-type lectins in immunity and homeostasis. Nat. Rev. Immunol. 2018, 18, 374–389. [Google Scholar] [CrossRef]
- Mokhtari, Y.; Pourbagheri-Sigaroodi, A.; Zafari, P.; Bagheri, N.; Ghaffari, S.H.; Bashash, D. Toll-like receptors (TLRs): An old family of immune receptors with a new face in cancer pathogenesis. J. Cell. Mol. Med. 2021, 25, 639–651. [Google Scholar] [CrossRef] [PubMed]
- Kawai, T.; Akira, S. The role of pattern-recognition receptors in innate immunity: Update on Toll-like receptors. Nat. Immunol. 2010, 11, 373–384. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Zen, K. Toll-Like Receptors Regulate the Development and Progression of Renal Diseases. Kidney Dis. 2021, 7, 14–23. [Google Scholar] [CrossRef] [PubMed]
- Onomoto, K.; Onoguchi, K.; Yoneyama, M. Regulation of RIG-I-like receptor-mediated signaling: Interaction between host and viral factors. Cell. Mol. Immunol. 2021, 18, 539–555. [Google Scholar] [CrossRef]
- Rehwinkel, J.; Gack, M.U. RIG-I-like receptors: Their regulation and roles in RNA sensing. Nat. Rev. Immunol. 2020, 20, 537–551. [Google Scholar] [CrossRef]
- Kanneganti, T.D.; Lamkanfi, M.; Nunez, G. Intracellular NOD-like receptors in host defense and disease. Immunity 2007, 27, 549–559. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.; Gibson, S.A.; Ting, J.P.Y. Gut microbiota, NLR proteins, and intestinal homeostasis. J. Exp. Med. 2020, 217. [Google Scholar] [CrossRef] [PubMed]
- Saur, I.M.L.; Panstruga, R.; Schulze-Lefert, P. NOD-like receptor-mediated plant immunity: From structure to cell death. Nat. Rev. Immunol. 2021, 21, 305–318. [Google Scholar] [CrossRef]
- Abrahams, V.M. The role of the Nod-like receptor family in trophoblast innate immune responses. J. Reprod. Immunol. 2011, 88, 112–117. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Sun, L.; Chen, Z.J. Regulation and function of the cGAS-STING pathway of cytosolic DNA sensing. Nat. Immunol. 2016, 17, 1142–1149. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Wu, J.; Du, F.; Chen, X.; Chen, Z.J. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science 2013, 339, 786–791. [Google Scholar] [CrossRef] [PubMed]
- Wan, D.; Jiang, W.; Hao, J. Research Advances in How the cGAS-STING Pathway Controls the Cellular Inflammatory Response. Front. Immunol. 2020, 11, 615. [Google Scholar] [CrossRef]
- O’Neill, L.A.; Kishton, R.J.; Rathmell, J. A guide to immunometabolism for immunologists. Nat. Rev. Immunol. 2016, 16, 553–565. [Google Scholar] [CrossRef]
- Baron, M.H.; Isern, J.; Fraser, S.T. The embryonic origins of erythropoiesis in mammals. Blood 2012, 119, 4828–4837. [Google Scholar] [CrossRef] [PubMed]
- Cedar, H.; Bergman, Y. Epigenetics of haematopoietic cell development. Nat. Rev. Immunol. 2011, 11, 478–488. [Google Scholar] [CrossRef]
- Crisan, M.; Dzierzak, E. The many faces of hematopoietic stem cell heterogeneity. Development 2016, 143, 4571–4581. [Google Scholar] [CrossRef]
- Cheng, H.; Zheng, Z.; Cheng, T. New paradigms on hematopoietic stem cell differentiation. Protein Cell 2020, 11, 34–44. [Google Scholar] [CrossRef]
- Bedoui, S.; Gebhardt, T.; Gasteiger, G.; Kastenmüller, W. Parallels and differences between innate and adaptive lymphocytes. Nat. Immunol. 2016, 17, 490–494. [Google Scholar] [CrossRef]
- Psaila, B.; Mead, A.J. Single-cell approaches reveal novel cellular pathways for megakaryocyte and erythroid differentiation. Blood 2019, 133, 1427–1435. [Google Scholar] [CrossRef]
- Kabelitz, D. Expression and function of Toll-like receptors in T lymphocytes. Curr. Opin. Immunol. 2007, 19, 39–45. [Google Scholar] [CrossRef]
- Crellin, N.K.; Trifari, S.; Kaplan, C.D.; Satoh-Takayama, N.; Di Santo, J.P.; Spits, H. Regulation of cytokine secretion in human CD127(+) LTi-like innate lymphoid cells by Toll-like receptor 2. Immunity 2010, 33, 752–764. [Google Scholar] [CrossRef] [PubMed]
- Pluddemann, A.; Mukhopadhyay, S.; Gordon, S. Innate immunity to intracellular pathogens: Macrophage receptors and responses to microbial entry. Immunol. Rev. 2011, 240, 11–24. [Google Scholar] [CrossRef]
- Gurtner, A.; Gonzalez-Perez, I.; Arnold, I.C. Intestinal eosinophils, homeostasis and response to bacterial intrusion. Semin. Immunopathol. 2021, 43, 295–306. [Google Scholar] [CrossRef] [PubMed]
- Shamri, R.; Xenakis, J.J.; Spencer, L.A. Eosinophils in innate immunity: An evolving story. Cell Tissue Res. 2011, 343, 57–83. [Google Scholar] [CrossRef] [PubMed]
- Kvarnhammar, A.M.; Cardell, L.O. Pattern-recognition receptors in human eosinophils. Immunology 2012, 136, 11–20. [Google Scholar] [CrossRef] [PubMed]
- Pellefigues, C.; Mehta, P.; Chappell, S.; Yumnam, B.; Old, S.; Camberis, M.; Le Gros, G. Diverse innate stimuli activate basophils through pathways involving Syk and IkappaB kinases. Proc. Natl. Acad. Sci. USA 2021, 118. [Google Scholar] [CrossRef]
- Qiu, H.N.; Wong, C.K.; Chu, I.M.; Hu, S.; Lam, C.W. Muramyl dipeptide mediated activation of human bronchial epithelial cells interacting with basophils: A novel mechanism of airway inflammation. Clin. Exp. Immunol. 2013, 172, 81–94. [Google Scholar] [CrossRef]
- Bieneman, A.P.; Chichester, K.L.; Chen, Y.H.; Schroeder, J.T. Toll-like receptor 2 ligands activate human basophils for both IgE-dependent and IgE-independent secretion. J. Allergy Clin. Immunol. 2005, 115, 295–301. [Google Scholar] [CrossRef]
- Jeon, J.H.; Ahn, K.B.; Kim, S.K.; Im, J.; Yun, C.H.; Han, S.H. Bacterial flagellin induces IL-6 expression in human basophils. Mol. Immunol. 2015, 65, 168–176. [Google Scholar] [CrossRef]
- Yousefi, S.; Morshed, M.; Amini, P.; Stojkov, D.; Simon, D.; von Gunten, S.; Kaufmann, T.; Simon, H.U. Basophils exhibit antibacterial activity through extracellular trap formation. Allergy 2015, 70, 1184–1188. [Google Scholar] [CrossRef] [PubMed]
- Espinosa-Riquer, Z.P.; Segura-Villalobos, D.; Ramirez-Moreno, I.G.; Perez Rodriguez, M.J.; Lamas, M.; Gonzalez-Espinosa, C. Signal Transduction Pathways Activated by Innate Immunity in Mast Cells: Translating Sensing of Changes into Specific Responses. Cells 2020, 9, 2411. [Google Scholar] [CrossRef] [PubMed]
- Agier, J.; Pastwinska, J.; Brzezinska-Blaszczyk, E. An overview of mast cell pattern recognition receptors. Inflamm. Res. 2018, 67, 737–746. [Google Scholar] [CrossRef]
- Koller, B.; Bals, R.; Roos, D.; Korting, H.C.; Griese, M.; Hartl, D. Innate immune receptors on neutrophils and their role in chronic lung disease. Eur. J. Clin. Investig. 2009, 39, 535–547. [Google Scholar] [CrossRef] [PubMed]
- Mantovani, A.; Cassatella, M.A.; Costantini, C.; Jaillon, S. Neutrophils in the activation and regulation of innate and adaptive immunity. Nat. Rev. Immunol. 2011, 11, 519–531. [Google Scholar] [CrossRef]
- Rungelrath, V.; Kobayashi, S.D.; DeLeo, F.R. Neutrophils in innate immunity and systems biology-level approaches. Wiley Interdiscip. Rev. Syst. Biol. Med. 2020, 12, e1458. [Google Scholar] [CrossRef] [PubMed]
- Dzopalic, T.; Rajkovic, I.; Dragicevic, A.; Colic, M. The response of human dendritic cells to co-ligation of pattern-recognition receptors. Immunol. Res. 2012, 52, 20–33. [Google Scholar] [CrossRef] [PubMed]
- Choreno-Parra, J.A.; Weinstein, L.I.; Yunis, E.J.; Zuniga, J.; Hernandez-Pando, R. Thinking Outside the Box: Innate- and B Cell-Memory Responses as Novel Protective Mechanisms Against Tuberculosis. Front. Immunol. 2020, 11, 226. [Google Scholar] [CrossRef] [PubMed]
- Adib-Conquy, M.; Scott-Algara, D.; Cavaillon, J.M.; Souza-Fonseca-Guimaraes, F. TLR-mediated activation of NK cells and their role in bacterial/viral immune responses in mammals. Immunol. Cell Biol. 2014, 92, 256–262. [Google Scholar] [CrossRef]
- Stokic-Trtica, V.; Diefenbach, A.; Klose, C.S.N. NK Cell Development in Times of Innate Lymphoid Cell Diversity. Front. Immunol. 2020, 11, 813. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Wang, Y.; Li, Q.; Tian, K.; Xu, L.; Liu, G.; Guo, C. Exopolysaccharide, Isolated From a Novel Strain Bifidobacterium breve lw01 Possess an Anticancer Effect on Head and Neck Cancer—Genetic and Biochemical Evidences. Front. Microbiol. 2019, 10, 1044. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Gonzalez, I.; Ghaedi, M.; Steer, C.A.; Matha, L.; Vivier, E.; Takei, F. ILC2 memory: Recollection of previous activation. Immunol. Rev. 2018, 283, 41–53. [Google Scholar] [CrossRef] [PubMed]
- Eberl, G.; Colonna, M.; Di Santo, J.P.; McKenzie, A.N. Innate lymphoid cells. Innate lymphoid cells: A new paradigm in immunology. Science 2015, 348, aaa6566. [Google Scholar] [CrossRef]
- Pavlovic, M.; Gross, C.; Chili, C.; Secher, T.; Treiner, E. MAIT Cells Display a Specific Response to Type 1 IFN Underlying the Adjuvant Effect of TLR7/8 Ligands. Front. Immunol. 2020, 11, 2097. [Google Scholar] [CrossRef]
- Legoux, F.; Salou, M.; Lantz, O. MAIT Cell Development and Functions: The Microbial Connection. Immunity 2020, 53, 710–723. [Google Scholar] [CrossRef]
- Dias, J.; Leeansyah, E.; Sandberg, J.K. Multiple layers of heterogeneity and subset diversity in human MAIT cell responses to distinct microorganisms and to innate cytokines. Proc. Natl. Acad. Sci. USA 2017, 114, E5434–E5443. [Google Scholar] [CrossRef]
- Provine, N.M.; Klenerman, P. MAIT Cells in Health and Disease. Annu. Rev. Immunol. 2020, 38, 203–228. [Google Scholar] [CrossRef]
- Mattner, J.; Debord, K.L.; Ismail, N.; Goff, R.D.; Cantu, C., 3rd; Zhou, D.; Saint-Mezard, P.; Wang, V.; Gao, Y.; Yin, N.; et al. Exogenous and endogenous glycolipid antigens activate NKT cells during microbial infections. Nature 2005, 434, 525–529. [Google Scholar] [CrossRef]
- Van Kaer, L.; Parekh, V.V.; Wu, L. The Response of CD1d-Restricted Invariant NKT Cells to Microbial Pathogens and Their Products. Front. Immunol. 2015, 6, 226. [Google Scholar] [CrossRef]
- Kronenberg, M.; Kinjo, Y. Innate-like recognition of microbes by invariant natural killer T cells. Curr. Opin. Immunol. 2009, 21, 391–396. [Google Scholar] [CrossRef] [PubMed]
- Brennan, P.J.; Brigl, M.; Brenner, M.B. Invariant natural killer T cells: An innate activation scheme linked to diverse effector functions. Nat. Rev. Immunol. 2013, 13, 101–117. [Google Scholar] [CrossRef]
- Olszak, T.; An, D.; Zeissig, S.; Vera, M.P.; Richter, J.; Franke, A.; Glickman, J.N.; Siebert, R.; Baron, R.M.; Kasper, D.L.; et al. Microbial exposure during early life has persistent effects on natural killer T cell function. Science 2012, 336, 489–493. [Google Scholar] [CrossRef]
- Cheroutre, H.; Lambolez, F.; Mucida, D. The light and dark sides of intestinal intraepithelial lymphocytes. Nat. Rev. Immunol. 2011, 11, 445–456. [Google Scholar] [CrossRef]
- Hayday, A.; Theodoridis, E.; Ramsburg, E.; Shires, J. Intraepithelial lymphocytes: Exploring the Third Way in immunology. Nat. Immunol. 2001, 2, 997–1003. [Google Scholar] [CrossRef] [PubMed]
- Jiang, W.; Wang, X.; Zeng, B.; Liu, L.; Tardivel, A.; Wei, H.; Han, J.; MacDonald, H.R.; Tschopp, J.; Tian, Z.; et al. Recognition of gut microbiota by NOD2 is essential for the homeostasis of intestinal intraepithelial lymphocytes. J. Exp. Med. 2013, 210, 2465–2476. [Google Scholar] [CrossRef] [PubMed]
- Lepage, A.C.; Buzoni-Gatel, D.; Bout, D.T.; Kasper, L.H. Gut-derived intraepithelial lymphocytes induce long term immunity against Toxoplasma gondii. J. Immunol. 1998, 161, 4902–4908. [Google Scholar]
- Wesch, D.; Beetz, S.; Oberg, H.H.; Marget, M.; Krengel, K.; Kabelitz, D. Direct costimulatory effect of TLR3 ligand poly(I:C) on human gamma delta T lymphocytes. J. Immunol. 2006, 176, 1348–1354. [Google Scholar] [CrossRef] [PubMed]
- Divangahi, M.; Aaby, P.; Khader, S.A.; Barreiro, L.B.; Bekkering, S.; Chavakis, T.; van Crevel, R.; Curtis, N.; DiNardo, A.R.; Dominguez-Andres, J.; et al. Trained immunity, tolerance, priming and differentiation: Distinct immunological processes. Nat. Immunol. 2021, 22, 2–6. [Google Scholar] [CrossRef]
- Ifrim, D.C.; Quintin, J.; Joosten, L.A.; Jacobs, C.; Jansen, T.; Jacobs, L.; Gow, N.A.; Williams, D.L.; van der Meer, J.W.; Netea, M.G. Trained immunity or tolerance: Opposing functional programs induced in human monocytes after engagement of various pattern recognition receptors. Clin. Vaccine Immunol. 2014, 21, 534–545. [Google Scholar] [CrossRef]
- Eljaszewicz, A.; Ruchti, F.; Radzikowska, U.; Globinska, A.; Boonpiyathad, T.; Gschwend, A.; Morita, H.; Helbling, A.; Arasi, S.; Kahlert, H.; et al. Trained immunity and tolerance in innate lymphoid cells, monocytes, and dendritic cells during allergen-specific immunotherapy. J. Allergy Clin. Immunol. 2021, 147, 1865–1877. [Google Scholar] [CrossRef] [PubMed]
- Cheng, S.C.; Quintin, J.; Cramer, R.A.; Shepardson, K.M.; Saeed, S.; Kumar, V.; Giamarellos-Bourboulis, E.J.; Martens, J.H.; Rao, N.A.; Aghajanirefah, A.; et al. mTOR- and HIF-1alpha-mediated aerobic glycolysis as metabolic basis for trained immunity. Science 2014, 345, 1250684. [Google Scholar] [CrossRef]
- Saeed, S.; Quintin, J.; Kerstens, H.H.; Rao, N.A.; Aghajanirefah, A.; Matarese, F.; Cheng, S.C.; Ratter, J.; Berentsen, K.; van der Ent, M.A.; et al. Epigenetic programming of monocyte-to-macrophage differentiation and trained innate immunity. Science 2014, 345, 1251086. [Google Scholar] [CrossRef] [PubMed]
- Larabee, J.L.; Shakir, S.M.; Barua, S.; Ballard, J.D. Increased cAMP in monocytes augments Notch signaling mechanisms by elevating RBP-J and transducin-like enhancer of Split (TLE). J. Biol. Chem. 2013, 288, 21526–21536. [Google Scholar] [CrossRef] [PubMed]
- Serezani, C.H.; Ballinger, M.N.; Aronoff, D.M.; Peters-Golden, M. Cyclic AMP: Master regulator of innate immune cell function. Am. J. Respir. Cell Mol. Biol. 2008, 39, 127–132. [Google Scholar] [CrossRef] [PubMed]
- Biswas, S.K.; Lopez-Collazo, E. Endotoxin tolerance: New mechanisms, molecules and clinical significance. Trends Immunol. 2009, 30, 475–487. [Google Scholar] [CrossRef] [PubMed]
- Perez, V.; Tellechea, J.; Corpa, J.M.; Gutierrez, M.; Garcia Marin, J.F. Relation between pathologic findings and cellular immune responses in sheep with naturally acquired paratuberculosis. Am. J. Vet. Res. 1999, 60, 123–127. [Google Scholar]
- Danesh Pazhooh, R.; Rahnamay Farnood, P.; Asemi, Z.; Mirsafaei, L.; Yousefi, B.; Mirzaei, H. mTOR pathway and DNA damage response: A therapeutic strategy in cancer therapy. DNA Repair 2021, 104, 103142. [Google Scholar] [CrossRef]
- Slemc, L.; Kunej, T. Transcription factor HIF1A: Downstream targets, associated pathways, polymorphic hypoxia response element (HRE) sites, and initiative for standardization of reporting in scientific literature. Tumour Biol. 2016, 37, 14851–14861. [Google Scholar] [CrossRef]
- Canovas, B.; Nebreda, A.R. Diversity and versatility of p38 kinase signalling in health and disease. Nat. Rev. Mol. Cell Biol. 2021, 22, 346–366. [Google Scholar] [CrossRef]
- Roskoski, R., Jr. RAF protein-serine/threonine kinases: Structure and regulation. Biochem. Biophys. Res. Commun. 2010, 399, 313–317. [Google Scholar] [CrossRef]
- Peyrin-Biroulet, L.; Beisner, J.; Wang, G.; Nuding, S.; Oommen, S.T.; Kelly, D.; Parmentier-Decrucq, E.; Dessein, R.; Merour, E.; Chavatte, P.; et al. Peroxisome proliferator-activated receptor gamma activation is required for maintenance of innate antimicrobial immunity in the colon. Proc. Natl. Acad. Sci. USA 2010, 107, 8772–8777. [Google Scholar] [CrossRef]
- Dudek-Wicher, R.; Junka, A.; Paleczny, J.; Bartoszewicz, M. Clinical Trials of Probiotic Strains in Selected Disease Entities. Int. J. Microbiol. 2020, 2020, 8854119. [Google Scholar] [CrossRef] [PubMed]
- Maldonado Galdeano, C.; Cazorla, S.I.; Lemme Dumit, J.M.; Vélez, E.; Perdigón, G. Beneficial Effects of Probiotic Consumption on the Immune System. Ann. Nutr. Metab. 2019, 74, 115–124. [Google Scholar] [CrossRef]
- Khalesi, S.; Bellissimo, N.; Vandelanotte, C.; Williams, S.; Stanley, D.; Irwin, C. A review of probiotic supplementation in healthy adults: Helpful or hype? Eur. J. Clin. Nutr. 2019, 73, 24–37. [Google Scholar] [CrossRef]
- Torpee, S.; Kantachote, D.; Rattanachuay, P.; Chiayvareesajja, S.; Tantirungkij, M. Dietary supplementation with probiotic Rhodobacter sphaeroides SS15 extract to control acute hepatopancreatic necrosis disease (AHPND)-causing Vibrio parahaemolyticus in cultivated white shrimp. J. Invertebr. Pathol. 2021, 107585. [Google Scholar] [CrossRef]
- de Moreno de LeBlanc, A.; LeBlanc, J.G. Effect of probiotic administration on the intestinal microbiota, current knowledge and potential applications. World J. Gastroenterol. 2014, 20, 16518–16528. [Google Scholar] [CrossRef] [PubMed]
- Negi, S.; Das, D.K.; Pahari, S.; Nadeem, S.; Agrewala, J.N. Potential Role of Gut Microbiota in Induction and Regulation of Innate Immune Memory. Front. Immunol. 2019, 10, 2441. [Google Scholar] [CrossRef]
- Belkaid, Y.; Harrison, O.J. Homeostatic Immunity and the Microbiota. Immunity 2017, 46, 562–576. [Google Scholar] [CrossRef] [PubMed]
- Mileti, E.; Matteoli, G.; Iliev, I.D.; Rescigno, M. Comparison of the immunomodulatory properties of three probiotic strains of Lactobacilli using complex culture systems: Prediction for in vivo efficacy. PLoS ONE 2009, 4, e7056. [Google Scholar] [CrossRef] [PubMed]
- Llewellyn, A.; Foey, A. Probiotic Modulation of Innate Cell Pathogen Sensing and Signaling Events. Nutrients 2017, 9, 1156. [Google Scholar] [CrossRef] [PubMed]
- Castillo, N.A.; Perdigon, G.; de Moreno de Leblanc, A. Oral administration of a probiotic Lactobacillus modulates cytokine production and TLR expression improving the immune response against Salmonella enterica serovar Typhimurium infection in mice. BMC Microbiol. 2011, 11, 177. [Google Scholar] [CrossRef] [PubMed]
- Villena, J.; Vizoso-Pinto, M.G.; Kitazawa, H. Intestinal Innate Antiviral Immunity and Immunobiotics: Beneficial Effects against Rotavirus Infection. Front. Immunol. 2016, 7, 563. [Google Scholar] [CrossRef]
- Ganguli, K.; Meng, D.; Rautava, S.; Lu, L.; Walker, W.A.; Nanthakumar, N. Probiotics prevent necrotizing enterocolitis by modulating enterocyte genes that regulate innate immune-mediated inflammation. Am. J. Physiol. Gastrointest. Liver Physiol. 2013, 304, G132–G141. [Google Scholar] [CrossRef]
- Sham, H.P.; Yu, E.Y.; Gulen, M.F.; Bhinder, G.; Stahl, M.; Chan, J.M.; Brewster, L.; Morampudi, V.; Gibson, D.L.; Hughes, M.R.; et al. SIGIRR, a negative regulator of TLR/IL-1R signalling promotes Microbiota dependent resistance to colonization by enteric bacterial pathogens. PLoS Pathog. 2013, 9, e1003539. [Google Scholar] [CrossRef] [PubMed]
- Salva, S.; Alvarez, S. The Role of Microbiota and Immunobiotics in Granulopoiesis of Immunocompromised Hosts. Front. Immunol. 2017, 8, 507. [Google Scholar] [CrossRef]
- Salva, S.; Marranzino, G.; Villena, J.; Agüero, G.; Alvarez, S. Probiotic Lactobacillus strains protect against myelosuppression and immunosuppression in cyclophosphamide-treated mice. Int. Immunopharmacol. 2014, 22, 209–221. [Google Scholar] [CrossRef]
- Herrera, M.; Salva, S.; Villena, J.; Barbieri, N.; Marranzino, G.; Alvarez, S. Dietary supplementation with Lactobacilli improves emergency granulopoiesis in protein-malnourished mice and enhances respiratory innate immune response. PLoS ONE 2014, 9, e90227. [Google Scholar] [CrossRef] [PubMed]
- de Moreno de LeBlanc, A.; Dogi, C.A.; Galdeano, C.M.; Carmuega, E.; Weill, R.; Perdigon, G. Effect of the administration of a fermented milk containing Lactobacillus casei DN-114001 on intestinal microbiota and gut associated immune cells of nursing mice and after weaning until immune maturity. BMC Immunol. 2008, 9, 27. [Google Scholar] [CrossRef] [PubMed]
- de Moreno de LeBlanc, A.; Maldonado Galdeano, C.; Dogi, C.A.; Carmuega, E.; Weill, R.; Perdigon, G. Adjuvant effect of a probiotic fermented milk in the protection against Salmonella enteritidis serovar typhimurium infection: Mechanisms involved. Int. J. Immunopathol. Pharmacol. 2010, 23, 1235–1244. [Google Scholar] [CrossRef] [PubMed]
- Mortaz, E.; Adcock, I.M.; Folkerts, G.; Barnes, P.J.; Paul Vos, A.; Garssen, J. Probiotics in the management of lung diseases. Mediat. inflamm. 2013, 2013, 751068. [Google Scholar] [CrossRef]
- Kawahara, T.; Takahashi, T.; Oishi, K.; Tanaka, H.; Masuda, M.; Takahashi, S.; Takano, M.; Kawakami, T.; Fukushima, K.; Kanazawa, H.; et al. Consecutive oral administration of Bifidobacterium longum MM-2 improves the defense system against influenza virus infection by enhancing natural killer cell activity in a murine model. Microbiol. Immunol. 2015, 59, 1–12. [Google Scholar] [CrossRef]
- Garcia-Castillo, V.; Tomokiyo, M.; Raya Tonetti, F.; Islam, M.A.; Takahashi, H.; Kitazawa, H.; Villena, J. Alveolar Macrophages Are Key Players in the Modulation of the Respiratory Antiviral Immunity Induced by Orally Administered Lacticaseibacillus rhamnosus CRL1505. Front. Immunol. 2020, 11, 568636. [Google Scholar] [CrossRef]
- Koizumi, S.; Wakita, D.; Sato, T.; Mitamura, R.; Izumo, T.; Shibata, H.; Kiso, Y.; Chamoto, K.; Togashi, Y.; Kitamura, H.; et al. Essential role of Toll-like receptors for dendritic cell and NK1.1(+) cell-dependent activation of type 1 immunity by Lactobacillus pentosus strain S-PT84. Immunol. Lett. 2008, 120, 14–19. [Google Scholar] [CrossRef] [PubMed]
- Forsythe, P. Probiotics and lung immune responses. Ann. Am. Thorac. Soc. 2014, 11 (Suppl. 1), S33–S37. [Google Scholar] [CrossRef]
- Dong, H.; Rowland, I.; Yaqoob, P. Comparative effects of six probiotic strains on immune function in vitro. Br. J. Nutr. 2012, 108, 459–470. [Google Scholar] [CrossRef] [PubMed]
- Du, B.; Meenu, M.; Liu, H.; Xu, B. A Concise Review on the Molecular Structure and Function Relationship of beta-Glucan. Int. J. Mol. Sci. 2019, 20, 4032. [Google Scholar] [CrossRef] [PubMed]
- Goodridge, H.S.; Wolf, A.J.; Underhill, D.M. Beta-glucan recognition by the innate immune system. Immunol. Rev. 2009, 230, 38–50. [Google Scholar] [CrossRef] [PubMed]
- Stier, H.; Bischoff, S.C. Influence of Saccharomyces boulardii CNCM I-745on the gut-associated immune system. Clin. Exp. Gastroenterol. 2016, 9, 269–279. [Google Scholar] [CrossRef]
- Sen, S.; Mansell, T.J. Yeasts as probiotics: Mechanisms, outcomes, and future potential. Fungal Genet. Biol. 2020, 137, 103333. [Google Scholar] [CrossRef] [PubMed]
- Brown, G.D.; Herre, J.; Williams, D.L.; Willment, J.A.; Marshall, A.S.; Gordon, S. Dectin-1 mediates the biological effects of beta-glucans. J. Exp. Med. 2003, 197, 1119–1124. [Google Scholar] [CrossRef]
- Iliev, I.D.; Funari, V.A.; Taylor, K.D.; Nguyen, Q.; Reyes, C.N.; Strom, S.P.; Brown, J.; Becker, C.A.; Fleshner, P.R.; Dubinsky, M.; et al. Interactions between commensal fungi and the C-type lectin receptor Dectin-1 influence colitis. Science 2012, 336, 1314–1317. [Google Scholar] [CrossRef] [PubMed]
- Rizzetto, L.; Ifrim, D.C.; Moretti, S.; Tocci, N.; Cheng, S.C.; Quintin, J.; Renga, G.; Oikonomou, V.; De Filippo, C.; Weil, T.; et al. Fungal Chitin Induces Trained Immunity in Human Monocytes during Cross-talk of the Host with Saccharomyces cerevisiae. J. Biol. Chem. 2016, 291, 7961–7972. [Google Scholar] [CrossRef] [PubMed]
- Mattia Pia, A.; Giuseppe, S.; Daniela, F. β-Glucans and Probiotics. Am. J. Immunol. 2017, 13, 34–44. [Google Scholar] [CrossRef]
- Salazar, N.; Ruas-Madiedo, P.; Kolida, S.; Collins, M.; Rastall, R.; Gibson, G.; de Los Reyes-Gavilán, C.G. Exopolysaccharides produced by Bifidobacterium longum IPLA E44 and Bifidobacterium animalis subsp. lactis IPLA R1 modify the composition and metabolic activity of human faecal microbiota in pH-controlled batch cultures. Int. J. Food Microbiol. 2009, 135, 260–267. [Google Scholar] [CrossRef] [PubMed]
- Goh, Y.J.; Klaenhammer, T.R. Genetic mechanisms of prebiotic oligosaccharide metabolism in probiotic microbes. Annu. Rev. Food Sci. Technol. 2015, 6, 137–156. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Lai, T.; Yao, M.; Zhang, M.; Yang, Z. Interaction of the Exopolysaccharide from Lactobacillus plantarum YW11 with Casein and Bioactivities of the Polymer Complex. Foods 2021, 10, 1153. [Google Scholar] [CrossRef]
- Angelin, J.; Kavitha, M. Exopolysaccharides from probiotic bacteria and their health potential. Int. J. Biol. Macromol. 2020, 162, 853–865. [Google Scholar] [CrossRef]
- Ciszek-Lenda, M.; Nowak, B.; Srottek, M.; Gamian, A.; Marcinkiewicz, J. Immunoregulatory potential of exopolysaccharide from Lactobacillus rhamnosus KL37: Effects on the production of inflammatory mediators by mouse macrophages. Int. J. Exp. Pathol. 2011, 92, 382–391. [Google Scholar] [CrossRef]




| Hallmarks | Factor Measured | Trained Immunity | References |
|---|---|---|---|
| Immunologic | IL-6 | Induction | [130,132] |
| TNF-α | Induction | [130,132] | |
| PPARG (NR1C3) | Maintained | [133] | |
| Metabolic | Glucose | Consummation increase/aerobic glycolysis | [132] |
| Lactate | Production * | [132] | |
| Oxygen | Reduction of consummation | [132] | |
| mTOR | Phosphorylation/activation | [132] | |
| NAD+/NADH ratio | Enhanced | [132] | |
| Akt | Phosphorylation/activation | [132] | |
| p38 kinase | Phosphorylation/activation | [12] | |
| Raf-1 pathway | Activation | [12] | |
| Epigenetic | H3K4 | Methylation enhanced | [12,132] |
| H3K27 | Acetylation modulation | [132] | |
| Sirtuin-1 | Inhibition | [132] | |
| HIF1 | Enhanced | [132] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cortes-Perez, N.G.; de Moreno de LeBlanc, A.; Gomez-Gutierrez, J.G.; LeBlanc, J.G.; Bermúdez-Humarán, L.G. Probiotics and Trained Immunity. Biomolecules 2021, 11, 1402. https://doi.org/10.3390/biom11101402
Cortes-Perez NG, de Moreno de LeBlanc A, Gomez-Gutierrez JG, LeBlanc JG, Bermúdez-Humarán LG. Probiotics and Trained Immunity. Biomolecules. 2021; 11(10):1402. https://doi.org/10.3390/biom11101402
Chicago/Turabian StyleCortes-Perez, Naima G., Alejandra de Moreno de LeBlanc, Jorge G. Gomez-Gutierrez, Jean Guy LeBlanc, and Luis G. Bermúdez-Humarán. 2021. "Probiotics and Trained Immunity" Biomolecules 11, no. 10: 1402. https://doi.org/10.3390/biom11101402
APA StyleCortes-Perez, N. G., de Moreno de LeBlanc, A., Gomez-Gutierrez, J. G., LeBlanc, J. G., & Bermúdez-Humarán, L. G. (2021). Probiotics and Trained Immunity. Biomolecules, 11(10), 1402. https://doi.org/10.3390/biom11101402







