Advances and Prospects of Phenolic Acids Production, Biorefinery and Analysis
Abstract
1. Introduction
2. Chemical Structure and Overview of Phenolic Acids
3. Phenolic Acids in Plants
3.1. Biosynthesis
3.2. The regulation of Phenolic Acid Biosynthesis
3.3. Biotic and Abiotic Stress
3.4. Factors Influencing the Amount of Accumulated Phenolic Acids
4. Production and Extraction of Phenolic Acids from Plants and Algae
4.1. Cereals
4.2. Seeds and Oilseeds
4.3. Fruits and Berries
4.4. Vegetables
4.5. Nuts
4.6. Spices and Medicinal Herbs
4.7. Tea, Cacao, and Coffee
4.8. Algae
4.9. Extraction of Phenolic Acids from Agro-Industrial Waste
5. Production and Extraction of Phenolic Acids from Fungi
5.1. Mushrooms
5.2. Lichens
6. Production of Phenolic Acids Using Non-Modified and Engineered Microorganisms
6.1. Biosynthesis of Phenolic Acids from Organic Compounds Using Non-Modified Microorganisms
6.2. Biosynthesis of Phenolic Acids from Organic Wastes Using Non-Modified Microorganisms
6.3. Food and Beverage Enrichment with Phenolic Acids
6.4. Engineered Microorganisms for Phenolic Acid Production
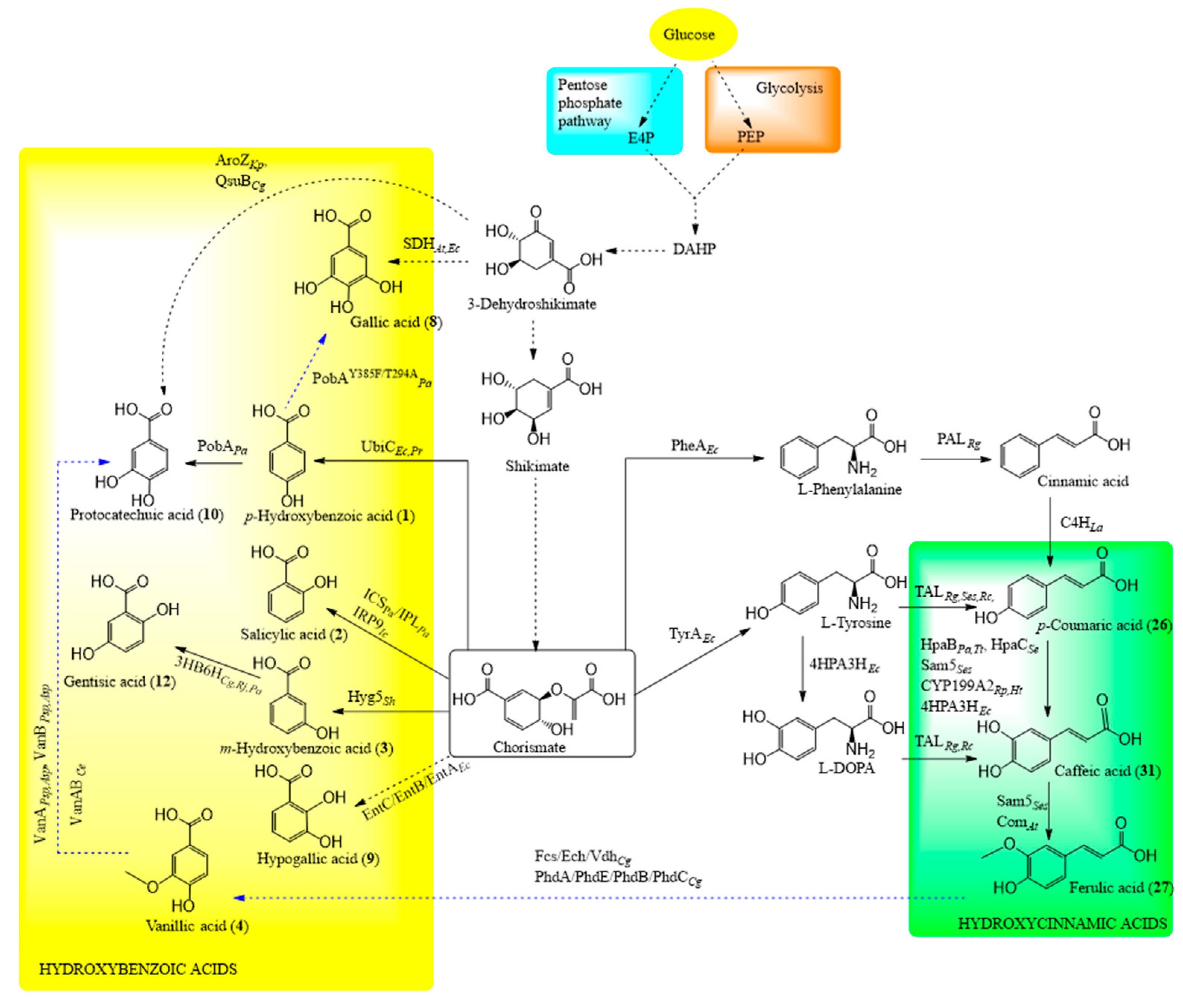
6.4.1. Shikimate Pathway in Engineered Microorganisms
6.4.2. Biosynthesis of Hydroxycinnamic Acids
6.4.3. Biosynthesis of Hydroxybenzoic Acids
7. Techniques for Extraction and Analysis of Phenolic Acids
7.1. Preparation of Material
7.2. Extraction
7.3. Separation
7.4. Analysis of Phenolic Acids
7.5. Alternative Methods for Quantification of Phenolic Acids: Biosensors
7.5.1. Enzyme-Based Biosensors
7.5.2. Transcription Factor-Based Biosensors
8. Future Perspectives and Limitations of Phenolic acid Production
8.1. Improved Phenolic Acid Production through Engineering Microorganisms
8.2. Transporters
8.3. Product Removal
8.4. Availability of Tools
8.5. By-Products
8.6. Enzyme Choice and Engineering
8.7. Fermentation Conditions and Substrates
8.8. Application of Other Methods
9. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Kumar, N.; Goel, N. Phenolic acids: Natural versatile molecules with promising therapeutic applications. Biotechnol. Rep. 2019, 24, e00370. [Google Scholar] [CrossRef] [PubMed]
- Khadem, S.; Marles, R.J. Monocyclic phenolic acids; hydroxy- and polyhydroxybenzoic acids: Occurrence and recent bioactivity studies. Molecules 2010, 15, 7985–8005. [Google Scholar] [CrossRef] [PubMed]
- Martins, S.; Mussatto, S.I.; Martínez-Avila, G.; Montañez-Saenz, J.; Aguilar, C.N.; Teixeira, J.A. Bioactive phenolic compounds: Production and extraction by solid-state fermentation. A review. Biotechnol. Adv. 2011, 29, 365–373. [Google Scholar] [CrossRef]
- Mandal, S.M.; Chakraborty, D.; Dey, S. Phenolic acids act as signaling molecules in plant-microbe symbioses. Plant Signal. Behav. 2010, 5, 359–368. [Google Scholar] [CrossRef] [PubMed]
- Barros, J.; Escamilla-Trevino, L.; Song, L.; Rao, X.; Serrani-Yarce, J.C.; Palacios, M.D.; Engle, N.; Choudhury, F.K.; Tschaplinski, T.J.; Venables, B.J.; et al. 4-Coumarate 3-hydroxylase in the lignin biosynthesis pathway is a cytosolic ascorbate peroxidase. Nat. Commun. 2019, 10, 1994. [Google Scholar] [CrossRef] [PubMed]
- Kulbat, K. The role of phenolic compounds in plant resistance. Biotechnol. Food Sci. 2016, 80, 97–108. [Google Scholar]
- Cory, H.; Passarelli, S.; Szeto, J.; Tamez, M.; Mattei, J. The role of polyphenols in human health and food systems: A mini-review. Front. Nutr. 2018, 5, 87. [Google Scholar] [CrossRef]
- Martin, K.R.; Appel, C.L. Polyphenols as dietary supplements: A double-edged sword. Nutr. Diet. Suppl. 2010, 2, 1–12. [Google Scholar] [CrossRef]
- Crozier, A.; Jaganath, I.B.; Clifford, M.N. Dietary phenolics: Chemistry, bioavailability and effects on health. Nat. Prod. Rep. 2009, 26, 965–1096. [Google Scholar] [CrossRef]
- Bhattacharya, A.; Sood, P.; Citovsky, V. The roles of plant phenolics in defence and communication during Agrobacterium and Rhizobium infection. Mol. Plant Pathol. 2010, 11, 705–719. [Google Scholar] [CrossRef]
- Pereira, D.M.; Valentão, P.; Pereira, J.A.; Andrade, P.B. Phenolics: From chemistry to biology. Molecules 2009, 14, 2202–2211. [Google Scholar] [CrossRef]
- de Camargo, A.C.; Regitano-d’Arce, M.A.B.; Rasera, G.B.; Canniatti-Brazaca, S.G.; do Prado-Silva, L.; Alvarenga, V.O.; Sant’Ana, A.S.; Shahidi, F. Phenolic acids and flavonoids of peanut by-products: Antioxidant capacity and antimicrobial effects. Food Chem. 2017, 237, 538–544. [Google Scholar] [CrossRef] [PubMed]
- Binutu, O.A.; Cordell, G.A. Gallic acid derivatives from mezoneuron benthamianum leaves. Pharm. Biol. 2011, 38, 284–286. [Google Scholar] [CrossRef]
- Pei, K.; Ou, J.; Huang, J.; Ou, S. p-Coumaric acid and its conjugates: Dietary sources, pharmacokinetic properties and biological activities. J. Sci. Food Agric. 2016, 96, 2952–2962. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, J.F.S.; Luthria, D.L.; Sasaki, T.; Heyerick, A. Flavonoids from Artemisia annua L. as antioxidants and their potential synergism with artemisinin against malaria and cancer. Molecules 2010, 15, 3135–3170. [Google Scholar] [CrossRef]
- Wani, B.A.; Bodha, R.H.; Wani, A.H. Nutritional and medicinal importance of mushrooms. J. Med. Plants Res. 2010, 4, 2598–2604. [Google Scholar]
- Lodovici, M.; Guglielmi, F.; Meoni, M.; Dolara, P. Effect of natural phenolic acids on DNA oxidation in vitro. Food Chem. Toxicol. 2001, 39, 1205–1210. [Google Scholar] [CrossRef]
- Itoh, A.; Isoda, K.; Kondoh, M.; Kawase, M.; Watari, A.; Kobayashi, M.; Yagi, K. Hepatoprotective effect of syringic acid and vanillic acid on CCl4-induced liver injury. Biol. Pharm. Bull. 2010, 33, 983–987. [Google Scholar] [CrossRef]
- Muthukumaran, J.; Srinivasan, S.; Venkatesan, R.S.; Ramachandran, V.; Muruganathan, U. Syringic acid, a novel natural phenolic acid, normalizes hyperglycemia with special reference to glycoprotein components in experimental diabetic rats. J. Acute Dis. 2013, 2, 304–309. [Google Scholar] [CrossRef]
- Pinto, P.C.R.; Costa, C.E.; Rodrigues, A.E. Oxidation of lignin from Eucalyptus globulus pulping liquors to produce syringaldehyde and vanillin. Ind. Eng. Chem. Res. 2013, 52, 4421–4428. [Google Scholar] [CrossRef]
- Mansouri, M.T.; Naghizadeh, B.; Ghorbanzadeh, B.; Farbood, Y.; Sarkaki, A.; Bavarsad, K. Gallic acid prevents memory deficits and oxidative stress induced by intracerebroventricular injection of streptozotocin in rats. Pharmacol. Biochem. Behav. 2013, 111, 90–96. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.J.; Moh, S.H.; Son, D.H.; You, S.; Kinyua, A.W.; Ko, C.M.; Song, M.; Yeo, J.; Choi, Y.-H.; Kim, K.W. Gallic acid promotes wound healing in normal and hyperglucidic conditions. Molecules 2016, 21, 899. [Google Scholar] [CrossRef] [PubMed]
- Park, H.J.; Cho, J.H.; Hong, S.H.; Kim, D.H.; Jung, H.Y.; Kang, I.K.; Cho, Y.J. Whitening and anti-wrinkle activities of ferulic acid isolated from Tetragonia tetragonioides in B16F10 melanoma and CCD-986sk fibroblast cells. J. Nat. Med. 2018, 72, 127–135. [Google Scholar] [CrossRef] [PubMed]
- Kumar, K.J.S.; Vani, M.G.; Wang, S.-Y.; Liao, J.-W.; Hsu, L.-S.; Yang, H.-L.; Hseu, Y.-C. In vitro and in vivo studies disclosed the depigmenting effects of gallic acid: A novel skin lightening agent for hyperpigmentary skin diseases. BioFactors 2013, 39, 259–270. [Google Scholar] [CrossRef]
- GMI. Salicylic acid Market Size, Industry Analysis Report, Regional Outlook, Application Development Potential, Price Trends, Competitive Market Share & Forecast, 2020–2026; Global Market Insights: Selbyville, DE, USA, 2020. [Google Scholar]
- GMI. Natural Ferulic Acid Market Size by Purity, Industry Analysis Report, Regional Outlook, Application Potential, Price Trends, Competitive Market Share & Forecast, 2019–2025; Global Market Insight: Selbyville, DE, USA, 2019. [Google Scholar]
- Ou, S.; Kwok, K.C. Ferulic acid: Pharmaceutical functions, preparation and applications in foods. J. Sci. Food Agric. 2004, 84, 1261–1269. [Google Scholar] [CrossRef]
- PMR. Gallic Acid Market-Global Industry Analysis 2014–2018 and Forecast 2019–2029; Persistence Market Research: New York, NY, USA, 2020. [Google Scholar]
- IMR. Global Vanillic Acid (CAS 121-34-6) Market Analysis 2013-2018 and Forecast 2019-2024; Index Market Research: New York, NY, USA, 2019. [Google Scholar]
- Solopova, A.; van Tilburg, A.Y.; Foito, A.; Allwood, J.W.; Stewart, D.; Kulakauskas, S.; Kuipers, O.P. Engineering Lactococcus lactis for the production of unusual anthocyanins using tea as substrate. Metab. Eng. 2019, 54, 160–169. [Google Scholar] [CrossRef]
- Gottardi, M.; Knudsen, J.D.; Prado, L.; Oreb, M.; Branduardi, P.; Boles, E. De novo biosynthesis of trans-cinnamic acid derivatives in Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 2017, 101, 4883–4893. [Google Scholar] [CrossRef]
- Russell, W.R.; Labat, A.; Scobbie, L.; Duncan, G.J.; Duthie, G.G. Phenolic acid content of fruits commonly consumed and locally produced in Scotland. Food Chem. 2009, 115, 100–104. [Google Scholar] [CrossRef]
- Kornsteiner, M.; Wagner, K.-H.; Elmadfa, I. Tocopherols and total phenolics in 10 different nut types. Food Chem. 2006, 98, 381–387. [Google Scholar] [CrossRef]
- Bach, F.; Zielinski, A.A.F.; Helm, C.V.; Maciel, G.M.; Pedro, A.C.; Stafussa, A.P.; Ávila, S.; Haminiuk, C.W.I. Bio compounds of edible mushrooms: In vitro antioxidant and antimicrobial activities. LWT 2019, 107, 214–220. [Google Scholar] [CrossRef]
- Karamaæ, M.; Kosiñska, A.; Pegg, R.B. Comparison of radical-scavenging activities. Pol. J. Food Nutr. Sci. 2005, 55, 165–170. [Google Scholar]
- Ali, H.M.; Abo-Shady, A.; Eldeen, H.A.S.; Soror, H.A.; Shousha, W.G.; Abdel-Barry, O.A.; Saleh, A.M. Structural features, kinetics and SAR study of radical scavenging and antioxidant activities of phenolic and anilinic compounds. Chem. Cent. J. 2013, 7, 53. [Google Scholar] [CrossRef] [PubMed]
- Amorati, R.; Pedulli, G.F.; Cabrin, L.; Zambonin, L.; Landi, L. Solvent and pH effects on the antioxidant activity of caffeic and other phenolic acids. J. Agric. 2006, 54, 2932–2937. [Google Scholar] [CrossRef] [PubMed]
- Kaewnarin, K.; Suwannarach, N.; Kumla, J.; Lumyong, S. Phenolic profile of various wild edible mushroom extracts from Thailand and their antioxidant properties, anti-tyrosinase and hyperglycaemic inhibitory activities. J. Funct. Foods 2016, 27, 352–364. [Google Scholar] [CrossRef]
- Palafox-Carlos, H.; Gil-Chávez, J.; Sotelo-Mundo, R.R.; Namiesnik, J.; Gorinstein, S.; González-Aguilar, G.A. Antioxidant interactions between major phenolic compounds found in ‘Ataulfo’ mango pulp: Chlorogenic, gallic, protocatechuic and vanillic acids. Molecules 2012, 17, 12657–12664. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.-Y.; Milbury, P.E.; Lapsley, K.; Blumberg, J.B. Flavonoids from almond skins are bioavailable and act synergistically with vitamins C and E to enhance hamster and human LDL resistance to oxidation. J. Nutr. 2005, 135, 1366–1373. [Google Scholar] [CrossRef]
- Yildiz, O.; Can, Z.; Laghari, A.Q.; Şahin, H.; Malkoç, M. Wild edible mushrooms as a natural source of phenolics and antioxidants. J. Food Biochem. 2015, 39, 148–154. [Google Scholar] [CrossRef]
- Bontpart, T.; Marlin, T.; Vialet, S.; Guiraud, J.L.; Pinasseau, L.; Meudec, E.; Sommerer, N.; Cheynier, V.; Terrier, N. Two shikimate dehydrogenases, VvSDH3 and VvSDH4, are involved in gallic acid biosynthesis in grapevine. J. Exp. Bot. 2016, 67, 3537–3550. [Google Scholar] [CrossRef]
- Heleno, S.A.; Martins, A.; Queiroz, M.J.R.P.; Ferreira, I.C.F.R. Bioactivity of phenolic acids: Metabolites versus parent compounds: A review. Food Chem. 2015, 173, 501–513. [Google Scholar] [CrossRef]
- Santos-Sánchez, N.F.; Salas-Coronado, R.; Hernández-Carlos, B.; Villanueva-Cañongo, C. Shikimic acid pathway in biosynthesis of phenolic compounds. In Plant Physiological Aspects of Phenolic Compounds; Soto-Hernández, M., García-Mateos, R., Palma-Tenango, M., Eds.; IntechOpen: London, UK, 2019. [Google Scholar]
- Serrano, M.; Wang, B.; Aryal, B.; Garcion, C.; Abou-Mansour, E.; Heck, S.; Geisler, M.; Mauch, F.; Nawrath, C.; Métraux, J.P. Export of salicylic acid from the chloroplast requires the multidrug and toxin extrusion-like transporter EDS5. Plant Physiol. 2013, 162, 1815–1821. [Google Scholar] [CrossRef]
- Werner, R.A.; Rossmann, A.; Schwarz, C.; Bacher, A.; Schmidt, H.-L.; Eisenreich, W. Biosynthesis of gallic acid in Rhus typhina: Discrimination between alternative pathways from natural oxygen isotope abundance. Phytochemistry 2004, 65, 2809–2813. [Google Scholar] [CrossRef] [PubMed]
- Widhalm, J.R.; Dudareva, N. A Familiar ring to it: Biosynthesis of plant benzoic acids. Mol. Plant 2015, 8, 83–97. [Google Scholar] [CrossRef] [PubMed]
- Boatright, J.; Negre, F.; Chen, X.; Kish, C.M.; Wood, B.; Peel, G.; Orlova, I.; Gang, D.; Rhodes, D.; Dudareva, N. Understanding in vivo benzenoid metabolism in petunia petal tissue. Plant Physiol. 2004, 135, 1993–2011. [Google Scholar] [CrossRef] [PubMed]
- Jarvis, A.P.; Schaaf, O.; Oldham, N.J. 3-hydroxy-3-phenylpropanoic acid is an intermediate in the biosynthesis of benzoic acid and salicylic acid but benzaldehyde is not. Planta 2000, 212, 119–126. [Google Scholar] [CrossRef] [PubMed]
- Malinowski, J.; Krzymowska, M.; Godoń, K.; Hennig, J.; Podstolski, A. A new catalytic activity from tobacco converting 2-coumaric acid to salicylic aldehyde. Physiol. Plant. 2007, 129, 461–471. [Google Scholar] [CrossRef]
- El-Basyquni, S.Z.; Chen, D.; Ibrahim, R.K.; Neish, A.C.; Towers, G.H.N. The biosynthesis of hydroxybenzoic acids in higher plants. Phytochemistry 1964, 3, 485–492. [Google Scholar] [CrossRef]
- Batista, A.N.L.; Batista, J.M., Jr.; Souza-Moreira, T.M.; Valentini, S.R.; Kato, M.J.; Zanelli, C.F.; Furlan, M. Biosynthetic insights into p-hydroxybenzoic acid-derived benzopyrans in Piper gaudichaudianum. J. Braz. Chem. Soc. 2018, 29, 1105–1114. [Google Scholar] [CrossRef]
- Hagel, P.; Kindl, H. p-Hydroxybenzoate synthase: A complex associated with mitochondrial membranes of roots Cucumis sativus. Febs Lett. 1975, 59, 120–124. [Google Scholar] [CrossRef]
- Ribnicky, D.M.; Shulaev, V.; Raskin, I. Intermediates of salicylic acid biosynthesis in tobacco. Plant Physiol. 1998, 118, 565–572. [Google Scholar] [CrossRef]
- Torrens-Spence, M.P.; Bobokalonova, A.; Carballo, V.; Glinkerman, C.M.; Pluskal, T.; Shen, A.; Weng, J.-K. PBS3 and EPS1 complete salicylic acid biosynthesis from isochorismate in Arabidopsis. Mol. Plant 2019, 12, 1577–1586. [Google Scholar] [CrossRef]
- Catinot, J.; Buchala, A.; Abou-Mansour, E.; Métraux, J.-P. Salicylic acid production in response to biotic and abiotic stress depends on isochorismate in Nicotiana benthamiana. Febs Lett. 2008, 582, 473–478. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Halitschke, R.; Yin, C.; Liu, C.-J.; Gan, S.-S. Salicylic acid 3-hydroxylase regulates Arabidopsis leaf longevity by mediating salicylic acid catabolism. Proc. Natl. Acad. Sci. USA 2013, 110, 14807–14812. [Google Scholar] [CrossRef] [PubMed]
- Czichi, U.; Kindl, H. Phenylalanine ammonia lyase and cinnamic acid hydroxylases as assembled consecutive enzymes on microsomal membranes of cucumber cotyledons: Cooperation and subcellular distribution. Planta 1977, 134, 133–143. [Google Scholar] [CrossRef] [PubMed]
- Ipson, B.R.; Fisher, A.L. Roles of the tyrosine isomers meta-tyrosine and ortho-tyrosine in oxidative stress. Ageing Res. Rev. 2016, 27, 93–107. [Google Scholar] [CrossRef]
- Neish, A.C. Formation of m- and p-coumaric acids by enzymatic deamination of the corresponding isomers of tyrosine. Phytochemistry 1961, 1, 1–24. [Google Scholar] [CrossRef]
- Huang, T.; Rehak, L.; Jander, G. meta-Tyrosine in Festuca rubra ssp. commutata (Chewings fescue) is synthesized by hydroxylation of phenylalanine. Phytochemistry 2012, 75, 60–66. [Google Scholar] [CrossRef]
- Vialart, G.; Hehn, A.; Olry, A.; Ito, K.; Krieger, C.; Larbat, R.; Paris, C.; Shimizu, B.; Sugimoto, Y.; Mizutani, M.; et al. A 2-oxoglutarate-dependent dioxygenase from Ruta graveolens L. exhibits p-coumaroyl CoA 2′-hydroxylase activity (C2′H): A missing step in the synthesis of umbelliferone in plants. Plant J. 2011, 70, 460–470. [Google Scholar] [CrossRef]
- Taura, F.; Iijima, M.; Yamanaka, E.; Takahashi, H.; Kenmoku, H.; Saeki, H.; Morimoto, S.; Asakawa, Y.; Kurosaki, F.; Morita, H. A novel class of plant type III polyketide synthase involved in orsellinic acid biosynthesis from Rhododendron dauricum. Front. Plant Sci. 2016, 7, 1452. [Google Scholar] [CrossRef]
- Dey, T.B.; Chakraborty, S.; Jain, K.K.; Sharma, A.; Kuhad, R.C. Antioxidant phenolics and their microbial production by submerged and solid state fermentation process: A review. Trends Food Sci. Technol. 2016, 53, 60–74. [Google Scholar]
- Herrmann, K.; Nagel, C.W. Occurrence and content of hydroxycinnamic and hydroxybenzoic acid compounds in foods. Crit. Rev. Food Sci. Nutr. 1989, 28, 315–347. [Google Scholar] [CrossRef]
- Cai, Z.; Kastell, A.; Knorr, D.; Smetanska, I. Exudation: An expanding technique for continuous production and release of secondary metabolites from plant cell suspension and hairy root cultures. Plant Cell Rep. 2012, 31, 461–477. [Google Scholar] [CrossRef] [PubMed]
- Enebe, M.C.; Babalola, O.O. The impact of microbes in the orchestration of plants’ resistance to biotic stress: A disease management approach. Appl. Microbiol. Biotechnol. 2019, 103, 9–25. [Google Scholar] [CrossRef] [PubMed]
- Nanda, A.K.; Andrio, E.; Marino, D.; Pauly, N.; Dunand, C. Reactive oxygen species during plant-microorganism early interactions. J. Integr. Plant Biol. 2010, 52, 195–204. [Google Scholar] [CrossRef] [PubMed]
- Gonugunta, V.K.; Srivastava, N.; Raghavendra, A.S. Cytosolic alkalinization is a common and early messenger preceding the production of ROS and NO during stomatal closure by variable signals, including abscisic acid, methyl jasmonate and chitosan. Plant Signal. Behav. 2009, 4, 561–564. [Google Scholar] [CrossRef]
- El-Shetehy, M.; Wang, C.; Shine, M.B.; Yu, K.; Kachroo, A.; Kachroo, P. Nitric oxide and reactive oxygen species are required for systemic acquired resistance in plants. Plant Signal. Behav. 2015, 10, e998544. [Google Scholar] [CrossRef]
- Zheng, X.Y.; Zhou, M.; Yoo, H.; Pruneda-Paz, J.L.; Spivey, N.W.; Kay, S.A.; Dong, X. Spatial and temporal regulation of biosynthesis of the plant immune signal salicylic acid. Proc. Natl. Acad. Sci. USA 2015, 112, 9166–9173. [Google Scholar] [CrossRef]
- Forouhar, F.; Yang, Y.; Kumar, D.; Chen, Y.; Fridman, E.; Park, S.W.; Chiang, Y.; Acton, T.B.; Montelione, G.T.; Pichersky, E.; et al. Structural and biochemical studies identify tobacco SABP2 as a methyl salicylate esterase and implicate it in plant innate immunity. PNAS 2005, 102, 1773–1778. [Google Scholar] [CrossRef]
- Park, S.W.; Kaimoyo, E.; Kumar, D.; Mosher, S.; Klessig, D.F. Methyl salicylate is a critical mobile signal for plant systemic acquired resistance. Science 2007, 318, 113–116. [Google Scholar] [CrossRef]
- Zander, M.; Thurow, C.; Gatz, C. TGA transcription factors activate the salicylic acid-suppressible branch of the ethylene-induced defense program by regulating ORA59 expression. Plant Physiol. 2014, 165, 1671–1683. [Google Scholar] [CrossRef]
- Schmiesing, A.; Emonet, A.; Gouhier-Darimont, C.; Reymond, P. Arabidopsis MYC transcription factors are the target of hormonal salicylic acid/jasmonic acid cross talk in response to Pieris brassicae egg extract. Plant Physiol. 2016, 170, 2432–2443. [Google Scholar] [CrossRef]
- Caarls, L.; Van der Does, D.; Hickman, R.; Jansen, W.; Van Verk, M.C.; Proietti, S.; Lorenzo, O.; Solano, R.; Pieterse, C.M.J.; Van Wees, S.C.M. Assessing the role of ethylene response factor transcriptional repressors in salicylic acid-mediated suppression of jasmonic acid-responsive genes. Plant Cell Physiol. 2017, 58, 266–278. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.F.; Chen, C.T.; Kao, C.H. Salicylic acid inhibits the biosynthesis of ethylene in detached rice leaves. Plant Growth Regul. 1993, 12, 79–82. [Google Scholar] [CrossRef]
- Zhu, J.-K. Abiotic stress signaling and responses in plants. Cell 2016, 167, 313–324. [Google Scholar] [CrossRef]
- Tuteja, N. Abscisic acid and abiotic stress signaling. Plant Signal. Behav. 2007, 2, 135–138. [Google Scholar] [CrossRef] [PubMed]
- Jain, A.; Singh, A.; Singh, S.; Singh, H.B. Phenols enhancement effect of microbial consortium in pea plants restrains Sclerotinia sclerotiorum. Biol. Control 2015, 89, 23–32. [Google Scholar] [CrossRef]
- Linić, I.; Šamec, D.; Grúz, J.; Bok, V.V.; Strnad, M.; Salopek-Sondi, B. Involvement of phenolic acids in short-term adaptation to salinity stress is species-specific among Brassicaceae. Plants 2019, 8, 155. [Google Scholar] [CrossRef]
- Jahangir, M.; Abdel-Farid, I.B.; Choi, Y.H.; Verpoorte, R. Metal ion-inducing metabolite accumulation in Brassica rapa. J. Plant Physiol. 2008, 165, 1429–1437. [Google Scholar] [CrossRef]
- Stelzner, J.; Roemhild, R.; Garibay-Hernández, A.; Harbaum-Piayda, B.; Mock, H.-P.; Wolfgang Bilger, W. Hydroxycinnamic acids in sunflower leaves serve as UV-A screening pigments. Photochem. Photobiol. Sci. 2019, 18, 1649–1659. [Google Scholar] [CrossRef]
- Jacobo-Velázquez, D.A.; Martínez-Hernández, G.B.; Del, C.; Rodríguez, S.; Cao, C.M.; Cisneros-Zevallos, L. Plants as biofactories: Physiological role of reactive oxygen species on the accumulation of phenolic antioxidants in carrot tissue under wounding and hyperoxia stress. J. Agric. Food Chem. 2011, 59, 6583–6593. [Google Scholar] [CrossRef]
- Heredia, J.B.; Cisneros-Zevallos, L. The effect of exogenous ethylene and methyl jasmonate on pal activity, phenolic profiles and antioxidant capacity of carrots (Daucus carota) under different wounding intensities. Postharvest Biol. Technol. 2009, 51, 242–249. [Google Scholar] [CrossRef]
- Ayaz, F.A.; Kadioglu, A.; Turgut, R. Water stress effects on the content of low molecular weight carbohydrates and phenolic acids in Ctenanthe setosa (Rosc.) Eichler. Can. J. Plant Sci. 2000, 80, 373–378. [Google Scholar] [CrossRef]
- Haskins, F.A.; Williams, L.G.; Gorz, H.J. Light-induced trans to cis conversion of β-d-glucosyl o-hydroxycinnamic acid in Melilotus alba leaves. Plant Physiol. 1964, 39, 777–781. [Google Scholar] [CrossRef] [PubMed]
- Değirmencioğlu, N.; Gürbüz, O.; Karatepe, G.E.; Irkin, R. Influence of hot air drying on phenolic compounds and antioxidant capacity of blueberry (Vaccinium myrtillus) fruit and leaf. J. Appl. Bot. Food Qual. 2017, 90, 115–125. [Google Scholar]
- Brereton, N.J.B.; Berthod, N.; Lafleur, B.; Pedneault, K.; Pitre, F.E.; Labrecque, M. Extractable phenolic yield variation in five cultivars of mature short rotation coppice willow from four plantations in Quebec. Ind. Crop. Prod. 2017, 97, 525–535. [Google Scholar] [CrossRef]
- Cardozo, E.L.; Ferrarese-Filho, O.; Filho, L.C.; Ferrarese, M.L.L.; Donaduzzi, C.M.; Sturion, J.A. Methylxanthines and phenolic compounds in mate (Ilex paraguariensis St. Hil.) progenies grown in Brazil. J. Food Compos. Anal. 2007, 20, 553–558. [Google Scholar] [CrossRef]
- Kivilompolo, M.; Hyotylainen, T. Comprehensive two-dimensional liquid chromatography in analysis of Lamiaceae herbs: Characterisation and quantification of antioxidant phenolic acids. J. Chromatogr. A 2007, 1145, 155–164. [Google Scholar] [CrossRef]
- Złotek, U.; Gawlik-Dziki, U.; Dziki, D.; Świeca, M.; Nowak, R.; Martinez, E. Influence of drying temperature on phenolic acids composition and antioxidant activity of sprouts and leaves of white and red Quinoa. J. Chem. 2019, 2019, 8. [Google Scholar] [CrossRef]
- Prabakaran, M.; Lee, J.-H.; Ahmad, A.; Kim, S.-H.; Woo, K.-S.; Kim, M.-J.; Chung, I.-M. Effect of storage time and temperature on phenolic compounds of soybean (Glycine max L.) flour. Molecules 2018, 23, 2269. [Google Scholar] [CrossRef]
- Khattab, R.; Eskin, M.; Aliani, M.; Thiyam, U. Determination of sinapic acid derivatives in canola extracts using high-performance liquid chromatography. J. Am. Oil Chem. Soc. 2010, 87, 147–155. [Google Scholar] [CrossRef]
- Ozer, H.K. Phenolic compositions and antioxidant activities of Maya nut (Brosimum alicastrum): Comparison with commercial nuts. Int. J. Food Prop. 2017, 20, 2772–2781. [Google Scholar] [CrossRef]
- Sampaio, C.R.P.; Hamerski, F.; Ribani, R.H. Antioxidant phytochemicals of Byrsonima ligustrifolia throughout fruit developmental stages. J. Funct. Foods 2015, 18, 400–410. [Google Scholar] [CrossRef]
- Mattila, P.; Hellström, J.; Törrönen, R. Phenolic acids in berries, fruits, and beverages. J. Agric. Food Chem. 2006, 54, 7193–7199. [Google Scholar] [CrossRef] [PubMed]
- Mallek-Ayadi, S.; Bahloul, N.; Kechaou, N. Characterization, phenolic compounds and functional properties of Cucumis melo L. peels. Food Chem. 2017, 221, 1691–1697. [Google Scholar] [CrossRef] [PubMed]
- Zadernowski, R.; Czaplicki, S.; Naczk, M. Phenolic acid profiles of mangosteen fruits (Garcinia mangostana). Food Chem. 2009, 112, 685–689. [Google Scholar] [CrossRef]
- Chon, S.-U.; Kim, Y.-M. Herbicidal potential and quantification of suspected allelochemicals from four grass crop extracts. J. Agron. Crop Sci. 2004, 190, 145–150. [Google Scholar] [CrossRef]
- Caldas, T.W.; Mazza, K.E.L.; Teles, A.S.C.; Mattos, G.N.; Brígidac, A.I.S.; Conte-Junior, C.A.; Borguinic, R.G.; Godoy, R.L.O.; Cabral, L.M.C.; Tonon, R.V. Phenolic compounds recovery from grape skin using conventional and non-conventional extraction methods. Ind. Crop. Prod. 2018, 111, 86–91. [Google Scholar] [CrossRef]
- Foo, S.C.; Yusoff, F.M.; Ismail, M.; Basri, M.; Yau, S.K.; Khong, N.M.H.; Chan, K.W.; Ebrahimi, M. Antioxidant capacities of fucoxanthin-producing algae as influenced by their carotenoid and phenolic contents. J. Biotechnol. 2017, 241, 175–183. [Google Scholar] [CrossRef]
- Shan, B.; Cai, Y.Z.; Sun, M.; Corke, H. Antioxidant capacity of 26 spice extracts and characterization of their phenolic constituents. J. Agric. Food Chem. 2005, 53, 7749–7759. [Google Scholar] [CrossRef]
- Bae, I.K.; Ham, H.M.; Jeong, M.H.; Kim, D.H.; Kim, H.J. Simultaneous determination of 15 phenolic compounds and caffeine in teas and mate using RP-HPLC/UV detection: Method development and optimization of extraction process. Food Chem. 2015, 172, 469–475. [Google Scholar] [CrossRef]
- Deng, G.F.; Shen, C.; Xu, X.R.; Kuang, R.D.; Guo, Y.J.; Zeng, L.S.; Gao, L.L.; Lin, X.; Xie, J.F.; Xia, E.Q.; et al. Potential of fruit wastes as natural resources of bioactive compounds. Int. J. Mol. Sci. 2012, 13, 8308–8323. [Google Scholar] [CrossRef]
- Silva-Beltrán, N.P.; Chaidez-Quiroz, C.; López-Cuevas, O.; Ruiz-Cruz, S.; López-Mata, M.A.; Del-Toro-Sánchez, C.L.; Marquez-Rios, E.; Ornelas-Paz, J.d.J. Phenolic compounds of potato peel extracts: Their antioxidant activity and protection against human enteric viruses. J. Microbiol. Biotechnol. 2017, 27, 234–241. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Cruz, O.; Paredes-López, O. Phytochemical profile and nutraceutical potential of chia seeds (Salvia hispanica L.) by ultra high performance liquid chromatography. J. Chromatogr. A 2014, 1346, 43–48. [Google Scholar]
- Prakash, D.; Singh, B.N.; Upadhyay, G. Antioxidant and free radical scavenging activities of phenols from onion (Allium cepa). Food Chem. 2007, 102, 1389–1393. [Google Scholar] [CrossRef]
- Arruda, H.S.; Pereira, G.A.; de Morais, D.R.; Eberlin, M.N.; Pastore, G.M. Determination of free, esterified, glycosylated and insoluble-bound phenolics composition in the edible part of araticum fruit (Annona crassiflora Mart.) and its by-products by HPLC-ESI-MS/MS. Food Chem. 2018, 245, 738–749. [Google Scholar] [CrossRef] [PubMed]
- Zgórka, G.; Głowniak, K. Variation of free phenolic acids in medicinal plants belonging to the Lamiaceae family. J. Pharm. Biomed. Anal. 2001, 26, 79–87. [Google Scholar] [CrossRef]
- Lu, M.; Yuan, B.; Zeng, M.; Chen, J. Antioxidant capacity and major phenolic compounds of spices commonly consumed in China. Food Res. Int. 2011, 44, 530–536. [Google Scholar] [CrossRef]
- Chandrasekara, N.; Shahidi, F. Effect of roasting on phenolic content and antioxidant activities of whole cashew nuts, kernels, and testa. J. Agric. Food Chem. 2011, 59, 5006–5014. [Google Scholar] [CrossRef] [PubMed]
- Karasová, G.; Lehotay, J. Chromatographic determination of derivatives of p-hydroxybenzoic acid in Melissa officinalis by HPLC. J. Liq. Chromatogr. Relat. Technol. 2005, 28, 2421–2431. [Google Scholar] [CrossRef]
- Budrat, P.; Shotipruk, A. Enhanced recovery of phenolic compounds from bitter melon (Momordica charantia) by subcritical water extraction. Sep. Purif. Technol. 2009, 66, 125–129. [Google Scholar] [CrossRef]
- Aresta, A.; Zambonin, C. Simultaneous determination of salicylic, 3-methyl salicylic, 4-methyl salicylic, acetylsalicylic and benzoic acids in fruit, vegetables and derived beverages by SPME–LC–UV/DAD. J. Pharm. Biomed. Anal. 2016, 121, 63–68. [Google Scholar] [CrossRef]
- Tian, Y.; Liimatainen, J.; Alanne, A.-L.; Lindstedt, A.; Liu, P.; Sinkkonen, J.; Kallio, H.; Yang, B. Phenolic compounds extracted by acidic aqueous ethanol from berries and leaves of different berry plants. Food Chem. 2017, 220, 266–281. [Google Scholar] [CrossRef] [PubMed]
- Wojdyło, A.; Oszmiański, J.; Czemerys, R. Antioxidant activity and phenolic compounds in 32 selected herbs. Food Chem. 2007, 105, 940–949. [Google Scholar] [CrossRef]
- Prabakaran, M.; Kim, S.-H.; Hemapriya, V.; Chung, I.-M. Evaluation of polyphenol composition and anti-corrosion properties of Cryptostegia grandiflora plant extract on mild steel in acidic medium. J. Ind. Eng. Chem. 2016, 37, 47–56. [Google Scholar] [CrossRef]
- Prabakaran, M.; Kim, S.-H.; Sasireka, A.; Chandrasekaran, M.; Chung, I.-M. Polyphenol composition and antimicrobial activity of various solvent extracts from different plant parts of Moringa oleifera. Food Biosci. 2018, 26, 23–29. [Google Scholar] [CrossRef]
- Chung, I.M.; Kim, K.H.; Ahn, J.K.; Chun, S.C.; Kim, S.C.; Kim, J.T.; Kim, S.H. Screening of allelochemicals on barnyardgrass (Echinochloa crus-galli) and identification of potentially allelopathic compounds from rice (Oryza sativa) variety hull extracts. Crop Prot. 2002, 21, 913–920. [Google Scholar] [CrossRef]
- Barron, C.; Surget, A.; Rouau, X. Relative amounts of tissues in mature wheat (Triticum aestivum L.) grain and their carbohydrate and phenolic acid composition. J. Cereal Sci. 2007, 45, 88–96. [Google Scholar] [CrossRef]
- Kim, K.-H.; Tsao, R.; Yang, R.; Cui, S.W. Phenolic acid profiles and antioxidant activities of wheat bran extracts and the effect of hydrolysis conditions. Food Chem. 2006, 95, 466–473. [Google Scholar] [CrossRef]
- Lemos, M.R.B.; de Almeida Siqueira, E.M.; Arruda, S.F.; Zambiazi, R.C. The effect of roasting on the phenolic compounds and antioxidant potential of baru nuts [Dipteryx alata Vog.]. Food Res. Int. 2012, 48, 592–597. [Google Scholar] [CrossRef]
- Kerienė, I.; Mankevičienė, A.; Bliznikas, S.; Jablonskytė-Raščė, D.; Maikštėnienė, S.; Česnuliavičienė, R. Biologically active phenolic compounds in buckwheat, oats and winter spelt wheat. Zemdirb. -Agric. 2015, 102, 289–296. [Google Scholar]
- Brouns, F.; Hemery, Y.; Price, R.; Anson, N.M. Wheat aleurone: Separation, composition, health aspects, and potential food use. Crit. Rev. Food Sci. Nutr. 2012, 52, 553–568. [Google Scholar] [CrossRef]
- Mccallum, J.A.; Walker, J.R.L. Phenolic biosynthesis during grain development in wheat (Triticum aestivum L.) III. Changes in hydroxycinnamic acids during grain development. J. Cereal Sci. 1991, 13, 161–172. [Google Scholar] [CrossRef]
- Karamać, M.; Kosińska, A.; Estrella, I.; Hernández, T.; Dueñas, M. Antioxidant activity of phenolic compounds identified in sunflower seeds. Eur. Food Res. Technol. 2012, 235, 221–230. [Google Scholar] [CrossRef]
- Waszkowiak, K.; Gliszczyńska-Świgło, A.; Barthet, V.; Skręty, J. Effect of extraction method on the phenolic and cyanogenic glucoside profile of flaxseed extracts and their antioxidant capacity. J. Am. Oil Chem. Soc. 2015, 92, 1609–1619. [Google Scholar] [CrossRef] [PubMed]
- Jun, H.-I.; Wiesenborn, D.P.; Kim, Y.-S. Antioxidant activity of phenolic compounds from canola (Brassica napus) seed. Food Sci. Biotechnol. 2014, 23, 1753–1760. [Google Scholar] [CrossRef]
- Anwar, M.M.; Ali, S.E.; Nasra, E.H. Improving the nutritional value of canola seed by gamma irradiation. J. Radiat. Res. Appl. Sci. 2015, 8, 328–333. [Google Scholar] [CrossRef]
- Diaconeasa, Z.; Florica, R.; Rugina, D.; Lucian, C.; Socaciu, C. HPLC/PDA–ESI/MS identification of phenolic acids, flavonol glycosides and antioxidant potential in blueberry, blackberry, raspberries and cranberries. J. Food Nutr. Res. 2014, 2, 781–785. [Google Scholar] [CrossRef]
- Mahmood, T.; Anwar, F.; Abbas, M.; Saari, N. Effect of maturity on phenolics (phenolic acids and flavonoids) profile of strawberry cultivars and mulberry species from Pakistan. Int. J. Mol. Sci. 2012, 13, 4591–4607. [Google Scholar] [CrossRef]
- Mattila, P.; Hellström, J. Phenolic acids in potatoes, vegetables, and some of their products. J. Food Compos. Anal. 2007, 20, 152–160. [Google Scholar] [CrossRef]
- Akyol, H.; Riciputi, Y.; Capanoglu, E.; Caboni, M.F.; Verardo, V. Phenolic compounds in the potato and its byproducts: An overview. Int. J. Mol. Sci. 2016, 17, 835. [Google Scholar] [CrossRef]
- Ferreres, F.; Gil, M.I.; Castañer, M.; Tomás-Barberán, F.A. Phenolic metabolites in red pigmented lettuce (Lactuca sativa). Changes with minimal processing and cold storage. J. Agric. Food Chem. 1997, 45, 4249–4254. [Google Scholar] [CrossRef]
- Dao, L.; Friedman, M. Chlorogenic acid content of fresh and processed potatoes determined by ultraviolet spectrophotometry. J. Agric. Food Chem. 1992, 40, 2152–2156. [Google Scholar] [CrossRef]
- Robbins, K.S.; Gong, Y.; Wells, M.L.; Greenspan, P.; Pegg, R.B. Investigation of the antioxidant capacity and phenolic constituents of U.S. pecans. J. Funct. Foods 2015, 15, 11–22. [Google Scholar] [CrossRef]
- Alasalvar, C.; Bolling, B.W. Review of nut phytochemicals, fat-soluble bioactives, antioxidant components and health effect. Br. J. Nutr. 2015, 113, S68–S78. [Google Scholar] [CrossRef] [PubMed]
- Milbury, P.E.; Chen, C.-Y.; Dolnikowski, G.G.; Blumberg, J.B. Determination of flavonoids and phenolics and their distribution in almonds. J. Agric. Food Chem. 2006, 54, 5027–5033. [Google Scholar] [CrossRef] [PubMed]
- Bolling, B.W.; Blumberg, J.B.; Chen, C.-Y.O. The influence of roasting, pasteurisation, and storage on the polyphenol content and antioxidant capacity of California almond skins. Food Chem. 2010, 123, 1040–1047. [Google Scholar] [CrossRef] [PubMed]
- Talcott, S.T.; Duncan, C.E.; Pozo-Insfran, D.D.; Gorbet, D.W. Polyphenolic and antioxidant changes during storage of normal, mid, and high oleic acid peanuts. Food Chem. 2005, 89, 77–84. [Google Scholar] [CrossRef]
- Ulewicz-Magulska, B.; Wesolowski, M. Total phenolic contents and antioxidant potential of herbs used for medical and culinary purposes. Plant Foods Hum. Nutr. 2019, 74, 61–67. [Google Scholar] [CrossRef]
- Bahadori, M.B.; Asghari, B.; Dinparast, L.; Zengin, G.; Sarikurkcu, C.; Abbas-Mohammadi, M.; Bahadori, S. Salvia nemorosa L.: A novel source of bioactive agents with functional connections. LWT 2017, 75, 42–50. [Google Scholar] [CrossRef]
- Górnas, P.; Dwiecki, K.; Siger, A.; Tomaszewska-Gras, J.; Michalak, M.; Polewski, K. Contribution of phenolic acids isolated from green and roasted boiled-type coffee brews to total coffee antioxidant capacity. Eur. Food Res. Technol. 2016, 242, 641–653. [Google Scholar] [CrossRef]
- Perdani, C.G.; Pranowo, D.; Qonitatilah. Total phenols content of green coffee (Coffea arabica and Coffea canephora) in East Java. Iop Conf. Ser. Earth Environ. Sci. 2019, 230, 012093. [Google Scholar] [CrossRef]
- Sadh, P.K.; Duhan, S.; Duhan, J.S. Agro-industrial wastes and their utilization using solid state fermentation: A review. Bioresour. Bioprocess. 2018, 5, 1. [Google Scholar] [CrossRef]
- Pfaltzgraff, L.A.; De bruyn, M.; Cooper, E.C.; Budarin, V.; Clark, J.H. Food waste biomass: A resource for high-value chemicals. Green Chem. 2013, 15, 307–314. [Google Scholar] [CrossRef]
- Li, M.; Jia, Z.; Wan, G.; Wang, S.; Min, D. Enhancing isolation of p-coumaric and ferulic acids from sugarcane bagasse by sequential hydrolysis. Chem. Pap. 2020, 74, 499–507. [Google Scholar] [CrossRef]
- Alhamad, M.N.; Rababah, T.M.; Al-u’datt, M.; Ereifej, K.; Esoh, R.; Feng, H.; Yang, W. The physicochemical properties, total phenolic, antioxidant activities, and phenolic profile of fermented olive cake. Arab. J. Chem. 2017, 10, 136–140. [Google Scholar] [CrossRef]
- Wu, J.; Collins, S.R.A.; Elliston, A.; Wellner, N.; Dicks, J.; Roberts, I.N.; Waldron, K.W. Release of cell wall phenolic esters during hydrothermal pretreatment of rice husk and rice straw. Biotechnol. Biofuels 2018, 11, 162. [Google Scholar] [CrossRef] [PubMed]
- Quinn, L.A.; Tang, H.H. Antioxidant properties of phenolic compounds in macadamia nuts. J. Am. Oil Chem. Soc. 1996, 73, 1585–1588. [Google Scholar] [CrossRef]
- Amaya-Cruz, D.M.; Pérez-Ramírez, I.F.; Delgado-García, J.; Mondragón-Jacobo, C.; Dector-Espinoza, A.; Reynoso-Camacho, R. An integral profile of bioactive compounds and functional properties of prickly pear (Opuntia ficus indica L.) peel with different tonalities. Food Chem. 2019, 278, 568–578. [Google Scholar] [CrossRef]
- Gómez-Mejía, E.; Rosales-Conrado, N.; León-González, M.E.; Madrid, Y. Citrus peels waste as a source of value-added compounds: Extraction and quantification of bioactive polyphenols. Food Chem. 2019, 289–299. [Google Scholar] [CrossRef]
- Orhan, I.; Ustun, O. Determination of total phenol content, antioxidant activity and acetylcholinesterase inhibition in selected mushrooms from Turkey. J. Food Compos. Anal. 2011, 24, 386–390. [Google Scholar] [CrossRef]
- Tepsongkroh, B.; Jangchud, K.; Trakoontivakorn, G. Antioxidant properties and selected phenolic acids of five different tray-dried and freeze-dried mushrooms using methanol and hot water extraction. J. Food Meas. Charact. 2019, 13, 3097–3105. [Google Scholar] [CrossRef]
- Barros, L.; Dueñas, M.; Ferreira, I.C.F.R.; Baptista, P.; Santos-Buelga, C. Phenolic acids determination by HPLC–DAD–ESI/MS in sixteen different Portuguese wild mushrooms species. Food Chem. Toxicol. 2009, 47, 1076–1079. [Google Scholar] [CrossRef] [PubMed]
- Heleno, S.A.; Barros, L.; Martins, A.; Queiroz, M.J.R.P.; Santos-Buelga, C.; Ferreira, I.C.F.R. Fruiting body spores and in vitro produced mycelium of Ganoderma lucidum from Northeast Portugal: A comparative study of the antioxidant potential of phenolic and polysaccharidic extracts. Food Res. Int. 2012, 46, 135–140. [Google Scholar] [CrossRef]
- Reis, F.S.; Heleno, S.A.; Barros, L.; Sousa, M.J.; Martins, A.; SantosBuelga, C.; Ferreira, I.C.F.R. Toward the antioxidant and chemical characterization of mycorrhizal mushrooms from Northeast Portugal. J. Food Sci. 2011, 76, 824–830. [Google Scholar] [CrossRef] [PubMed]
- Vaz, J.A.; Barros, L.; Martins, A.; Morais, J.S.; Vasconcelos, M.H.; Ferreira, I.C.F.R. Phenolic profile of seventeen Portuguese wild mushrooms. Lwt-Food Sci. Technol. 2011, 44, 343–346. [Google Scholar] [CrossRef]
- Vaz, J.A.; Barros, L.; Martins, A.; Santos-Buelga, C.; Vasconcelos, M.H.; Ferreira, I.C.F.R. Chemical composition of wild edible mushrooms and antioxidant properties of their water soluble polysaccharidic and ethanolic fractions. Food Chem. 2011, 126, 610–616. [Google Scholar] [CrossRef]
- Alves, M.J.; Ferreira, I.C.F.R.; Froufe, H.J.C.; Abreu, R.M.V.; Martins, A.; Pintado, M. Antimicrobial activity of phenolic compounds identified in wild mushrooms, SAR analysis and docking studies. J. Appl. Microbiol. 2013, 115, 346–357. [Google Scholar] [CrossRef] [PubMed]
- Reis, F.S.; Martins, A.; Barros, L.; Ferreira, I.C.F.R. Antioxidant properties and phenolic profile of the most widely appreciated cultivated mushrooms: A comparative study between in vivo and in vitro samples. Food Chem. Toxicol. 2012, 50, 1201–1207. [Google Scholar] [CrossRef]
- Bertrand, R.L.; Sorensen, J.L. A comprehensive catalogue of polyketide synthase gene clusters in lichenizing fungi. J. Ind. Microbiol. Biotechnol. 2018, 45, 1067–1081. [Google Scholar] [CrossRef]
- Torres-Benítez, A.; Rivera-Montalvo, M.; Sepúlveda, B.; Olivio, N.; Castro, O.N.; Nagles, E.; Simirgiotis, M.J.; García-Beltrán, O.; Areche, C. Metabolomic analysis of two Parmotrema lichens: P. robustum (Degel.) hale and P. andinum (Mull. Arg.) hale using UHPLC-ESI-OT-MS-MS. Molecules 2017, 22, 1861. [Google Scholar]
- Jin, Y.; Ma, Y.; Xie, W.; Hou, L.; Xu, H.; Zhang, K.; Zhang, L.; Du, Y. UHPLC-Q-TOF-MS/MS-oriented characteristic components dataset and multivariate statistical techniques for the holistic quality control of Usnea. Rsc Adv. 2018, 8, 15487–15500. [Google Scholar] [CrossRef]
- Lopes, T.I.B.; Coelho, R.G.; Yoshida, N.C.; Honda, N.K. Radical-scavenging activity of orsellinates. Chem. Pharm. Bull. 2008, 56, 1551–1554. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, G.; Sachan, A.; Ghosh, S.; Mitra, A. Conversion of sinapic acid to syringic acid by a filamentous fungus Paecilomyces variotii. J. Gen. Appl. Microbiol. 2006, 52, 131–135. [Google Scholar] [CrossRef] [PubMed]
- Ishikura, T.; Nishida, H.; Tanno, K.; Miyachi, N.; Ozaki, A. Microbial production of salicylic acid from naphthalene. Agric. Biol. Chem. 1968, 32, 12–20. [Google Scholar]
- Gupta, D.; Sinha, S.N. Production of salicylic acid by a purple non sulfur bacterium Rubrivivax gelatinosus strain RASN4 from rhizosperic soil of paddy fields. J. Glob. Biosci. 2020, 9, 6718–6736. [Google Scholar]
- Mishra, S.; Kullu, M.; Sachan, A.; Vidyarthi, A.S.; Sachan, S.G. Bioconversion of ferulic acid to vanillic acid by Paenibacillus lactis SAMS-2001. Ann. Microbiol. 2016, 66, 875–882. [Google Scholar] [CrossRef]
- Vyrides, I.; Agathangelou, M.; Dimitriou, R.; Souroullas, K.; Salamex, A.; Ioannou, A.; Koutinas, M. Novel Halomonas sp. B15 isolated from Larnaca Salt Lake in Cyprus that generates vanillin and vanillic acid from ferulic acid. World J. Microbiol. Biotechnol. 2015, 31, 1291–1296. [Google Scholar] [CrossRef]
- Tang, P.L.; Hassan, O. Bioconversion of ferulic acid attained from pineapple peels and pineapple crown leaves into vanillic acid and vanillin by Aspergillus niger I-1472. Bmc Chem. 2020, 14, 7. [Google Scholar] [CrossRef]
- Topakas, E.; Kalogeris, E.; Kekos, D.; Macris, B.J.; Christakopoulos, P. Bioconversion of ferulic acid into vanillic acid by the thermophilic fungus Sporotrichum thermophile. Lwt Food Sci. Technol. 2003, 36, 561–565. [Google Scholar] [CrossRef]
- Skóra, J.; Sulyok, M.; Nowak, A.; Otlewska, A.; Gutarowska, B. Toxinogenicity and cytotoxicity of Alternaria, Aspergillus and Penicillium moulds isolated from working environments. Int. J. Environ. Sci. Technol. Vol. 2017, 14, 595–608. [Google Scholar] [CrossRef]
- Bajpai, B.; Patil, S. A new approach to microbial production of gallic acid. Braz. J. Microbiol. 2008, 39, 708–711. [Google Scholar] [CrossRef]
- Fathy, S.A.; Mahmoud, A.E.; Rashad, M.M.; Ezz, M.K.; Mohammed, A.T. Improving the nutritive value of olive pomace by solid state fermentation of Kluyveromyces marxianus with simultaneous production of gallic acid. Int. J. Recycl. Org. Waste Agric. 2018, 7, 135–141. [Google Scholar] [CrossRef]
- Limón, R.I.; Peñas, E.; Torino, M.I.; Martínez-Villaluenga, C.; Dueñas, M.; Frias, J. Fermentation enhances the content of bioactive compounds in kidney bean extracts. Food Chem. 2015, 72, 343–352. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, C.G.; Gonçalve, L.M.; Prietto, L.; Hackbart, H.S.; Furlong, E.B. Antioxidant activity and enzyme inhibition of phenolic acids from fermented rice bran with fungus Rizhopus oryzae. Food Chem. 2014, 146, 371–377. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, R.; Mukherjee, G.; Patra, K.C. Microbial transformation of tannin-rich substrate to gallic acid through co-culture method. Bioresour. Technol. 2005, 96, 949–953. [Google Scholar] [CrossRef]
- Jamal, P.; Idris, Z.M.; Alam, M.Z. Effects of physicochemical parameters on the production of phenolic acids from palm oil mill effluent under liquid-state fermentation by Aspergillus niger IBS-103ZA. Food Chem. 2011, 124, 1595–1602. [Google Scholar] [CrossRef]
- Sakamoto, T.; Nishimura, S.; Kato, T.; Sunagawa, Y.; Tsuchiyama, M.; Kawasaki, H. Efficient extraction of ferulic acid from sugar beet pulp using the culture supernatant of Penicillium chrysogenum. J. Appl. Glycosci. 2005, 52, 115–120. [Google Scholar] [CrossRef]
- Daumy, G.O.; McColl, A.S.; Andrews, G.C. Bioconversion of m-hydroxybenzoate to 2,3-dihydroxybenzoate by mutants of Pseudomonas testosteroni. J. Bacteriol. 1980, 141, 293–296. [Google Scholar] [CrossRef]
- Couto, S.R.; Sanroman, M.A. Application of solid-state fermentation to food industry–A review. J. Food Eng. 2006, 76, 291–302. [Google Scholar] [CrossRef]
- Mitchell, D.A.; von Meien, O.F.; Krieger, N.; Dalsenter, F.D.H. A review of recent developments in modeling of microbial growth kinetics and intraparticle phenomena in solid-state fermentation. Biochem. Eng. J. 2004, 17, 15–26. [Google Scholar] [CrossRef]
- Lambert, R.J.; Stratford, M. Weak-acid preservatives: Modelling microbial inhibition and response. J. Appl. Microbiol. 1999, 86, 157–164. [Google Scholar] [CrossRef]
- Mycroft, Z.; Gomis, M.; Mines, P.; Law, P.; Bugg, T.D.H. Biocatalytic conversion of lignin to aromatic dicarboxylic acids in Rhodococcus jostii RHA1 by re-routing aromatic degradation pathways. Green Chem. 2015, 17, 4974–4979. [Google Scholar] [CrossRef]
- Kumar, M.; Singh, J.; Singh, M.K.; Singhal, A.; Thakur, I.S. Investigating the degradation process of kraft lignin by β-proteobacterium, Pandoraea sp. ISTKB. Environ. Sci. Pollut. Res. 2015, 22, 15690–15702. [Google Scholar] [CrossRef] [PubMed]
- Peng, X.; Misawa, N.; Harayama, S. Isolation and characterization of thermophilic Bacilli degrading cinnamic, 4-coumaric, and ferulic acids. Appl. Environ. Microbiol. 2003, 69, 1417–1427. [Google Scholar] [CrossRef] [PubMed]
- Duan, J.; Huo, H.; Du, W.J.; Liang, J.D.; Wang, D.Q.; Yang, S.C. Biodegradation of kraft lignin by a newly isolated anaerobic bacterial strain, Acetoanaerobium sp. WJDL-Y2. Lett. Appl. Microbiol. 2015, 62, 55–62. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Z.; Shetty, K. Solid-state bioconversion of phenolics from cranberry pomace and role of Lentinus edodes beta-glucosidase. J. Agric. Food Chem. 2000, 48, 895–900. [Google Scholar] [CrossRef] [PubMed]
- Brosnan, B.; Coffey, A.; Arendt, E.K.; Furey, A. Rapid identification, by use of the LTQ Orbitrap hybrid FT mass spectrometer, of antifungal compounds produced by lactic acid bacteria. Anal. Bioanal. Chem. 2012, 403, 2983–2995. [Google Scholar] [CrossRef]
- Broberg, A.; Jacobsson, K.; Ström, K.; Schnürer, J. Metabolite profiles of lactic acid bacteria in grass silage. Appl. Environ. Microbiol. 2007, 73, 5547–5552. [Google Scholar] [CrossRef]
- Tian, R.-R.; Qiu-Hong Pan, Q.-H.; Zhan, J.-C.; Li, J.-M.; Wan, S.-B.; Zhang, Q.-H.; Huang, W.-D. Comparison of phenolic acids and flavan-3-ols during wine fermentation of grapes with different harvest times. Molecules 2009, 14, 827–838. [Google Scholar] [CrossRef]
- Klepacka, J.; Fornal, L. Ferulic acid and its position among the phenolic compounds of wheat. Crit. Rev. Food Sci. Nutr. 2006, 46, 639–647. [Google Scholar] [CrossRef]
- Antognoni, F.; Mandrioli, R.; Potente, G.; Taneyo Saa, D.L.; Gianotti, A. Changes in carotenoids, phenolic acids and antioxidant capacity in bread wheat doughs fermented with different lactic acid bacteria strains. Food Chem. 2019, 292, 211–216. [Google Scholar] [CrossRef]
- Dey, T.B.; Kuhad, R.C. Enhanced production and extraction of phenolic compounds from wheat by solid-state fermentation with Rhizopus oryzae RCK2012. Biotechnol. Rep. 2014, 4, 120–127. [Google Scholar]
- Rodriguez, A.; Chen, Y.; Khoomrung, S.; Özdemir, E.; Borodina, I.; Nielsen, J. Comparison of the metabolic response to over-production of p-coumaric acid in two yeast strains. Metab. Eng. 2017, 44, 265–272. [Google Scholar] [CrossRef] [PubMed]
- Vargas-Tah, A.; Martínez, L.M.; Hernández-Chávez, G.; Rocha, M.; Martínez, A.; Bolívar, F.; Gosset, G. Production of cinnamic and p-hydroxycinnamic acid from sugar mixtures with engineered Escherichia coli. Microb. Cell Factories 2015, 14, 6. [Google Scholar] [CrossRef] [PubMed]
- Kitade, Y.; Hashimoto, R.; Suda, M.; Hiraga, K.; Inui, M. Production of 4-hydroxybenzoic acid by an aerobic growth-arrested bioprocess using metabolically engineered Corynebacterium glutamicum. Appl. Environ. Biotechnol. 2018, 84, e02587-17. [Google Scholar]
- Porro, D.; Branduardi, P. Yeast cell factory: Fishing for the best one or engineering it? Microb. Cell Factories 2009, 8, 51. [Google Scholar] [CrossRef]
- Gottardi, M.; Reifenrath, M.; Boles, E.; Tripp, J. Pathway engineering for the production of heterologous aromatic chemicals and their derivatives in Saccharomyces cerevisiae: Bioconversion from glucose. Fems Yeast Res. 2017, 17, fox035. [Google Scholar] [CrossRef][Green Version]
- Lee, J.-H.; Wendisch, V.F. Biotechnological production of aromatic compounds of the extended shikimate pathway from renewable biomass. J. Biotechnol. 2017, 257, 211–221. [Google Scholar] [CrossRef]
- Meijnen, J.; Verhoef, S.; Briedjlal, A.A.; de Winde, J.H.; Ruijssenaars, H.J. Improved p-hydroxybenzoate production by engineered Pseudomonas putida S12 by using a mixed-substrate feeding strategy. Appl. Microbiol. Biotechnol. 2011, 90, 885–893. [Google Scholar] [CrossRef]
- Rodrigues, J.L.; Coutoa, M.R.; Araújo, R.G.; Prather, K.L.J.; Kluskens, L.; Rodrigues, L.R. Hydroxycinnamic acids and curcumin production in engineered Escherichia coli using heat shock promoters. Biochem. Eng. J. 2017, 125, 41–49. [Google Scholar] [CrossRef]
- Civolani, C.; Barghini, P.; Roncetti, A.R.; Ruzzi, M.; Schiesser, A. Bioconversion of ferulic acid into vanillic acid by means of a vanillate-negative mutant of Pseudomonas fluorescens strain BF13. Appl. Environ. Microbiol. 2000, 66, 2311–2317. [Google Scholar] [CrossRef]
- Yi, J.; Li, K.; Draths, K.M.; Frost, J.W. Modulation of phosphoenolpyruvate synthase expression increases shikimate pathway product yields in E. coli. Biotechnol. Prog. 2002, 1141–1148. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Li, J.; Qian, B.; Cheng, L.; Xu, S.; Wang, R. De Novo biosynthesis of p-coumaric acid in E. coli with a trans-cinnamic acid 4-hydroxylase from the Amaryllidaceae plant Lycoris aurea. Molecules 2018, 23, 3185. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Yu, T.; Li, X.; Chen, Y.; Campbell, K.; Nielsen, J.; Chen, Y. Rewiring carbon metabolism in yeast for high level production of aromatic chemicals. Nat. Commun. 2019, 10, 4976. [Google Scholar] [CrossRef] [PubMed]
- An, D.G.; Cha, M.N.; Nadarajan, S.P.; Kim, B.G.; Ahn, J.-H. Bacterial synthesis of four hydroxycinnamic acids. Appl. Biol. Chem. 2016, 59, 173–179. [Google Scholar] [CrossRef]
- Furuya, T.; Arai, Y.; Kino, K. Biotechnological production of caffeic acid by bacterial cytochrome P450 CYP199A2. Appl. Environ. Microbiol. 2012, 78, 6087–6094. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Yan, Y. Biosynthesis of caffeic acid in Escherichia coli using its endogenous hydroxylase complex. Microb. Cell Factories 2012, 11, 42–51. [Google Scholar] [CrossRef]
- Berner, M.; Krug, D.; Bihlmaier, C.; Vente, A.; Müller, R.; Bechthold, A. Genes and enzymes involved in caffeic acid biosynthesis in the Actinomycete Saccharothrix espanaensis. J. Bacteriol. 2006, 188, 2666–2673. [Google Scholar] [CrossRef]
- Rodrigues, J.L.; Araújo, R.G.; Prather, K.L.J.; Kluskens, L.D.; Rodrigues, L.R. Heterologous production of caffeic acid from tyrosine in Escherichia coli. Enzym. Microb. Technol. 2015, 71, 36–44. [Google Scholar] [CrossRef]
- Liu, L.; Liu, H.; Zhang, W.; Yao, M.; Li, B.; Liu, D.; Yuan, Y. Engineering the biosynthesis of caffeic acid in Saccharomyces cerevisiae with heterologous enzyme combinations. Engineering 2019, 5, 287–295. [Google Scholar] [CrossRef]
- Kim, Y.H.; Kwon, T.; Yang, H.J.; Kim, W.; Youn, H.; Lee, J.Y.; Youn, B. Gene engineering, purification, crystallization and preliminary X-ray diffraction of cytochrome P450 p-coumarate-3-hydroxylase (C3H), the Arabidopsis membrane protein. Protein Expr. Purif. 2011, 79, 149–155. [Google Scholar] [CrossRef]
- Kang, S.-Y.; Choi, O.; Lee, J.K.; Hwang, B.Y.; Uhm, T.-B.; Hong, Y.-S. Artificial biosynthesis of phenylpropanoic acids in a tyrosine overproducing Escherichia coli strain. Microb. Cell Factories 2012, 11, 153. [Google Scholar] [CrossRef] [PubMed]
- Nair, R.B.; Bastress, K.L.; Ruegger, M.O.; Denault, J.W.; Chapple, C. The Arabidopsis thaliana REDUCED EPIDERMAL FLUORESCENCE1 gene encodes an aldehyde dehydrogenase involved in ferulic acid and sinapic acid biosynthesis. Plant Cell 2004, 16, 544–554. [Google Scholar] [CrossRef] [PubMed]
- Kallscheuer, N.; Marienhagen, J. Corynebacterium glutamicum as platform for the production of hydroxybenzoic acids. Microb. Cell Factories 2018, 17, 70. [Google Scholar] [CrossRef] [PubMed]
- Muir, R.M.; Ibáñez, A.M.; Uratsu, S.L.; Ingham, E.S.; Leslie, C.A.; McGranahan, G.H.; Batra, N.; Goyal, S.; Joseph, J.; Jemmis, E.D.; et al. Mechanism of gallic acid biosynthesis in bacteria (Escherichia coli) and walnut (Juglans regia). Plant Mol. Biol. 2011, 75, 555–565. [Google Scholar] [CrossRef] [PubMed]
- Kambourakis, S.; Draths, K.M.; Frost, J.W. Synthesis of gallic acid and pyrogallol from glucose: replacing natural product isolation with microbial catalysis. J. Am. Chem. Soc. 2000, 122, 9042–9043. [Google Scholar] [CrossRef]
- Sun, X.; Lin, Y.; Yuan, Q.; Yan, Y. Biological production of muconic acid via a prokaryotic 2,3-dihydroxybenzoic acid decarboxylase. ChemSusChem 2014, 7, 2478–2481. [Google Scholar] [CrossRef]
- Okai, N.; Masuda, T.; Takeshima, Y.; Tanaka, K.; Yoshida, K.; Miyamoto, M.; Ogino, C.; Kondo, A. Biotransformation of ferulic acid to protocatechuic acid by Corynebacterium glutamicum ATCC 21420 engineered to express vanillate o-demethylase. Amb Express 2017, 7, 130. [Google Scholar] [CrossRef]
- Segura, A.; Bünz, P.V.; D’Argenio, D.A.; Ornston, L.N. Genetic analysis of a chromosomal region containing vanA and vanB, genes required for conversion of either ferulate or vanillate to protocatechuate in Acinetobacter. J. Biotechnol. 1999, 181, 3494–3504. [Google Scholar]
- Upadhyay, P.; Singh, N.K.; Tupe, R.; Odenath, A.; Lali, A. Biotransformation of corn bran derived ferulic acid to vanillic acid using engineered Pseudomonas putida KT2440. Prep. Biochem. Biotechnol. 2019, 50, 341–348. [Google Scholar] [CrossRef]
- Kamimura, N.; Goto, T.; Takahashi, K.; Kasai, D.; Otsuka, Y.; Nakamura, M.; Katayama, Y.; Fukuda, M.; Masai, E. A bacterial aromatic aldehyde dehydrogenase critical for the efficient catabolism of syringaldehyde. Sci. Rep. 2017, 7, 44422. [Google Scholar] [CrossRef]
- Seshime, Y.; Juvvadi, P.R.; Kitamoto, K.; Ebizuka, Y.; Nonaka, T.; Fujii, I. Aspergillus oryzae type III polyketide synthase CsyA is involved in the biosynthesis of 3,5-dihydroxybenzoic acid. Bioorg. Med. Chem. Lett. 2010, 20, 4785–4788. [Google Scholar] [CrossRef] [PubMed]
- Gao, L.; Cai, M.; Shen, W.; Zhou, X.; Zhang, Y. Engineered fungal polyketide biosynthesis in Pichia pastoris: A potential excellent host for polyketide production. Microb. Cell Factories 2013, 12, 77. [Google Scholar] [CrossRef] [PubMed]
- Xu, Q.; Bai, C.; Liu, Y.; Song, L.; Tian, L.; Yan, Y.; Zhou, J.; Zhou, X.; Zhang, Y.; Cai, M. Modulation of acetate utilization in Komagataella phaffii by metabolic engineering of tolerance and metabolism. Biotechnol. Biofuels 2019, 12, 61. [Google Scholar] [CrossRef] [PubMed]
- Kong, C.; Huang, H.; Xue, Y.; Liu, Y.; Peng, Q.; Liu, Q.; Xu, Q.; Zhu, Q.; Yin, Y.; Zhou, X.; et al. Heterologous pathway assembly reveals molecular steps of fungal terreic acid biosynthesis. Sci. Rep. 2018, 8, 2116. [Google Scholar] [CrossRef] [PubMed]
- Hitschler, J.; Boles, E. De novo production of aromatic m-cresol in Saccharomyces cerevisiae mediated by heterologous polyketide synthases combined with a 6-methylsalicylic acid decarboxylase. Metab. Eng. Commun. 2019, 9, e00093. [Google Scholar] [CrossRef]
- Yang, D.; Kim, W.J.; Yoo, S.M.; Choi, J.H.; Ha, S.H.; Leea, M.H.; Lee, S.Y. Repurposing type III polyketide synthase as a malonyl-CoA biosensor for metabolic engineering in bacteria. Proc. Natl. Acad. Sci. USA 2018, 115, 9835–9844. [Google Scholar] [CrossRef]
- Braesel, J.; Fricke, J.; Schwenk, D.; Hoffmeister, D. Biochemical and genetic basis of orsellinic acid biosynthesis and prenylation in a stereaceous basidiomycete. Fungal Genet. Biol. 2017, 98, 12–19. [Google Scholar] [CrossRef]
- Tan, Z.; Clomburg, J.M.; Gonzalez, R. Synthetic pathway for the production of olivetolic acid in Escherichia coli. ACS Synth. Biol. 2018, 7, 1886–1896. [Google Scholar] [CrossRef]
- Averesch, N.J.H.; Prima, A.; Krömer, J.O. Enhanced production of para-hydroxybenzoic acid by genetically engineered Saccharomyces cerevisiae. Bioprocess Biosyst. Eng. 2017, 2017, 1283–1289. [Google Scholar] [CrossRef]
- Purwanto, H.S.; Kang, M.-S.; Ferrer, L.; Han, S.-S.; Lee, J.-Y.; Kim, H.-S.; Lee, J.-H. Rational engineering of the shikimate and related pathways in Corynebacterium glutamicum for 4-hydroxybenzoate production. J. Biotechnol. 2018, 282, 92–100. [Google Scholar] [CrossRef]
- Lenzen, C.; Wynands, B.; Otto, M.; Bolzenius, J.; Mennicken, P.; Blank, L.M.; Wierckx, M. High-Yield Production of 4-hydroxybenzoate from glucose or glycerol by an engineered Pseudomonas taiwanensis VLB120. Front. Bioeng. Biotechnol. 2019, 7, 130. [Google Scholar] [CrossRef] [PubMed]
- Noda, S.; Shirai, T.; Oyama, S.; Kondo, A. Metabolic design of a platform Escherichia coli strain producing various chorismate derivatives. Metab. Eng. 2016, 33, 119–129. [Google Scholar] [CrossRef] [PubMed]
- Weber, C.; Brückner, C.; Weinreb, S.; Lehr, C.; Essl, C.; Boles, E. Biosynthesis of cis,cis-muconic acid and its aromatic precursors, catechol and protocatechuic acid, from renewable feedstocks by Saccharomyces cerevisiae. Appl. Environ. Microbiol. 2012, 78, 8421–8430. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Xie, D.; Frost, J.W. Benzene-free synthesis of catechol: interfacing microbial and chemical catalysis. J. Am. Chem. Soc. 2005, 127, 2874–2882. [Google Scholar] [CrossRef]
- Chen, Z.; Shen, X.; Wang, J.; Wang, J.; Yuan, Q.; Yan, Y. Rational engineering of p-hydroxybenzoate hydroxylase to enable efficient gallic acid synthesis via a novel artificial biosynthetic pathway. Biotechnol. Bioeng. 2017, 114, 2571–2580. [Google Scholar] [CrossRef]
- Noda, S.; Shirai, T.; Mori, Y.; Oyama, S.; Kondo, A. Engineering a synthetic pathway for maleate in Escherichia coli. Nat. Commun. 2017, 8, 1153. [Google Scholar] [CrossRef]
- Chen, Z.; Sun, X.; Li, Y.; Yan, Y.; Yuan, Q. Metabolic engineering of Escherichia coli for microbial synthesis of monolignols. Metab. Eng. 2017, 39, 102–109. [Google Scholar] [CrossRef]
- Rodriguez, A.; Kildegaard, K.R.; Li, M.; Borodina, I.; Nielsen, J. Establishment of a yeast platform strain for production of p-coumaric acid through metabolic engineering of aromatic amino acid biosynthesis. Metab. Eng. 2015, 31, 181–188. [Google Scholar] [CrossRef]
- Huang, Q.; Lin, Y.; Yan, Y. Caffeic acid production enhancement by engineering a phenylalanine over-producing Escherichia coli strain. Biotechnol. Bioeng. 2013, 110, 3188–3196. [Google Scholar] [CrossRef]
- Gao, Y.; Ma, S.; Wang, M.; Feng, X.-Y. Characterization of free, conjugated, and bound phenolic acids in seven commonly consumed vegetables. Molecules 2017, 22, 1878. [Google Scholar] [CrossRef]
- Xu, F.; Sun, R.-C.; Sun, J.-X.; Liu, C.-F.; He, B.-H.; Fan, J.-S. Determination of cell wall ferulic and p-coumaric acids in sugarcane bagasse. Anal. Chim. Acta 2005, 552, 207–217. [Google Scholar] [CrossRef]
- Nardini, M.; Cirillo, E.; Natella, F.; Mencarelli, D.; Comisso, A.; Scaccini, C. Detection of bound phenolic acids: Prevention by ascorbic acid and ethylenediaminetetraacetic acid of degradation of phenolic acids during alkaline hydrolysis. Food Chem. 2002, 79, 119–124. [Google Scholar] [CrossRef]
- Bouzid, O.; Navarro, D.; Roche, M.; Asther, M.; Haon, M.; Delattre, M.; Lorquin, J.; Labat, M.; Asther, M.; Lesage-Meessen, L. Fungal enzymes as a powerful tool to release simple phenolic compounds from olive oil by-product. Process Biochem. 2005, 40, 1855–1862. [Google Scholar] [CrossRef]
- Zhang, S.-B.; Zhai, H.-C.; Wang, L.; Yu, G.-H. Expression, purification and characterization of a feruloyl esterase A from Aspergillus flavus. Protein Expr. Purif. 2013, 92, 36–40. [Google Scholar] [CrossRef] [PubMed]
- Gopalan, N.; Nampoothiri, K.M. Biorefining of wheat bran for the purification of ferulic acid. Biocatal. Agric. Biotechnol. 2018, 15, 304–310. [Google Scholar] [CrossRef]
- Xu, Z.; Wang, T.; Zhang, S. Extracellular secretion of feruloyl esterase derived from Lactobacillus crispatus in Escherichia coli and its application for ferulic acid production. Bioresour. Technol. 2019, 288, 121526. [Google Scholar] [CrossRef]
- Torres-Mancera, M.T.; Cordova-López, J.; Rodríguez-Serrano, G.; Roussos, S.; Ramírez-Coronel, M.A.; Favela-Torres, E.; Saucedo-Castañeda, G. Enzymatic extraction of hydroxycinnamic acids from coffee pulp. Food Technol. Biotechnol. 2011, 49, 369–373. [Google Scholar]
- Curiel, J.; Betancor, L.; de las Rivas, B.; Muñoz, R.; Guisan, J.M.; Fernández-Lorente, G. Hydrolysis of tannic acid catalyzed by immobilized−stabilized derivatives of tannase from Lactobacillus plantarum. J. Agric. Food Chem. 2010, 58, 6403–6409. [Google Scholar] [CrossRef]
- Wu, H.; Li, H.; Xue, Y.; Luo, G.; Gan, L.; Liu, J.; Mao, L.; Long, M. High efficiency co-production of ferulic acid and xylooligosaccharides from wheat bran by recombinant xylanase and feruloyl esterase. Biochem. Eng. J. 2017, 120, 41–48. [Google Scholar] [CrossRef]
- Yu, P.; Maenz, D.D.; Mckinnon, J.J.; Racz, V.J.; Christensen, D.A. Release of ferulic acid from oat hulls by Aspergillus ferulic acid esterase and Trichoderma xylanase. J. Agrocultural Food Chem. 2002, 50, 1625–1630. [Google Scholar] [CrossRef]
- Chemat, F.; Fabiano-Tixier, A.S.; Vian, M.A.; Allaf, T.; Vorobiev, E. Solvent-free extraction of food and natural products. Trac Trends Anal. Chem. 2015, 71, 157–168. [Google Scholar] [CrossRef]
- Carabias-Martínez, R.; Rodríguez-Gonzalo, E.; Revilla-Ruiz, P.; Hernández-Méndez, J. Pressurized liquid extraction in the analysis of food and biological samples. J. Chromatogr. 2005, 1089, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Chemat, F.; Rombaut, N.; Sicaire, A.-G.; Meullemiestre, A.; Fabiano-Tixier, A.-S.; Abert-Vian, M. Ultrasound assisted extraction of food and natural products. Mechanisms, techniques, combinations, protocols and applications. A review. Ultrason. Sonochem. 2017, 34, 540–560. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.; Yan, J.; Liu, R.; Marcone, M.F.; Aisa, H.A.; Tsao, R. Optimization of microwave-assisted extraction of phenolics from potato and its downstream waste using orthogonal array design. Food Chem. 2012, 133, 1292–1298. [Google Scholar] [CrossRef]
- Virot, M.; Tomao, V.; Le Bourvellec, C.; Renard, C.M.C.G.; Chemat, F. Towards the industrial production of antioxidants from food processing by-products with ultrasound-assisted extraction. Ultrason. Sonochem. 2010, 17, 1066–1074. [Google Scholar] [CrossRef]
- Ma, Y.-Q.; Chen, J.-C.; Liu, D.-H.; Ye, X.-Q. Simultaneous extraction of phenolic compounds of citrus peel extracts: Effect of ultrasound. Ultrason. Sonochem. 2009, 16, 57–62. [Google Scholar] [CrossRef]
- Verberne, M.C.; Brouwer, N.; Delbianco, F.; Linthorst, H.J.; Bol, J.F.; Verpoorte, R. Method for the extraction of the volatile compound salicylic acid from tobacco leaf material. Phytochem. Anal. 2002, 13, 45–50. [Google Scholar] [CrossRef]
- Liazid, A.; Palma, M.; Brigui, J.; Barroso, C.G. Investigation on phenolic compounds stability during microwave-assisted extraction. J. Chromatogr. A 2007, 1140, 29–34. [Google Scholar] [CrossRef]
- Qiao, L.; Ye, X.; Sun, Y.; Ying, J.; Shen, Y.; Chen, J. Sonochemical effects on free phenolic acids under ultrasound treatment in a model system. Ultrason. Sonochem. 2013, 20, 1017–1025. [Google Scholar] [CrossRef]
- Christian, K.R.; Jackson, J. Changes in total phenolic and monomeric anthocyanin composition and antioxidant activity of three varieties of sorrel (Hibiscus sabdariffa) during maturity. J. Food Compos. Anal. 2009, 22, 663–667. [Google Scholar] [CrossRef]
- Waksmundzka – Hajnos, M.; Oniszczuk, A.; Szewczyk, K.; Wianowska, D. Effect of sample-preparation methods on the HPLC quantitation of some phenolic acids in plant materials. Acta Chromatogr. 2007, 19, 227–237. [Google Scholar]
- Yan-Ying, Y.; Wei, Z.; Shu-Wen, C. Extraction of ferulic acid and caffeic acid with ionic liquids. Chin. J. Anal. Chem. 2007, 35, 1726–1730. [Google Scholar] [CrossRef]
- Wang, X.H.; Wang, G.Y.; Hou, Y.Y.; Qin, L. Extraction of ferulic acid and vanilla acid by hydrophobic ionic liquid 1-hexyl-3-methylimidazolium hexafluorophosphate. J. Food Sci. Technol. 2018, 55, 3508–3517. [Google Scholar] [CrossRef] [PubMed]
- Žiaková, A.; Brandšteterová, E.; Blahová, B. Matrix solid-phase dispersion for the liquid chromatographic determination of phenolic acids in Melissa officinalis. J. Chromatogr. A 2003, 983, 271–275. [Google Scholar] [CrossRef]
- Rahman, M. Application of computational methods in isolation of plant secondary metabolites. In Computational Phytochemistry; Sarker, S.D., Nahar, L., Eds.; Elsevier BV: London, UK, 2018; pp. 107–139. [Google Scholar]
- Zhang, L.; Zhang, W.; Chen, W.; Chen, G. Simultaneous determination of five bioactive constituents in Rhizoma Chuanxiong by capillary electrophoresis with a carbon nanotube-polydimethylsiloxane composite electrode. J. Pharm. Biomed. Anal. 2016, 131, 107–112. [Google Scholar] [CrossRef]
- Emwas, A.H.; Roy, R.; McKay, R.T.; Tenori, L.; Saccenti, E.; Gowda, G.A.N.; Raftery, D.; Alahmari, F.; Jaremko, L.; Jaremko, M.; et al. NMR spectroscopy for metabolomics research. Metabolites 2019, 9, 123. [Google Scholar] [CrossRef]
- Monošík, R.; Streďanský, M.; Šturdík, E. Biosensors — classification, characterization. Acta Chim. Slovaca 2012, 5, 109–120. [Google Scholar]
- Diaconu, M.; Litescu, S.C.; Radu, G.L. Laccase–MWCNT–chitosan biosensor—A new tool for total polyphenolic content evaluation from in vitro cultivated plants. Sens. Actuators B Chem. 2010, 145, 800–806. [Google Scholar] [CrossRef]
- ElKaoutit, M.; Naranjo-Rodriguez, I.; Temsamani, K.; de la Vega, M.D.; de Cisneros, J.L. Dual laccase–tyrosinase based sonogel–carbon biosensor for monitoring polyphenols in beers. J. Agric. Food Chem. 2007, 55, 8011–8018. [Google Scholar] [CrossRef]
- Jarosz-Wilkołazka, A.; Ruzgas, T.; Gorton, L. Use of laccase-modified electrode for amperometric detection of plant flavonoids. Enzym. Microb. Technol. 2004, 35, 238–241. [Google Scholar] [CrossRef]
- Carralero Sanz, V.; Luz Mena, M.; González-Cortés, A.; Yáñez-Sedeño, P.; Pingarrón, J.M. Development of a tyrosinase biosensor based on gold nanoparticles-modified glassy carbon electrodes: Application to the measurement of a bioelectrochemical polyphenols index in wines. Anal. Chim. Acta 2005, 528, 1–8. [Google Scholar] [CrossRef]
- Mello, L.D.; Sotomayor, M.D.P.T.; Kubota, L.T. HRP-based amperometric biosensor for the polyphenols determination in vegetables extract. Sens. Actuators B Chem. 2003, 96, 636–645. [Google Scholar] [CrossRef]
- Ambri, F.; D’Ambrosio, V.; Blasi, R.D.; Maury, J.; Jacobsen, S.A.B.; McCloskey, D.; Jensen, M.K.; Keasling, J.D. High-resolution scanning of optimal biosensor reporter promoters in yeast. Acs Synth. Biol. 2020, 9, 218–226. [Google Scholar] [CrossRef] [PubMed]
- Rogers, J.K.; Taylor, N.D.; Church, G.M. Biosensor-based engineering of biosynthetic pathways. Curr. Opin. Biotechnol. 2016, 42, 84–91. [Google Scholar] [CrossRef] [PubMed]
- Alvarez-Gonzalez, G.; Dixon, N. Genetically encoded biosensors for lignocellulose valorization. Biotechnol. Biofuels 2019, 12, 246. [Google Scholar] [CrossRef] [PubMed]
- Siedler, S.; Khatri, N.K.; Zsohar, A.; Kjærbølling, I.; Vogt, M.; Hammar, P.; Nielsen, C.F.; Marienhagen, J.; Sommer, M.O.A.; Joensson, H.N. Development of a bacterial biosensor for rapid screening of yeast p-coumaric acid production. Acs Synth. Biol. 2017, 6, 1860–1869. [Google Scholar] [CrossRef]
- Frei, C.S.; Qian, S.; Cirino, P.C. New engineered phenolic biosensors based on the AraC regulatory protein. Protein Eng. Des. Sel. 2018, 31, 213–220. [Google Scholar] [CrossRef]
- Qian, S.; Li, Y.; Cirino, P.C. Biosensor-guided improvements in salicylate production by recombinant Escherichia coli. Microb. Cell Factories 2019, 18, 18. [Google Scholar] [CrossRef]
- Meyer, A.J.; Segall-Shapiro, T.H.; Glassey, E.; Zhang, J.; Voigt, C.A. Escherichia coli “Marionette” strains with 12 highly optimized small-molecule sensors. Nat. Chem. Biol. 2019, 15, 196–204. [Google Scholar] [CrossRef]
- Machado, L.F.M.; Dixon, N. Development and substrate specificity screening of an in vivo biosensor for the detection of biomass derived aromatic chemical building blocks. Chem. Commun. 2016, 52, 11402–11405. [Google Scholar] [CrossRef]
- Jha, R.K.; Kern, T.L.; Fox, D.T.; Strauss, C.E.M. Engineering an Acinetobacter regulon for biosensing and high-throughput enzyme screening in E. coli via flow cytometry. Nucleic Acids Res. 2014, 42, 8150–8160. [Google Scholar] [CrossRef] [PubMed]
- Machado, L.F.M.; Currin, A.; Dixon, N. Directed evolution of the PcaV allosteric transcription factor to generate a biosensor for aromatic aldehydes. J. Biol. Eng. 2019, 13, 91. [Google Scholar] [CrossRef] [PubMed]
- Jha, R.K.; Bingen, J.M.; Johnson, C.W.; Kern, T.L.; Khanna, P.; Trettel, D.S.; Strauss, C.E.M.; Beckham, G.T.; Dale, T. A protocatechuate biosensor for Pseudomonas putida KT2440 via promoter and protein evolution. Metab. Eng. Commun. 2018, 6, 33–38. [Google Scholar] [CrossRef] [PubMed]
- Jha, R.K.; Chakraborti, S.; Kern, T.L.; Fox, D.T.; Strauss, C.E.M. Rosetta comparative modeling for library design: Engineering alternative inducer specificity in a transcription factor. Proteins 2015, 83, 1327–1340. [Google Scholar] [CrossRef] [PubMed]
- Strachan, C.R.; Singha, R.; VanInsberghe, D.; Ievdokymenko, K.; Budwill, K.; Mohn, W.W.; Eltis, L.D.; Hallam, S.J. Metagenomic scaffolds enable combinatorial lignin transformation. Proc. Natl. Acad. Sci. USA 2014, 111, 10143–10148. [Google Scholar] [CrossRef]
- Varman, A.M.; Follenfant, R.; Liu, F.; Davis, R.W.; Lin, Y.K.; Singh, S. Hybrid phenolic-inducible promoters towards construction of self-inducible systems for microbial lignin valorization. Biotechnol. Biofuels 2018, 11, 182. [Google Scholar] [CrossRef]
- Kunjapur, A.M.; Prather, K.L.J. Development of a vanillate biosensor for the vanillin biosynthesis pathway in E. coli. Acs Synth. Biol. 2019, 8, 1958–1967. [Google Scholar] [CrossRef]
- Jha, R.K.; Narayanan, N.; Pandey, N.; Bingen, J.M.; Kern, T.L.; Johnson, C.W.; Strauss, C.E.M.; Beckham, G.T.; Hennelly, S.P.; Dale, T. Sensor-enabled alleviation of product inhibition in chorismate pyruvate-lyase. Acs Synth. Biol. 2019, 8, 775–786. [Google Scholar] [CrossRef]
- Chen, J.X.; Steel, H.; Wu, Y.-H.; Wang, Y.; Xu, J.; Rampley, C.P.N.; Thompson, I.P.; Papachristodoulou, A.; Huang, W.E. Development of aspirin-inducible biosensors in Escherichia coli and SimCells. Appl. Environ. Microbiol. 2019, 85, e02959-18. [Google Scholar] [CrossRef]
- Xue, H.; Shi, H.; Yu, Z.; He, S.; Liu, S.; Hou, Y.; Pan, X.; Wang, H.; Zheng, P.; Cui, C.; et al. Design, construction, and characterization of a set of biosensors for aromatic compounds. Acs Synth. Biol. 2014, 3, 1011–1014. [Google Scholar] [CrossRef]
- Kawaguchi, H.; Katsuyama, Y.; Danyao, D.; Kahar, P.; Nakamura-Tsuruta, S.; Teramura, H.; Wakai, K.; Yoshihara, K.; Hiromichi, M.; Ogino, C.; et al. Caffeic acid production by simultaneous saccharification and fermentation of kraft pulp using recombinant Escherichia coli. Appl. Microbiol. Biotechnol. 2017, 101, 5279–5290. [Google Scholar] [CrossRef] [PubMed]
- Verhoef, S.; Ballerstedt, H.; Volkers, R.J.; de Winde, J.H.; Ruijssenaars, H.J. Comparative transcriptomics and proteomics of p-hydroxybenzoate producing Pseudomonas putida S12: Novel responses and implications for strain improvement. Appl. Microbiol. Biotechnol. 2010, 87, 679–690. [Google Scholar] [CrossRef] [PubMed]
- Wagner, S.; Klepsch, M.M.; Schlegel, S.; Appel, A.; Draheim, R.; Tarry, M.; Högbom, M.; van Wijk, K.J.; Slotboom, D.J.; Persson, J.O.; et al. Tuning Escherichia coli for membrane protein overexpression. Proc. Natl. Acad. Sci. USA 2008, 105, 14371–14376. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.; Plan, M.R.; Winter, G.; Kromer, J.O. Metabolic engineering of Pseudomonas putida KT2440 for the production of para-hydroxy benzoic acid. Front. Bioeng. Biotechnol. 2016, 4, 90. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Li, W.; Li, X.; Sun, X.; Yuan, Q. Constructing an efficient salicylate biosynthesis platform by Escherichia coli chromosome integration. J. Biotechnol. 2019, 298, 5–10. [Google Scholar] [CrossRef]
- Verhoef, S.; Gao, G.; Ruijssenaars, H.J.; de Winde, J.H. Crude glycerol as feedstock for the sustainable production of p-hydroxybenzoate by Pseudomonas putida S12. New Biotechnol. 2014, 31, 114–119. [Google Scholar] [CrossRef]
- Lütke-Eversloh, T.; Stephanopoulos, G. L-Tyrosine production by deregulated strains of Escherichia coli. Appl. Microbiol. Biotechnol. 2007, 75, 103–110. [Google Scholar] [CrossRef]
- Wang, Z.; Liu, J.; Chen, L.; Zeng, A.-P.; Solem, C.; Jensen, P.R. Alterations in the transcription factors GntR1 and RamA enhance the growth and central metabolism of Corynebacterium glutamicum. Metab. Eng. 2018, 48, 1–12. [Google Scholar] [CrossRef]
- Tinikul, R.; Chenprakhon, P.; Maenpuen, S.; Chaiyen, P. Biotransformation of plant-derived phenolic acids. Biotechnol. J. 2018, 13, e1700632. [Google Scholar] [CrossRef]
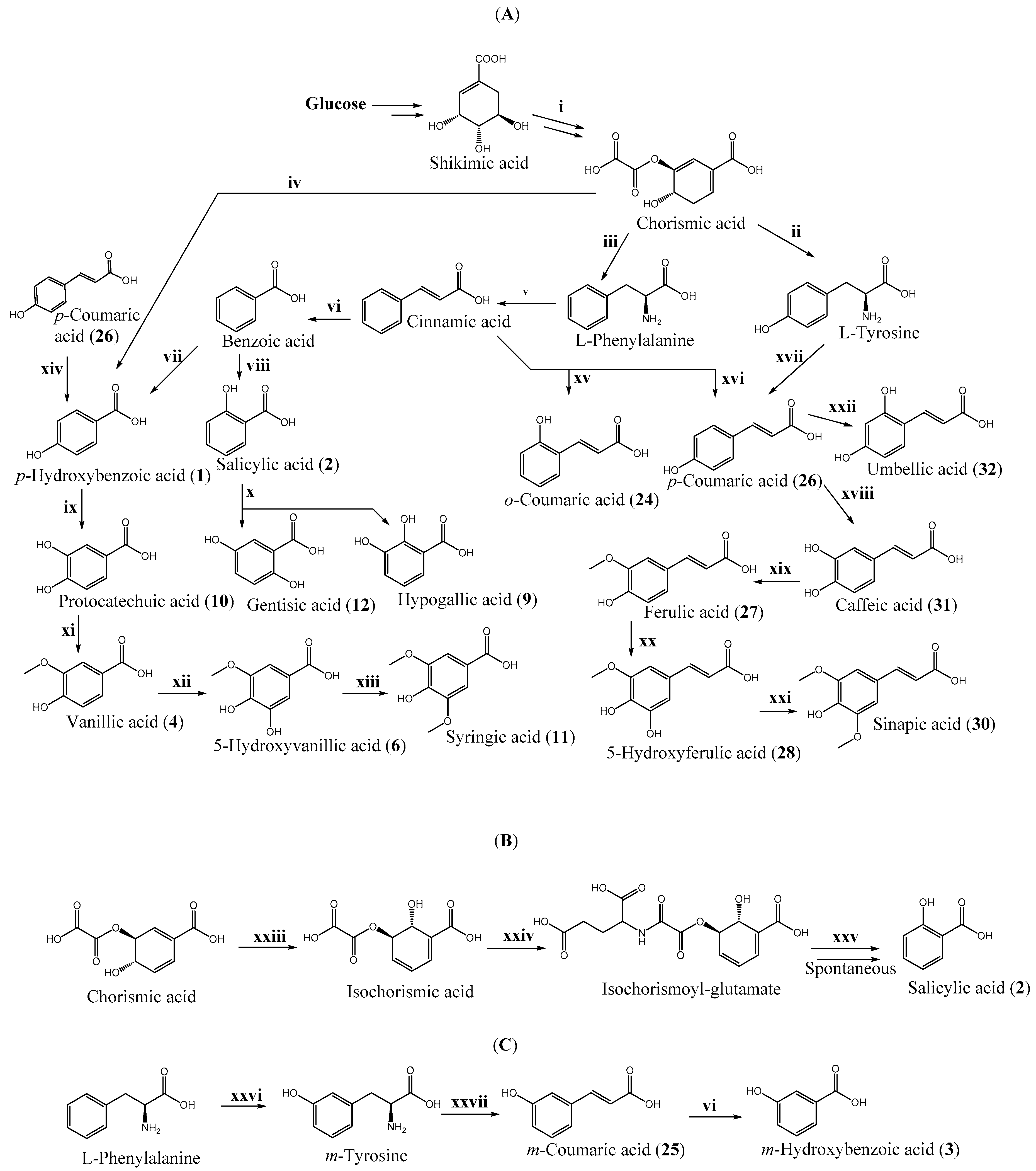
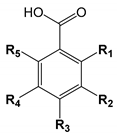 | ||||||
| Number | Hydroxybenzoic Acid | R1 | R2 | R3 | R4 | R5 |
| 1 | p-Hydroxybenzoic acid | OH | ||||
| 2 | Salicylic acid | OH | ||||
| 3 | m-Hydroxybenzoic acid | OH | ||||
| 4 | Vanillic acid | CH3O | OH | |||
| 5 | Isovanillic acid | OH | CH3O | |||
| 6 | 5-Hydroxyisovanillic acid | OH | CH3O | OH | ||
| 7 | o-Vanillic acid | OH | CH3O | |||
| 8 | Gallic acid | OH | OH | OH | ||
| 9 | Hypogallic acid | OH | OH | |||
| 10 | Protocatechuic acid | OH | OH | |||
| 11 | Syringic acid | CH3O | OH | CH3O | ||
| 12 | Gentisic acid | OH | OH | |||
| 13 | α-Resorcylic acid | OH | OH | |||
| 14 | β-Resorcylic acid | OH | OH | |||
| 15 | γ-Resorcylic acid | OH | OH | |||
| 16 | Orsellinic acid | OH | OH | CH3 | ||
| 17 | p-Orsellinic acid | OH | CH3 | OH | ||
| 18 | o,o-Dimethylorsellinic acid | CH3O | CH3O | CH3 | ||
| 19 | 3-Methylsalicylic acid | OH | CH3 | |||
| 20 | 4-Methylsalicylic acid | OH | CH3 | |||
| 21 | 6-Methylsalicylic acid | OH | CH3 | |||
| 22 | Everninic acid | OH | CH3O | CH3 | ||
| 23 | Olivetolic acid | OH | OH | C5H11 | ||
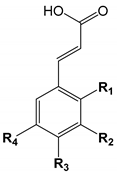 | |||||
| Number | Hydroxycinnamic Acids | R1 | R2 | R3 | R4 |
| 24 | o-Coumaric acid | OH | |||
| 25 | m-Coumaric acid | OH | |||
| 26 | p-Coumaric acid | OH | |||
| 27 | Ferulic acid | CH3O | OH | ||
| 28 | 5-Hydroxyferulic acid | CH3O | OH | OH | |
| 29 | Isoferulic acid | OH | CH3O | ||
| 30 | Sinapic acid | CH3O | OH | CH3O | |
| 31 | Caffeic acid | OH | OH | ||
| 32 | Umbellic acid | OH | OH | ||
| Phenolic Acid | Plant Type and Part | Extraction Method | Solvent | Hydrolysis Applied *a | Yield (μg/g Dry Weight) | Reference |
|---|---|---|---|---|---|---|
| p-Hydroxybenzoic acid (1) | Maya nut (Brosimum. alicastrum) | Ultrasound-assisted extraction | Methanol–acetic acid | Alkaline | 326.2 | [95] |
| Pulp from Byrsonima ligustrifolia fruits | Ultrasound-assisted extraction | Methanol, acetone | Alkaline, acidic | 1000–2820 | [96] | |
| Raspberry (Rubus idaeus L.) fruits | Percolation | Water–ethyl acetate | Acidic, alkaline | 709.62 | [32] | |
| Strawberry (Fragaria ananassa) fruits | Ultrasound-assisted extraction | Methanol with butylated hydroxyanisole −10% acetic acid | Acidic | 44–63 b | [97] | |
| m-Hydroxybenzoic acid (3) | Melon (Cucumis melo L.) peels | Maceration | Ethanol–water | - | 334.5 | [98] |
| Mangosteen (Garcinia mangostana L.) fruit peel | Maceration | Water–methanol | - | 0.07 | [99] | |
| Salicylic acid (2) | Wheat (Triticum aestivum L.) straws | Maceration | Water | - | 190.1 | [100] |
| Fresh red quinoa (Chenopodium quinoa) leaves | Accelerated solvent extraction | Methanol–water | - | 0.48 | [92] | |
| Vanillic acid (4) | Blueberry (Vaccinium myrtillus L.) leaves | Ultrasound-assisted extraction | Methanol–formic acid, acetone–formic acid | - | 1156.80 | [88] |
| Maya nut (Brosimum. alicastrum) | Sonication | Methanol–acetic acid | Alkaline | 103 | [95] | |
| Grape (Citrus paradisi) pomace | Ultrasound-assisted extraction | Ethanol–water | - | 86 | [101] | |
| Isovanillic acid (5) | Melon (C. melo L.) peels | Maceration | Ethanol–water | - | 237 | [98] |
| Gallic acid (8) | Microalgae (Ophiocordyceps sinensis) | Maceration | Methanol | - | 9489 | [102] |
| Clove (Eugenia caryophylata Thunb.) leaves | Maceration | Methanol–water | - | 7385 | [103] | |
| Black tea (Clonorchis sinensis) leaves | Percolation | Ethanol–water | 6550 | [104] | ||
| Chinese olive (Canarium album) peel | Maceration | Methanol–acetic acid–water mixture | 3696 b | [105] | ||
| Pulp from B. ligustrifolia fruits | Ultrasound-assisted extraction | Methanol, acetone, acetic acid | Acidic, alkaline | 1485-30603 | [96] | |
| Potatoes (Solanum tuberosum L.) peels | Maceration, ultrasound-assisted extraction | Ethanol–acetic acid | Acidic | 2330 | [106] | |
| Raspberry (R. idaeus L.) fruits | Maceration | Water | Acidic, alkaline | 1669 | [32] | |
| Protocatechuic acid (10) | Chia (Salvia hispanica L.) seeds | Maceration | Methanol–water | - | 759 | [107] |
| Cocao (Theobroma cacao) powder | Maceration | Methanol, acetic acid, butylated hydroxyanisole | Acidic, alkaline | 400 | [97] | |
| Red onion (Allium cepa L.) outer layer | Maceration | Methanol–water | - | 354 | [108] | |
| Araticum (Annona crassiflora Mart.) fruit peels | Ultrasound-assisted extraction | Methanol–acetone–water | - | 318 | [109] | |
| Hyssop (Hyssopus officinalis L.) herb | Maceration | Methanol | - | 310 | [110] | |
| Star anise (Illicium verum) fruits | Maceration | Ethanol | - | 209.7 | [111] | |
| Syringic acid (11) | Blueberry (V. myrtillus L.) fruits | Ultrasound-assisted extraction | Methanol–formic acid, acetone–formic acid | Acidic | 5627.47 | [88] |
| Cashew (Anacardium occidentale L.) nut testa (defatted) | Maceration | Ethanol–water | Acidic, alkaline | 2507 | [112] | |
| Lemon balm (Melissa officinalis L.) plants | Percolation | Methanol–water | - | 540.8 | [113] | |
| Raspberry (R. idaeus L.) fruits | Percolation | Water–ethyl acetate | Acidic, alkaline | 113.41 | [32] | |
| Gentisic acid (12) | Lavender (Lavandula officinalis L.) flowers | Maceration | Methanol | - | 8600 | [110] |
| Bitter melon (Momordica charantia L.) fruits | Accelerated solvent extraction | Water | - | 5910 | [114] | |
| Strawberry (Fragaria ananassa L.) fruits | Percolation | Water–ethyl acetate | Acidic, alkaline | 120 | [32] | |
| 4-Methylsalicylic acid (20) | Blueberry (V. myrtillus L.) fruits | Maceration | Sodium carbonate solution | Acidic | 24 | [115] |
| Beans (Vicia faba L.) | Maceration | Sodium carbonate solution | Acidic | 0.92 | [115] | |
| 3-Methylsalicylic acid (19) | Beans (V. faba L.) | Maceration | Sodium carbonate solution | Acidic | 4.37 | [115] |
| Blueberry (V. myrtillus L.) fruits | Maceration | Sodium carbonate solution | Acidic | 0.8 | [115] | |
| p-Coumaric acid (26) | Chokeberry (Aronia melonocarpa L.) fruits | Ultrasound-assisted extraction | Acidic water–ethanol | - | 4020 b | [116] |
| Pulp from B. ligustrifolia fruits | Ultrasound-assisted extraction | Methanol, acetone, acetic acid | Alkaline, acidic | 2898–5265 | [96] | |
| Leaves of walnut tree (Juglans regia L.) | Ultrasound-assisted extraction | Methanol | - | 1250 | [117] | |
| Strawberry (Fragaria ananassa L.) fruits | Maceration | Water | Acidic, alkaline | 1108 | [32] | |
| o-Coumaric acid (24) | Leaves of rubber vine (Conradina grandiflora) | Maceration | Methanol | - | 45.55 | [118] |
| Leaves of moringa (Moringa oleifera L.) | Maceration | Methanol, ethanol, ethyl acetate, water, and acetone | - | 37 | [119] | |
| Barley (Hordeum vulgare L.) straws | Maceration | Water | - | 1.8 | [100] | |
| m- Coumaric acid (25) | Rice (Labelle) hull | Maceration | Methanol–water | - | 432 | [120] |
| Melon (C. melo L.) peels | Maceration | Ethanol–water | - | 199.1 | [98] | |
| Barley (Hordeum vulgare L.) straws | Maceration | Water | - | 3.1 | [100] | |
| Leaves of rubber vine (C. grandiflora) | Maceration | Methanol | - | 1.02 | [118] | |
| Caffeic acid (31) | Roasted coffee beans c | Maceration | Hot water | - | 17,400 | [97] |
| Leaves of tansy (Tanacetum vulgare L.) | Ultrasound-assisted extraction | Methanol | - | 8940 | [117] | |
| Potatoes (S. tuberosum L.) peels | Maceration, ultrasound-assisted extraction | Ethanol–acetic acid | Acidic | 3320 | [106] | |
| Basil (Ocimum basilicum L.)herb | Maceration | Methanol | - | 2600 | [110] | |
| Mate (Ilex paraguariensis St. Hil.) leaves | Percolation | Ethanol–water | - | 760 | [104] | |
| Blackcurrant (Ribes nigrum L.) fruits | Maceration | Water | Acidic, alkaline | 537 | [32] | |
| Ferulic acid (27) | Wheat (T. aestivum L.) aleurone | Maceration | Sodium hydroxide solution in water | Alkaline | >7000 | [121] |
| Wheat (T. aestivum L.) bran | Maceration | Methanol–water | - | 2020 | [122] | |
| Mate (I. paraguariensis St.Hil.) leaves | Percolation | Ethanol–water | - | 1360 | [104] | |
| Hyssop (Hyssopus officinalis L.) herbs | Maceration | Methanol | - | 460 | [110] | |
| Baru (Dipteryx alata Vog.) nuts | Maceration | Methanol–hydrochloric acid solution | - | 454 | [123] | |
| White onion (A. cepa L.) outer layer | Maceration | Methanol–water | - | 116 | [108] | |
| Sinapic acid (30) | Mate (I. paraguariensis) leaves | Percolation | Ethanol–water | - | 1870 | [104] |
| Defatted canola (Brassica napus L.) seeds | Ultrasound-assisted extraction | Methanol–water | - | 590 b | [94] | |
| Strawberry (F. ananassa L.) fruits | Maceration | Water | Acidic | 445 | [32] |
| Phenolic Acid | Mushroom | Extraction Method | Solvent | Yield (μg/g DW) | Reference |
|---|---|---|---|---|---|
| Protocatechuic acid (10) | Ramaria botrytis | Maceration | Methanol | 342.7 | [156] |
| Morchella esculent | Ultrasound-assisted extraction | Methanol | 17.15 | [41] | |
| Ganoderma lucidum | Ultrasound-assisted extraction | Methanol | 3.01 | [41] | |
| p-Hydroxybenzoic acid (1) | Agaricus brasiliensis | Maceration | Ethanol–water | 332.76 | [34] |
| Agaricus silvicola | Maceration | Methanol | 238.7 | [156] | |
| R. botrytis | Maceration | Methanol | 14.00 | [156] | |
| G. lucidum | Ultrasound-assisted extraction | Methanol | 5.22 | [41] | |
| Gallic acid (8) | A. brasiliensis | Maceration | Ethanol–water | 491.89 | [34] |
| M. esculent | Ultrasound-assisted extraction | Methanol | 0.7818 | [41] | |
| Rugiboletus extremiorientalis | Ultrasound-assisted extraction | Water | 0.03654 | [38] | |
| p-Coumaric acid (26) | A. silvicola | Maceration | Methanol | 45.72 | [156] |
| A. brasiliensis | Maceration | Ethanol–water | 24.47 | [34] | |
| Agaricus bisporus (white) | Ultrasound-assisted extraction | Methanol–water mixture | 2.31 | [162] | |
| G. lucidum | Ultrasound-assisted extraction | Methanol | 1.39 | [41] | |
| Ferulic acid (27) | A. brasiliensis | Maceration | Ethanol–water | 752.54 | [34] |
| M. esculent | Ultrasound-assisted extraction | Methanol | 0.075 | [41] | |
| R. extremiorientalis | Ultrasound-assisted extraction | Water | 0.001 | [38] | |
| Vanillic acid (4) | G. lucidum | Ultrasound-assisted extraction | Methanol | 15.96 | [41] |
| R. extremiorientalis | Ultrasound-assisted extraction | Water | 0.0113 | [38] | |
| Syringic acid (11) | G. lucidum | Ultrasound-assisted extraction | Methanol | 2.34 | [41] |
| R. extremiorientalis | Ultrasound-assisted extraction | Water | 0.0016 | [38] | |
| Sinapic acid (30) | R. extremiorientalis | Ultrasound-assisted extraction | Water | 0.0022 | [38] |
| Gentisic acid (12) | A. brasiliensis | Maceration | Ethanol–water | 27.73 | [34] |
| Product | Initial Concentration | Final Concentration or Yield | Raw Material or Substrate | Fungi/Bacteria | Reference |
|---|---|---|---|---|---|
| p-Hydroxybenzoic acid (1) | 3.48 ± 0.10 μg/g | 21.80 ± 1.5 μg/g | Hemicelluloses from kidney bean extract | B. subtilis | [177] |
| 6.2 ± 1.8 mg/gdry weight | 22.3 mg/gdry weigt | Lignin from rice bran | R. oryzae | [178] | |
| Salicylic acid (2) | 0 g/L | ≈15 g/L | Naphtalene (2%) | P. aeruginosa | [168] |
| 0 mg/L | 27.3 mg/L (0.13%) | Sucrose (80 mM/L) from RM2 medium | R. gelatinosus RASN4 | [169] | |
| Gallic acid (8) | 0 g/g of biomass accumulated | 7.35 g/g of biomass accumulated | Tannic acid | A. fischeri MTCC 150 | [175] |
| 0% | 94.8% | Tannic acid | R. oryzae (RO IIT RB-13, NRRL 21498) and A. foetidus (GMRB013 MTCC 3557) | [179] | |
| n.d. | 154.5 mg/gdry weight | Lignin from rice bran | R. oryzae | [178] | |
| 13.2 μg/mL | 160 μg/mL | Palm oil mill effluent * | A. niger IBS-103ZA | [180] | |
| Orsellinic acid (16) | 0 mg/g | 33 mg/g | Dextrose from Sabouraud medium | P. polonicum C3 | [174] |
| Protocatechuic acid (10) | 8.7 ± 1.2 mg/gdry weight | 13.6 mg/gdry weight | Lignin from rice bran | R. oryzae | [178] |
| Vanillic acid (4) | 0 mg/L | 250 mg/L | Ferulic acid, 4 mmol/L | S. thermophile | [173] |
| 0 mg/L | 365 mg/L (36.5%) | Ferulic acid, 1 g/L | H. elognata | [171] | |
| 0% | 57.3% | Ferulic acid | P. lactis SAMS-2001 | [170] | |
| Syringic acid (11) | 2.6 ± 0.6 mg/gdry weight | 12.7 mg/gdry weight | Lignin from rice bran | R. oryzae | [178] |
| 0 mg/L | 85 mg/L | Sinapic acid (5 mM) solution in minimal medium | P. variotii | [167] | |
| p-Coumaric acid (26) | 71.8 μg/mL | 146 μg/mL | Palm oil mill effluent * | A. niger IBS-103ZA | [180] |
| Caffeic acid (31) | 1.6 ± 0.2 mg/gdry weight | 28.7 mg/gdry weight | Lignin from rice bran | R. oryzae | [178] |
| 286 μg/mL | 340 μg/mL | Palm oil mill effluent * | A. niger IBS-103ZA | [180] | |
| Ferulic acid (27) | 159 μg/mL | 225 μg/mL | Palm oil mill effluent * | A. niger IBS-103ZA | [180] |
| 10.56 ± 2.46 | 69.98 ± 13.75 μg/g | Hemicelluloses from kidney bean extract | B. subtilis | [177] | |
| 0% | 85% of alcaline extracted compounds | Pectin in sugar beet pulp | P. chrysogenum 31B | [181] | |
| Hypogallic acid (9) | 0 mM | 2.9 mM (50.4% yield) | m-Hydroxybenzoate | P. testosteroni | [182] |
| Target Molecule of Phenolic Acid | Sensing Element a | Output Element b | Dynamic Range (At the Concentration of Analyte) | Reference |
|---|---|---|---|---|
| Protocatechuic acid (10) | PcaU | Engineered PpcaU | 14 (20 mM) | [287] |
| PcaUAM | Engineered PpcaU, P3B5B | 1.5; 1.8 (1 mM) | [285] | |
| PcaV | Engineered PPv | 3 (1 mM) | [288] | |
| PcaU | Engineered PpcaU | ≈12 (0.003mM) | [289] | |
| Engineered PobR | PpobR | 64 (10 mM) | [290] | |
| Vanillic acid (4) | VanR | PTEF1 | ≈8 (4 mM) | [279] |
| EmrR | PEmrR | 1 (50 μM) | [291] | |
| Engineered EmrR | Engineered PEmrR, | 9.5 (5 mM) | [292] | |
| Pvtac, | 6.8 (5 mM) | |||
| Pvtrc and Pvtic | 2.1 (5 mM) | |||
| VanR-VanO | Engineered PVanAB | 14 (1mM) | [293] | |
| VanRam | PVanCC | 2.3 (100 μM) | [285] | |
| p-Hydroxybenzoic acid (1) | EmrR | PEmrR | 0.5 (1 mM) | [291] |
| PobR variant | Ppob | 64 (1 mM) | [290] | |
| PobR variant | Ppob | ≈12 (30 mM) | [294] | |
| PcaV | Engineered PPv | 3.6 (1 mM) | [288] | |
| m-Hydroxybenzoic acid (3) | PcaV | Engineered PPv | 2.8 (1 mM) | [288] |
| Salicylic acid (2) | AraC-TAL | PBAD | 218 (5 mM) | [283] |
| NahRAM | PsalTCC | 2.1 (100 μM) | [285] | |
| Engineered AraC | PBAD | ≈200 (5 mM) | [284] | |
| SalR | Psal | 10 uM | [295] | |
| 4-Methylsalicylic acid (20) | NahR, NahF-R | PJ23114 | ≈2 (1 mM) | [296] |
| 3-Methylsalicylic acid (19) | XylS | Pm | ≈2 (1 mM) | [296] |
| NahF-R | PJ23114 | ≈2 (1 mM) | ||
| Hypogallic acid (9) | NahR, NahF-R | PJ23114 | ≈2 (1 mM) | [296] |
| p-Coumaric acid (26) | EmrR | Engineered PEmrR: Pvtac, Pvtrc | 10.4 and 8.5 (1 mM) | [292] |
| AraC-TAL | PBAD | 2.3 (5 mM) | [283] | |
| FerC | Engineered PLC | 25 (1 mM) | [286] | |
| PadR | PpadC | ≈130 (2 mM) | [282] | |
| Ferulic acid (27) | FerC | Engineered PLC | 26.2 (1 mM) | [286] |
| Caffeic acid (31) | FerC | Engineered PLC | 11.2 (1 mM) | [286] |
| Sinapic acid (30) | FerC | Engineered PLC | 15.4 (1 mM) | [286] |
| Umbellic acid (32) | FerC | Engineered PLC | 9.6 (1 mM) | [286] |
| 5-Hydroxyferulic acid (28) | FerC | Engineered PLC | 14.8 (1 mM) | [286] |
| Isoferullic acid (29) | FerC | Engineered PLC | 33.5 (1 mM) | [286] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Valanciene, E.; Jonuskiene, I.; Syrpas, M.; Augustiniene, E.; Matulis, P.; Simonavicius, A.; Malys, N. Advances and Prospects of Phenolic Acids Production, Biorefinery and Analysis. Biomolecules 2020, 10, 874. https://doi.org/10.3390/biom10060874
Valanciene E, Jonuskiene I, Syrpas M, Augustiniene E, Matulis P, Simonavicius A, Malys N. Advances and Prospects of Phenolic Acids Production, Biorefinery and Analysis. Biomolecules. 2020; 10(6):874. https://doi.org/10.3390/biom10060874
Chicago/Turabian StyleValanciene, Egle, Ilona Jonuskiene, Michail Syrpas, Ernesta Augustiniene, Paulius Matulis, Andrius Simonavicius, and Naglis Malys. 2020. "Advances and Prospects of Phenolic Acids Production, Biorefinery and Analysis" Biomolecules 10, no. 6: 874. https://doi.org/10.3390/biom10060874
APA StyleValanciene, E., Jonuskiene, I., Syrpas, M., Augustiniene, E., Matulis, P., Simonavicius, A., & Malys, N. (2020). Advances and Prospects of Phenolic Acids Production, Biorefinery and Analysis. Biomolecules, 10(6), 874. https://doi.org/10.3390/biom10060874





