Comparative Characterization of Plasmodium falciparum Hsp70-1 Relative to E. coli DnaK Reveals the Functional Specificity of the Parasite Chaperone
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
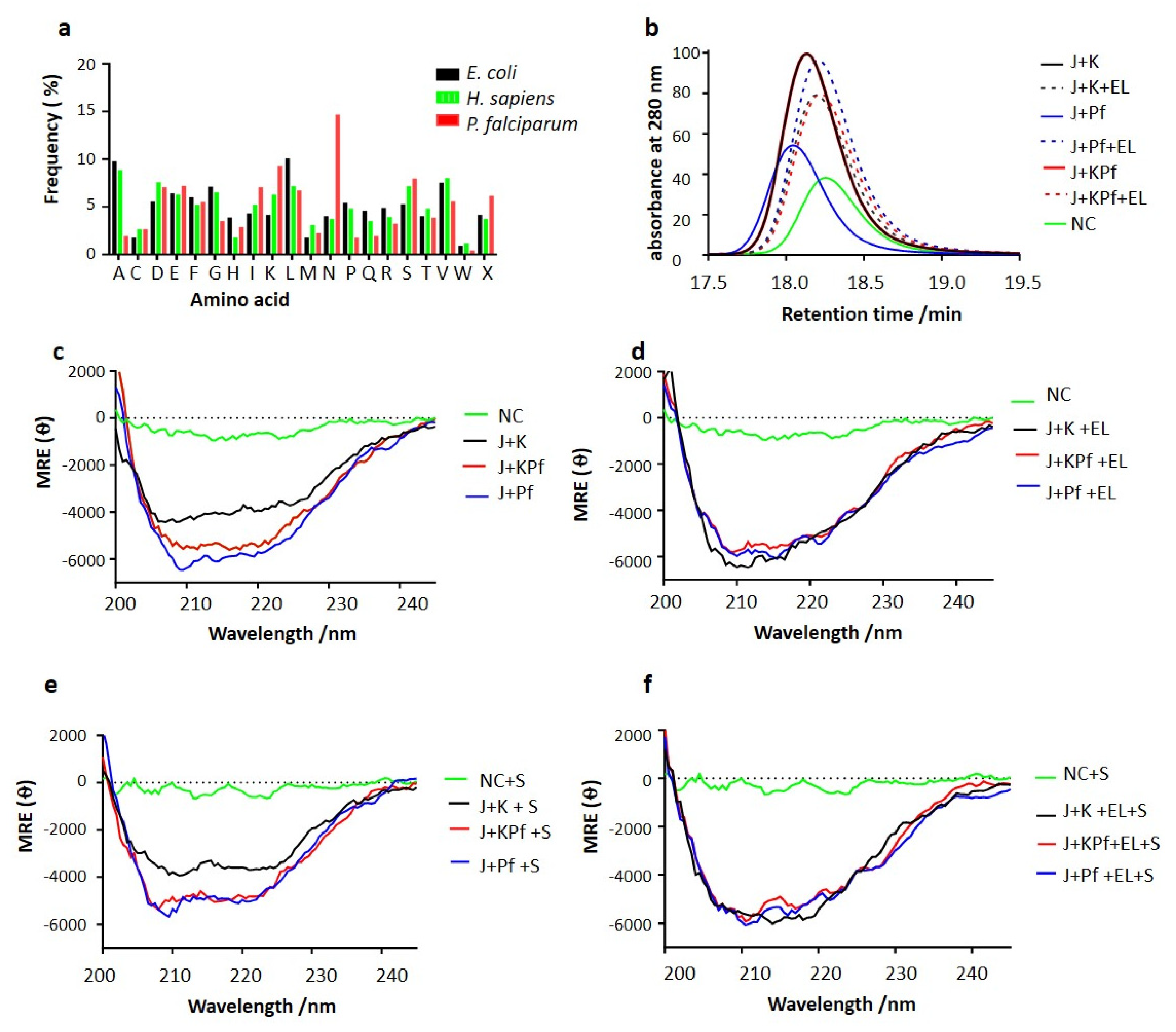
2.2. Comparative Analysis of Amino Acid Composition of AdoMetDC/ODC and Three-Dimensional Modelling of Hsp70s
2.3. Expression and Purification of Recombinant Molecular Chaperones
2.4. Co-expression of PfAdoMetDC with Supplementary Molecular Chaperones
2.5. Analysis of the Secondary Structures of the Recombinant Proteins
2.6. Fluorescence-Based Analysis of the Tertiary Structural Organization of the Recombinant Proteins
2.7. Evaluation of ATPase Activities of DnaK, PfHsp70-1 and KPf
2.8. Determination of the Nucleotide Binding Affinities of PfHsp70-1, DnaK and KPf
2.9. Investigation of Self-Association of Hsp70 Proteins
2.10. Investigation of Interaction of PfHsp40 with DnaK, PfHsp70-1 and KPf
2.11. Interaction of DnaK, PfHsp70-1 and KPf with Model Peptide Substrates
3. Results
3.1. Secondary and Tertiary Structural Analysis of DnaK, KPf and PfHsp70-1
3.2. PfHsp40 Stimulates the ATPase Activities of All Three Hsp70s
3.3. KPf Exhibits Higher Affinity for ATP than either DnaK or PfHsp70-1
3.4. Comparative Self-Association Capabilities
3.5. All Three Hsp70s Directly Interacted with PfHsp40
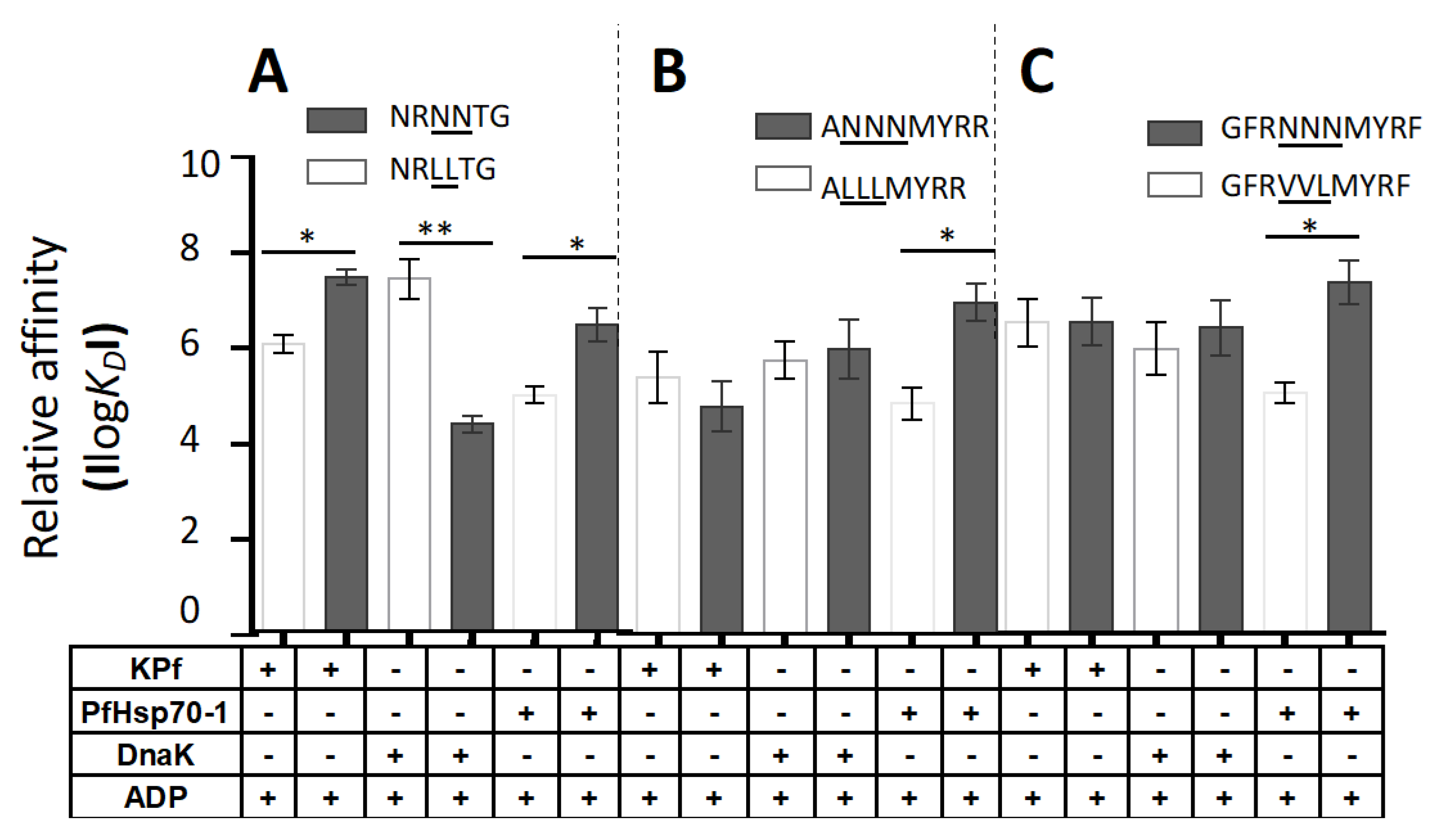
3.6. PfHsp70-1 Preferentially Bound to Asparagine-Enriched Peptide Substrates
3.7. SEC Analysis of Recombinant PfAdoMetDC Protein Co-Produced with Supplementary Molecular Chaperones
3.8. PfAdoMetDC Co-Produced with PfHsp70-1 and KPf Exhibits Unique Secondary Structural Features
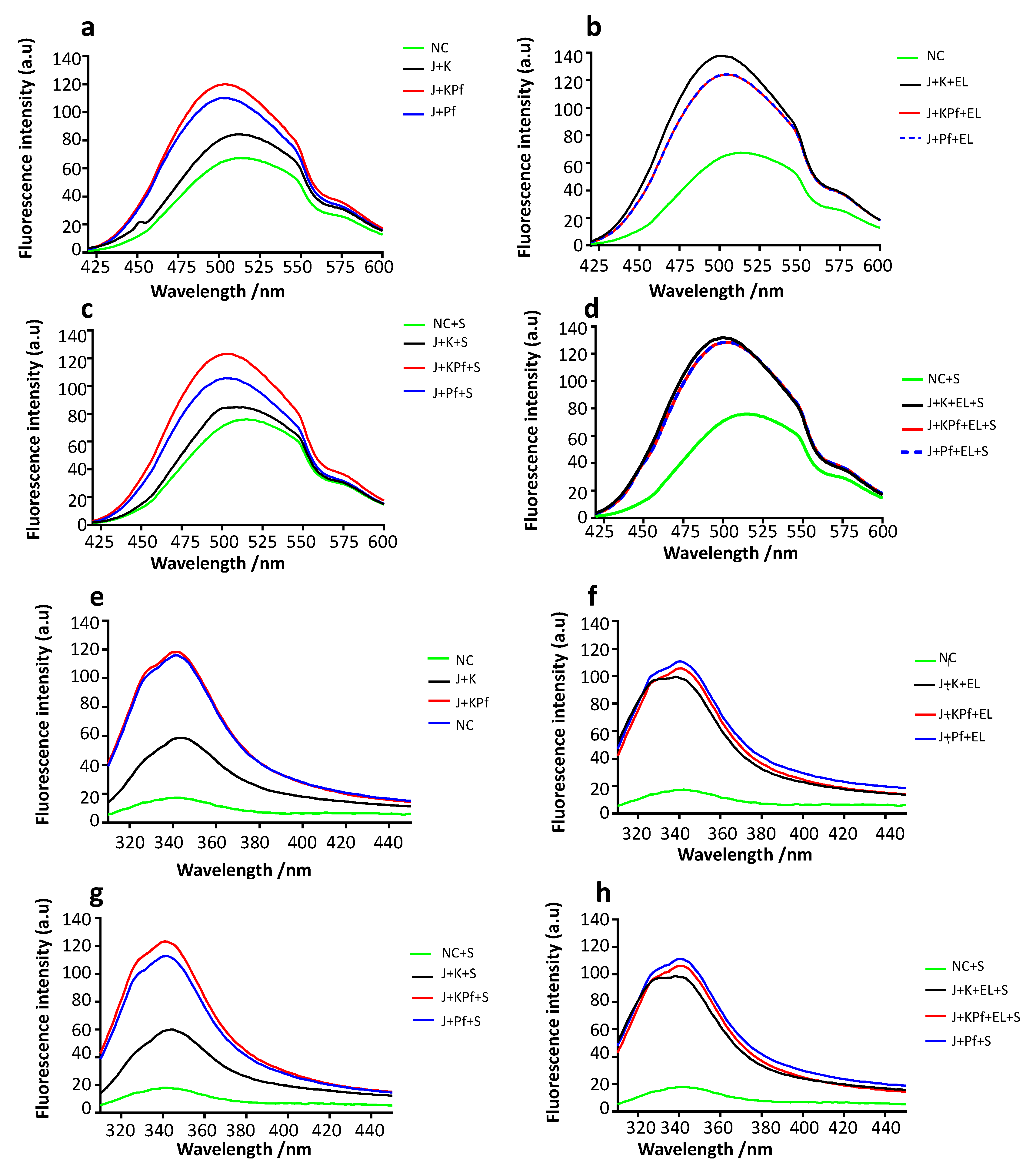
3.9. Confirmation of PfAdoMetDC Fold Using ANS Fluorescence-Based Assay
3.10. Analysis of Tertiary Structure of PfAdoMetDC using Intrinsic Tyrosine and Tryptophan Fluorescence
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Shonhai, A.; Boshoff, A.; Blatch, G.L. The structural and functional diversity of Hsp70 proteins fromPlasmodium falciparum. Protein Sci. 2007, 16, 1803–1818. [Google Scholar] [CrossRef]
- Chiang, A.N.; Valderramos, J.-C.; Balachandran, R.; Chovatiya, R.; Mead, B.P.; Schneider, C.; Bell, S.L.; Klein, M.G.; Huryn, D.M.; Chen, X.S.; et al. Select pyrimidinones inhibit the propagation of the malarial parasite, Plasmodium falciparum. Bioorganic Med. Chem. 2009, 17, 1527–1533. [Google Scholar] [CrossRef]
- Shonhai, A. Plasmodial heat shock proteins: Targets for chemotherapy. FEMS Immunol. Med. Microbiol. 2010, 58, 61–74. [Google Scholar] [CrossRef]
- Cockburn, I.L.; Pesce, E.-R.; Pryzborski, J.M.; Davies-Coleman, M.; Clark, P.G.; Keyzers, R.A.; Stephens, L.L.; Blatch, G.L. Screening for small molecule modulators of Hsp70 chaperone activity using protein aggregation suppression assays: Inhibition of the plasmodial chaperone PfHsp70-1. Boil. Chem. 2011, 392, 431–438. [Google Scholar] [CrossRef]
- Daniyan, M.; Przyborski, J.M.; Shonhai, A. Partners in Mischief: Functional Networks of Heat Shock Proteins of Plasmodium falciparum and Their Influence on Parasite Virulence. Biomolecules 2019, 9, 295. [Google Scholar] [CrossRef]
- Mayer, M.P.; Bukau, B. Hsp70 chaperones: Cellular functions and molecular mechanism. Cell. Mol. Life Sci. 2005, 62, 670–684. [Google Scholar] [CrossRef]
- Zhu, X.; Zhao, X.; Burkholder, W.F.; Gragerov, A.; Ogata, C.M.; Gottesman, M.E.; Hendrickson, W.A. Structural Analysis of Substrate Binding by the Molecular Chaperone DnaK. Science 1996, 272, 1606–1614. [Google Scholar] [CrossRef]
- Gragerov, A.; Zeng, L.; Zhao, X.; Burkholder, W.; Gottesman, M.E. Specificity of DnaK-peptide Binding. J. Mol. Boil. 1994, 235, 848–854. [Google Scholar] [CrossRef]
- Liberek, K.; Marszalek, J.; Ang, D.; Georgopoulos, C.; Zylicz, M. Escherichia coli DnaJ and GrpE heat shock proteins jointly stimulate ATPase activity of DnaK. Proc. Natl. Acad. Sci. USA 1991, 88, 2874–2878. [Google Scholar] [CrossRef]
- Rüdiger, S.G.; Schneider--Mergener, J.; Bukau, B. Its substrate specificity characterizes the DnaJ co-chaperone as a scanning factor for the DnaK chaperone. EMBO J. 2001, 20, 1042–1050. [Google Scholar] [CrossRef]
- Mayer, M.P. Hsp70 chaperone dynamics and molecular mechanism. Trends Biochem. Sci. 2013, 38, 507–514. [Google Scholar] [CrossRef]
- Bertelsen, E.B.; Chang, L.; Gestwicki, J.E.; Zuiderweg, E.R. Solution conformation of wild-type E. coli Hsp70 (DnaK) chaperone complexed with ADP and substrate. Proc. Natl. Acad. Sci. USA 2009, 106, 8471–8476. [Google Scholar] [CrossRef]
- Bukau, B.; Walker, G.C. Cellular defects caused by deletion of the Escherichia coli dnaK gene indicate roles for heat shock protein in normal metabolism. J. Bacteriol. 1989, 171, 2337–2346. [Google Scholar] [CrossRef] [PubMed]
- Bell, S.L.; Chiang, A.N.; Brodsky, J.L. Expression of a Malarial Hsp70 Improves Defects in Chaperone-Dependent Activities in ssa1 Mutant Yeast. PLoS ONE 2011, 6, e20047. [Google Scholar] [CrossRef]
- Shonhai, A.; Maier, A.G.; Przyborski, J.M.; Blatch, G.L. Intracellular protozoan parasites of humans: The role of molecular chaperones in development and pathogenesis. Protein Pept. Lett. 2011, 18, 143–157. [Google Scholar] [CrossRef] [PubMed]
- Chakafana, G.; Zininga, T.; Shonhai, A. Comparative structure-function features of Hsp70s of Plasmodium falciparum and human origins. Biophys. Rev. 2019, 11, 591–602. [Google Scholar] [CrossRef] [PubMed]
- Botha, M.; Chiang, A.N.; Needham, P.G.; Stephens, L.L.; Hoppe, H.C.; Külzer, S.; Przyborski, J.M.; Lingelbach, K.; Wipf, P.; Brodsky, J.L.; et al. Plasmodium falciparum encodes a single cytosolic type I Hsp40 that functionally interacts with Hsp70 and is upregulated by heat shock. Cell Stress Chaperones 2010, 16, 389–401. [Google Scholar] [CrossRef] [PubMed]
- Qiu, X.-B.; Shao, Y.-M.; Miao, S.; Wang, L. The diversity of the DnaJ/Hsp40 family, the crucial partners for Hsp70 chaperones. Cell. Mol. Life Sci. 2006, 63, 2560–2570. [Google Scholar] [CrossRef] [PubMed]
- Nicoll, W.; Botha, M.; McNamara, C.; Schlange, M.; Pesce, E.-R.; Boshoff, A.; Ludewig, M.; Zimmermann, R.; Cheetham, M.E.; Chapple, J.; et al. Cytosolic and ER J-domains of mammalian and parasitic origin can functionally interact with DnaK. Int. J. Biochem. Cell Boil. 2006, 39, 736–751. [Google Scholar] [CrossRef]
- Kampinga, H.H.; Craig, E. The HSP70 chaperone machinery: J proteins as drivers of functional specificity. Nat. Rev. Mol. Cell Boil. 2010, 11, 579–592. [Google Scholar] [CrossRef]
- Pallarès, I.; De Groot, N.S.; Iglesias, V.; Sant’Anna, R.; Biosca, A.; Fernàndez-Busquets, X.; Ventura, S. Discovering Putative Prion-Like Proteins in Plasmodium falciparum: A Computational and Experimental Analysis. Front. Microbiol. 2018, 9, 1737. [Google Scholar] [CrossRef] [PubMed]
- Singh, G.; Chandra, B.R.; Bhattacharya, A.; Akhouri, R.R.; Singh, S.K.; Sharma, A. Hyper-expansion of asparagines correlates with an abundance of proteins with prion-like domains in Plasmodium falciparum. Mol. Biochem. Parasitol. 2004, 137, 307–319. [Google Scholar] [CrossRef] [PubMed]
- Przyborski, J.M.; Diehl, M.; Blatch, G.L. Plasmodial HSP70s are functionally adapted to the malaria parasite life cycle. Front. Mol. Biosci. 2015, 2, 34. [Google Scholar] [CrossRef] [PubMed]
- Makhoba, X.H.; Burger, A.; Coertzen, D.; Zininga, T.; Birkholtz, L.-M.; Shonhai, A. Use of a Chimeric Hsp70 to Enhance the Quality of Recombinant Plasmodium falciparum S-Adenosylmethionine Decarboxylase Protein Produced in Escherichia coli. PLoS ONE 2016, 11, e0152626. [Google Scholar] [CrossRef] [PubMed]
- Mabate, B.; Zininga, T.; Ramatsui, L.; Makumire, S.; Achilonu, I.; Dirr, H.; Shonhai, A. Structural and biochemical characterization of Plasmodium falciparum Hsp70--x reveals functional versatility of its C-terminal EEVN motif. Proteins Struct. Funct. Bioinform. 2018, 86, 1189–1201. [Google Scholar] [CrossRef]
- Külzer, S.; Charnaud, S.; Dagan, T.; Riedel, J.; Mandal, P.; Pesce, E.R.; Blatch, G.L.; Crabb, B.S.; Gilson, P.R.; Przyborski, J.M. Plasmodium falciparum-encoded exported Hsp70/Hsp40 chaperone/co-chaperone complexes within the host erythrocyte. Cell. Microbiol. 2012, 14, 1784–1795. [Google Scholar] [CrossRef]
- Shonhai, A.; Boshoff, A.; Blatch, G.L. Plasmodium falciparum heat shock protein 70 is able to suppress the thermosensitivity of an Escherichia coli DnaK mutant strain. Mol. Genet. Genom. 2005, 274, 70–78. [Google Scholar] [CrossRef]
- Stephens, L.L.; Shonhai, A.; Blatch, G.L. Co-expression of the Plasmodium falciparum molecular chaperone, PfHsp70, improves the heterologous production of the antimalarial drug target GTP cyclohydrolase I, PfGCHI. Protein Expr. Purif. 2011, 77, 159–165. [Google Scholar] [CrossRef]
- Müller, S.; Coombs, G.H.; Walter, R.D. Targeting polyamines of parasitic protozoa in chemotherapy. Trends Parasitol. 2001, 17, 242–249. [Google Scholar] [CrossRef]
- Saibil, H.R.; Fenton, W.A.; Clare, D.K.; Horwich, A.L. Structure and Allostery of the Chaperonin GroEL. J. Mol. Boil. 2013, 425, 1476–1487. [Google Scholar] [CrossRef]
- Brinker, A.; Pfeifer, G.; Kerner, M.J.; Naylor, D.J.; Hartl, F.U.; Hayer-Hartl, M. Dual Function of Protein Confinement in Chaperonin-Assisted Protein Folding. Cell 2001, 107, 223–233. [Google Scholar] [CrossRef]
- Shonhai, A.; Botha, M.; De Beer, T.; Boshoff, A.; Blatch, G.L. Structure-Function Study of a Plasmodium falciparum Hsp70 Using Three Dimensional Modelling and in Vitro Analyses. Protein Pept. Lett. 2008, 15, 1117–1125. [Google Scholar] [CrossRef]
- Zininga, T.; Ramatsui, L.; Makhado, P.B.; Makumire, S.; Achilonu, I.; Hoppe, H.C.; Dirr, H.; Shonhai, A. (−)-Epigallocatechin-3-Gallate Inhibits the Chaperone Activity of Plasmodium falciparum Hsp70 Chaperones and Abrogates Their Association with Functional Partners. Molecules 2017, 22, 2139. [Google Scholar] [CrossRef] [PubMed]
- Williams, M.; Sprenger, J.; Human, E.; Al-Karadaghi, S.; Persson, L.; Louw, A.; Birkholtz, L.-M. Biochemical characterisation and novel classification of monofunctional S-adenosylmethionine decarboxylase of Plasmodium falciparum. Mol. Biochem. Parasitol. 2011, 180, 17–26. [Google Scholar] [CrossRef]
- Zininga, T.; Achilonu, I.; Hoppe, H.C.; Prinsloo, E.; Dirr, H.; Shonhai, A. Plasmodium falciparum Hsp70-z, an Hsp110 homologue, exhibits independent chaperone activity and interacts with Hsp70-1 in a nucleotide-dependent fashion. Cell Stress Chaperones 2016, 21, 499–513. [Google Scholar] [CrossRef] [PubMed]
- Zininga, T.; Achilonu, I.; Hoppe, H.C.; Prinsloo, E.; Dirr, H.; Shonhai, A. Overexpression, Purification and Characterisation of the Plasmodium falciparum Hsp70-z (PfHsp70-z) Protein. PLoS ONE 2015, 10, e0129445. [Google Scholar] [CrossRef]
- Whitmore, L.; Wallace, B. DICHROWEB, an online server for protein secondary structure analyses from circular dichroism spectroscopic data. Nucleic Acids Res. 2004, 32, W668–W673. [Google Scholar] [CrossRef]
- Singh, S.P.; Agnihotri, P.; Pratap, J.V. Characterization of a Novel Putative S-Adenosylmethionine Decarboxylase-Like Protein from Leishmania donovani. PLoS ONE 2013, 8, e65912. [Google Scholar] [CrossRef] [PubMed]
- Matulis, D.; Baumann, C.G.; Bloomfield, V.A.; Lovrien, R.E. 1-Anilino-8-naphthalene sulfonate as a protein conformational tightening agent. Biopolymers 1999, 49, 451–458. [Google Scholar] [CrossRef]
- Matambo, T.; Odunuga, O.O.; Boshoff, A.; Blatch, G.L. Overproduction, purification, and characterization of the Plasmodium falciparum heat shock protein 70. Protein Expr. Purif. 2004, 33, 214–222. [Google Scholar] [CrossRef] [PubMed]
- Misra, G.; Ramachandran, R. Hsp70-1 from Plasmodium falciparum: Protein stability, domain analysis and chaperone activity. Biophys. Chem. 2009, 142, 55–64. [Google Scholar] [CrossRef]
- Gong, W.; Hu, W.; Xu, L.; Wu, H.; Wu, S.; Zhang, H.; Wang, J.; Jones, G.W.; Perrett, S. The C-terminal GGAP motif of Hsp70 mediates substrate recognition and stress response in yeast. J. Boil. Chem. 2018, 293, 17663–17675. [Google Scholar] [CrossRef]
- Yu, H.Y.; Ziegelhoffer, T.; Craig, E. Functionality of Class A and Class B J-protein co-chaperones with Hsp70. FEBS Lett. 2015, 589, 2825–2830. [Google Scholar] [CrossRef] [PubMed]
- Slepenkov, S.V.; Witt, S. Kinetic Analysis of Interdomain Coupling in a Lidless Variant of the Molecular Chaperone DnaK: DnaK’s Lid Inhibits Transition to the Low Affinity State†. Biochemistry 2002, 41, 12224–12235. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.-C.; Zhou, C.-J.; Zhou, Z.-R.; Wu, M.; Cao, C.-Y.; Hu, H.-Y. The C-terminal Helices of Heat Shock Protein 70 Are Essential for J-domain Binding and ATPase Activation. J. Boil. Chem. 2012, 287, 6044–6052. [Google Scholar] [CrossRef] [PubMed]
- Aprile, F.A.; Dhulesia, A.; Stengel, F.; Roodveldt, C.; Benesch, J.L.P.; Tortora, P.; Robinson, C.V.; Salvatella, X.; Dobson, C.M.; Cremades, N. Hsp70 Oligomerization Is Mediated by an Interaction between the Interdomain Linker and the Substrate-Binding Domain. PLoS ONE 2013, 8, e67961. [Google Scholar] [CrossRef]
- Benaroudj, N.; Fouchaq, B.; Ladjimi, M.M. The COOH-terminal Peptide Binding Domain Is Essential for Self-association of the Molecular Chaperone HSC70. J. Boil. Chem. 1997, 272, 8744–8751. [Google Scholar] [CrossRef]
- Schönfeld, H.-J.; Schmidt, D.; Schröder, H.; Bukau, B. The DnaK Chaperone System ofEscherichia coli: Quaternary Structures and Interactions of the DnaK and GrpE Components. J. Boil. Chem. 1995, 270, 2183–2189. [Google Scholar] [CrossRef]
- Sarbeng, E.B.; Liu, Q.; Tian, X.; Yang, J.; Li, H.; Wong, J.L.; Zhou, L.; Liu, Q. A Functional DnaK Dimer Is Essential for the Efficient Interaction with Hsp40 Heat Shock Protein. J. Boil. Chem. 2015, 290, 8849–8862. [Google Scholar] [CrossRef]
- Kim, J.H.; Alderson, T.R.; Frederick, R.O.; Markley, J.L. Nucleotide-dependent interactions within a specialized Hsp70/Hsp40 complex involved in Fe–S cluster biogenesis. J. Am. Chem. Soc. 2014, 136, 11586–11589. [Google Scholar] [CrossRef]
- Han, W.; Christen, P. cis -Effect of DnaJ on DnaK in ternary complexes with chimeric DnaK/DnaJ-binding peptides. FEBS Lett. 2004, 563, 146–150. [Google Scholar] [CrossRef]
- Xu, X.; Sarbeng, E.B.; Vorvis, C.; Kumar, D.P.; Zhou, L.; Liu, Q. Unique Peptide Substrate Binding Properties of 110-kDa Heat-shock Protein (Hsp110) Determine Its Distinct Chaperone Activity. J. Boil. Chem. 2011, 287, 5661–5672. [Google Scholar] [CrossRef] [PubMed]
- Muralidharan, V.; Oksman, A.; Pal, P.; Lindquist, S.; Goldberg, D.E. Plasmodium falciparum heat shock protein 110 stabilizes the asparagine repeat-rich parasite proteome during malarial fevers. Nat. Commun. 2012, 3, 1310. [Google Scholar] [CrossRef]
- Royer, C.A. Probing protein folding and conformational transitions with fluorescence. Chem. Rev. 2006, 106, 1769–1784. [Google Scholar] [CrossRef] [PubMed]
- Zininga, T.; Pooe, O.; Makhado, P.B.; Ramatsui, L.; Prinsloo, E.; Achilonu, I.; Dirr, H.; Shonhai, A. Polymyxin B inhibits the chaperone activity of Plasmodium falciparum Hsp70. Cell Stress Chaperones 2017, 22, 707–715. [Google Scholar] [CrossRef] [PubMed]
- Zininga, T.; Shonhai, A. Small Molecule Inhibitors Targeting the Heat Shock Protein System of Human Obligate Protozoan Parasites. Int. J. Mol. Sci. 2019, 20, 5930. [Google Scholar] [CrossRef]
- Kityk, R.; Kopp, J.; Sinning, I.; Mayer, M.P. Structure and Dynamics of the ATP-Bound Open Conformation of Hsp70 Chaperones. Mol. Cell 2012, 48, 863–874. [Google Scholar] [CrossRef]
- Thompson, A.D.; Bernard, S.M.; Skiniotis, G.; Gestwicki, J.E. Visualization and functional analysis of the oligomeric states of Escherichia coli heat shock protein 70 (Hsp70/DnaK). Cell Stress Chaperones 2012, 17, 313–327. [Google Scholar] [CrossRef]
- Marcion, G.; Seigneuric, R.; Chavanne, E.; Artur, Y.; Briand, L.; Hadi, T.; Gobbo, J.; Garrido, C.; Neiers, F. C-terminal amino acids are essential for human heat shock protein 70 dimerization. Cell Stress Chaperones 2014, 20, 61–72. [Google Scholar] [CrossRef]
- Chakafana, G.; Zininga, T.; Shonhai, A. The Link That Binds: The Linker of Hsp70 as a Helm of the Protein’s Function. Biomolecules 2019, 9, 543. [Google Scholar] [CrossRef]

| Ligand | Analyte | Ka (Ms−1) | Kd (1−1) | KD (M) | X2 |
|---|---|---|---|---|---|
| DnaK | DnaK + ATP | 1.38 (±0.08) × 102 | 3.83 (±0.33) × 10−4 | 4.13 (±0.30) × 10−6* | 5.15 |
| DnaK | 1.27 (±0.07) × 103 | 6.51 (±0.01) × 10−2 | 4.67 (±0.27) × 10−5 | 4.21 | |
| DnaK + ADP | 1.22 (±0.13) × 102 | 6.49 (±0.09) × 10−3 | 4.60 (±0.09) × 10−5 | 5.41 | |
| KPf | KPf + ATP | 1.54 (±0.04) × 102 | 5.44 (±0.04) × 10−3 | 3.23 (±0.03) × 10−7* | 2.30 |
| KPf | 1.32 (±0.02) × 102 | 4.17 (±0.17) × 10−5 | 5.38 (±0.08) × 10−6 | 3.17 | |
| KPf + ADP | 1.42 (±0.02) × 102 | 5.23 (±0.03) × 10−3 | 4.65 (±1.5) × 10−5 | 2.37 | |
| PfHsp70-1 | PfHsp70-1 + ATP | 2.14 (±0.04) × 104 | 1.13 (±1.2) × 10−2 | 5.28 (±0.08) × 10−7* | 4.42 |
| PfHsp70-1 | 8.51 (±1.20) × 103 | 2.04 (±0.11) × 10−3 | 2.39 (±0.09) × 10−6 | 1.20 | |
| PfHsp70-1 + ADP | 1.00 (±0.17) × 102 | 1.71 (±0.07) × 10−4 | 1.71 (±0.10) × 10−6 | 4.45 |
| Ligand | Analyte | Ka (Ms−1) | Kd (1−1) | KD (M) | X2 |
|---|---|---|---|---|---|
| PfHsp70-1 | PfHsp40 +ATP | 1.81 (±0.01) × 102 | 1.32 (±0.02) × 10−4 | 2.08 (±0.80) × 10−7 ** | 2.12 |
| PfHsp40 +ADP | 1.53 (±0.03) × 103 | 8.66 (±0.06) × 10−6 | 9.98 (±0.18) × 10−6 | 3.08 | |
| PfHsp40 | 1.33 (±0.03) × 103 | 9.81(±0.10) × 10−6 | 1.88 (±0.08) × 10−5 | 7.80 | |
| KPf | PfHsp40 + ATP | 1.72 (±0.02) × 102 | 7.44 (±0.04) × 10−3 | 7.23 (±0.30) × 10−7** | 1.75 |
| PfHsp40 + ADP | 1.36 (±0.26) × 102 | 5.43 (±0.03) × 10−4 | 2.62 (±0.02) × 10−6 | 2.15 | |
| PfHsp40 | 1.24 (±0.04) × 104 | 6.54 (±0.04) × 10−5 | 2.53 (±0.03) × 10−5 | 2.13 | |
| DnaK | PfHsp40 + ATP | 1.22 (±0.02) × 102 | 7.44 (±0.04) × 10−4 | 7.23 (±0.03) × 10−6* | 1.66 |
| PfHsp40 + ADP | 1.16 (±0.06) × 102 | 5.43 (±0.32) × 10−3 | 2.62 (±0.02) × 10−5 | 1.57 | |
| PfHsp40 | 1.12 (±0.02) × 102 | 7.44 (±0.04) × 10−3 | 7.23 (±0.03) × 10−5 | 2.07 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lebepe, C.M.; Matambanadzo, P.R.; Makhoba, X.H.; Achilonu, I.; Zininga, T.; Shonhai, A. Comparative Characterization of Plasmodium falciparum Hsp70-1 Relative to E. coli DnaK Reveals the Functional Specificity of the Parasite Chaperone. Biomolecules 2020, 10, 856. https://doi.org/10.3390/biom10060856
Lebepe CM, Matambanadzo PR, Makhoba XH, Achilonu I, Zininga T, Shonhai A. Comparative Characterization of Plasmodium falciparum Hsp70-1 Relative to E. coli DnaK Reveals the Functional Specificity of the Parasite Chaperone. Biomolecules. 2020; 10(6):856. https://doi.org/10.3390/biom10060856
Chicago/Turabian StyleLebepe, Charity Mekgwa, Pearl Rutendo Matambanadzo, Xolani Henry Makhoba, Ikechukwu Achilonu, Tawanda Zininga, and Addmore Shonhai. 2020. "Comparative Characterization of Plasmodium falciparum Hsp70-1 Relative to E. coli DnaK Reveals the Functional Specificity of the Parasite Chaperone" Biomolecules 10, no. 6: 856. https://doi.org/10.3390/biom10060856
APA StyleLebepe, C. M., Matambanadzo, P. R., Makhoba, X. H., Achilonu, I., Zininga, T., & Shonhai, A. (2020). Comparative Characterization of Plasmodium falciparum Hsp70-1 Relative to E. coli DnaK Reveals the Functional Specificity of the Parasite Chaperone. Biomolecules, 10(6), 856. https://doi.org/10.3390/biom10060856





