Saturation Mutagenesis for Phenylalanine Ammonia Lyases of Enhanced Catalytic Properties
Abstract
1. Introduction
2. Materials and Methods
2.1. Saturation Mutagenesis Procedure:
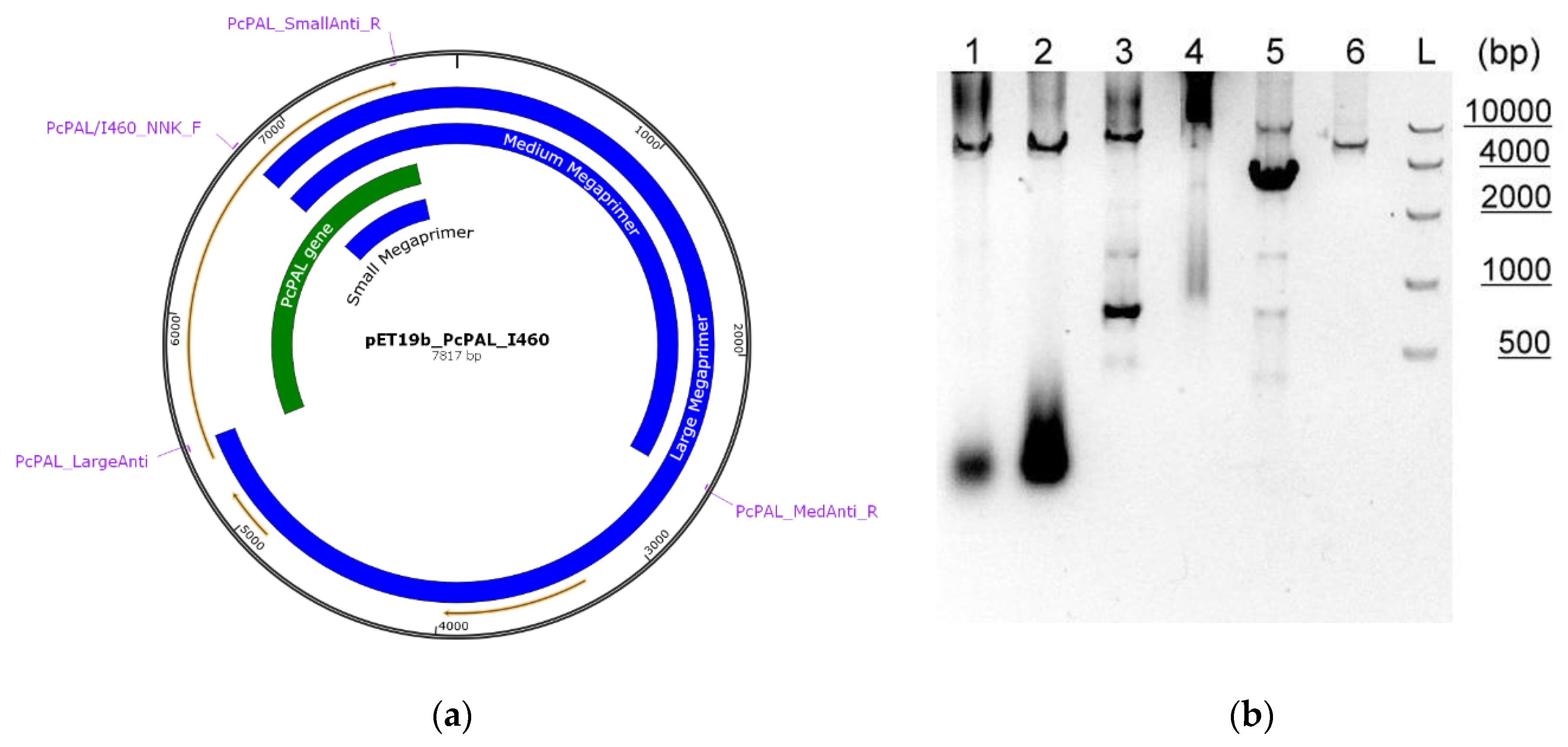
2.1.1. Procedure Using Megaprimers:
2.1.2. Procedure Using Partially Overlapping Primers:
2.2. Activity Screens of Transformant Libraries
2.3. Protein Expression, Isolation, and Thermal Unfolding Assessments
2.4. Enzyme Kinetics
2.5. HPLC Analysis for Conversion and Enantioselectivity Determinations
2.6. Analyzing Thermal Unfolding of PcPAL Mutants through nanoDSF Measurements
2.7. Molecular Docking
3. Results and Discussion
3.1. Site-Saturation Mutagenesis
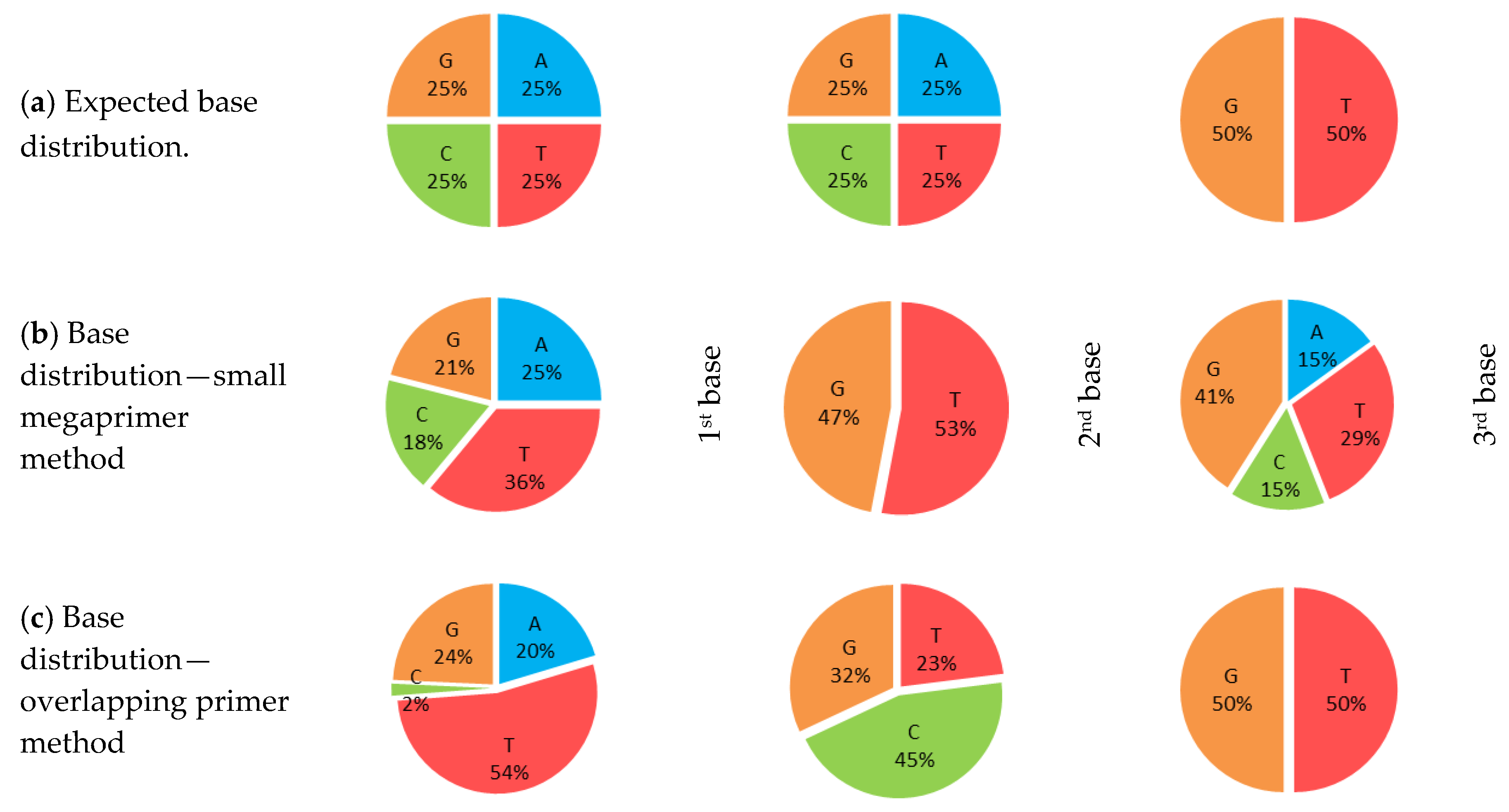
3.2. Quality of Transformant Libraries
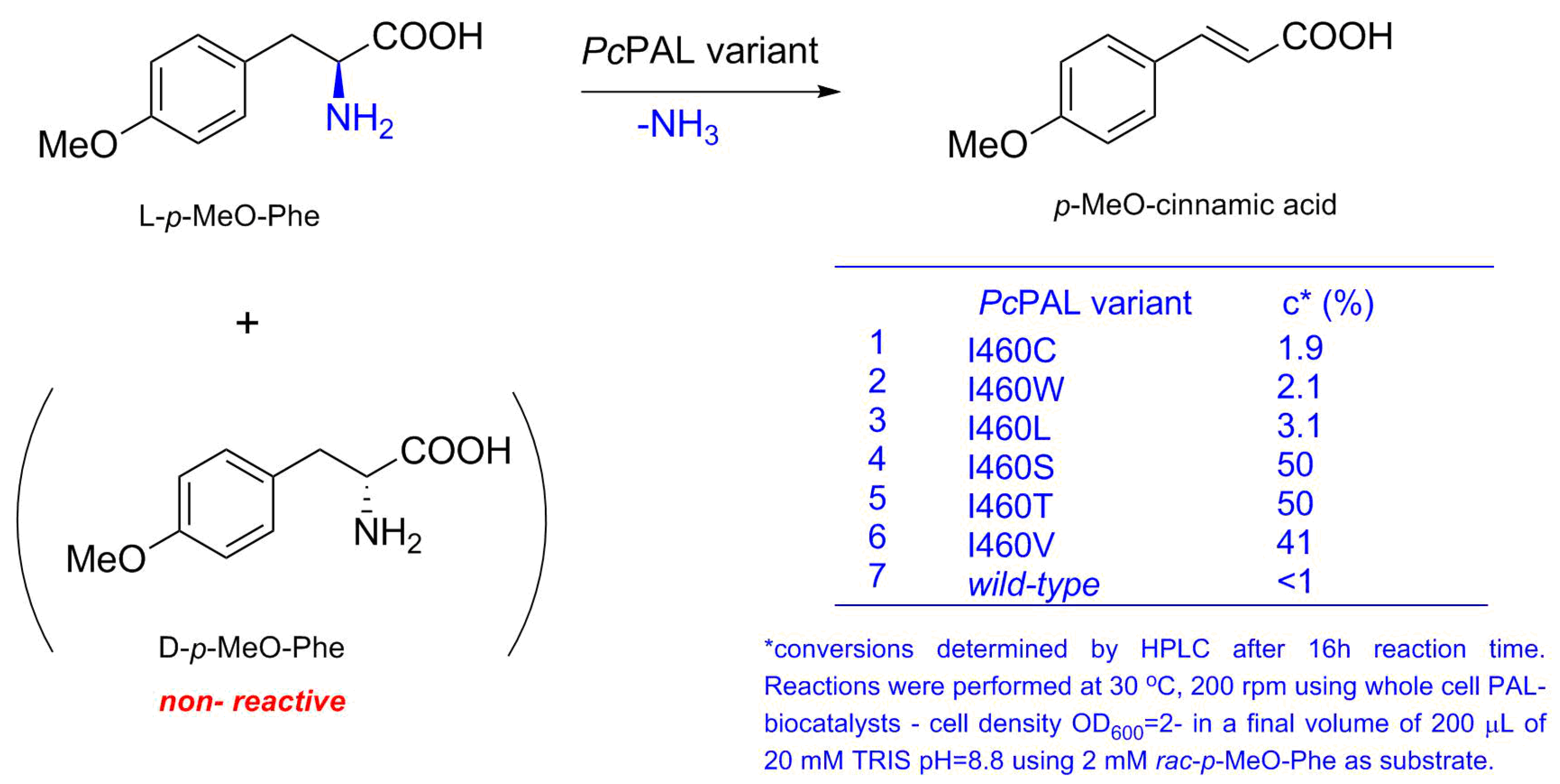
3.3. Screening of Transformant Libraries
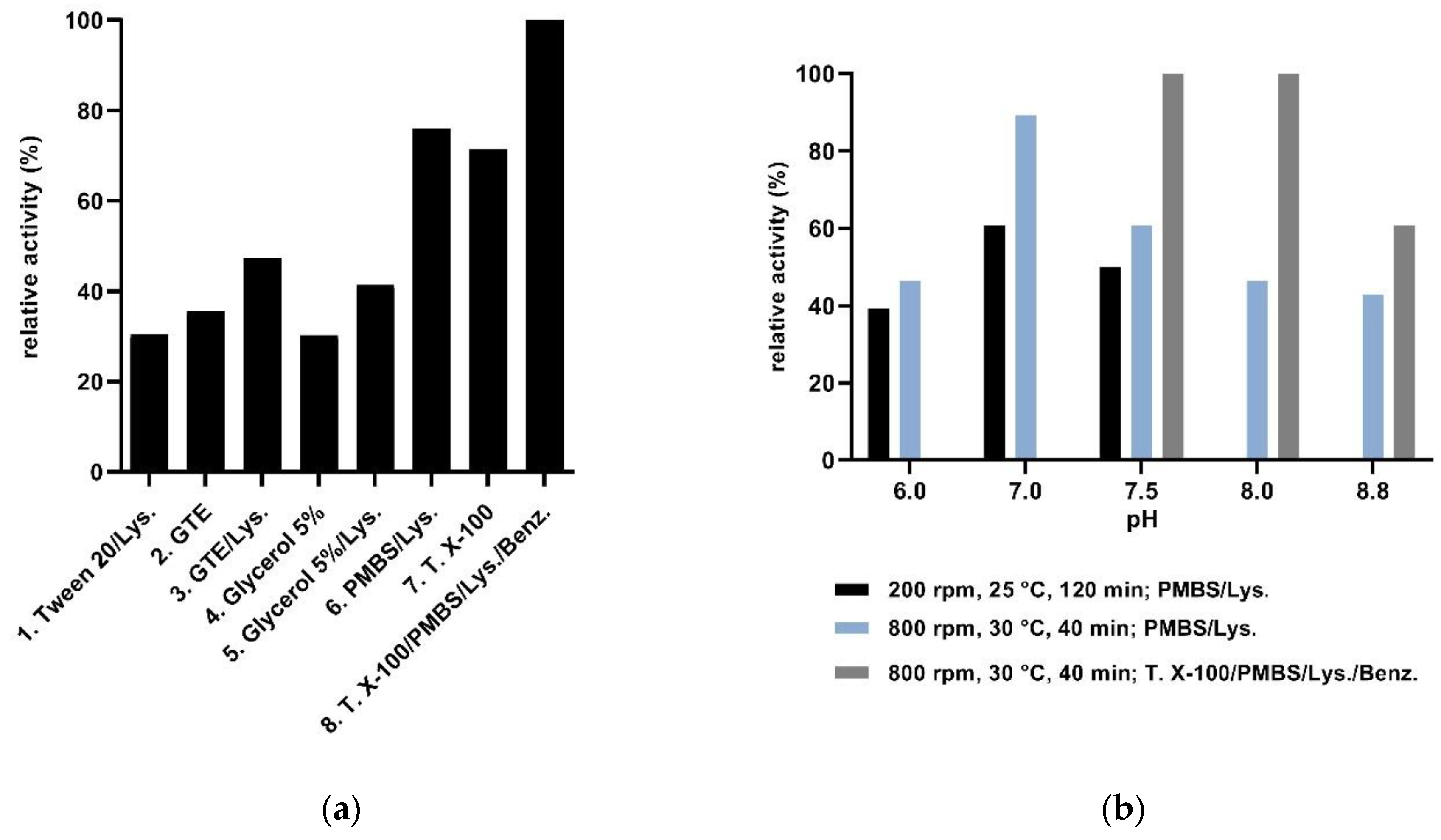
3.3.1. Assay Development
3.3.2. Activity Screens of Transformant Libraries
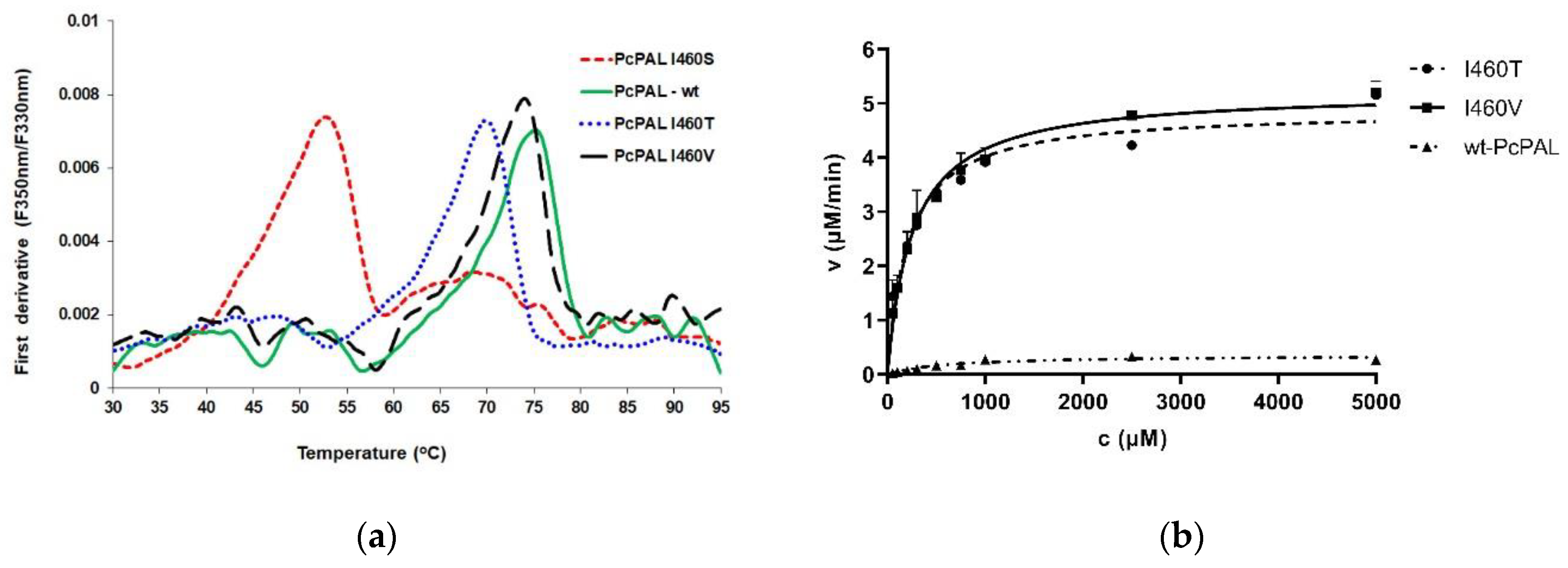
3.4. Functional and Structural Characterization of Improved PcPAL Variants
3.5. Molecular Docking
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Yamada, S.; Nabe, K.; Izuo, N. Production of L-phenylalanine from trans-cinnamic acid with Rhodotorula glutinis containing L-phenylalanine ammonia-lyase activity. Appl. Environ. Microbiol. 1981, 42, 773–778. [Google Scholar] [CrossRef] [PubMed]
- Renard, G.; Guilleux, J.C.; Bore, C.; Malta-Valette, V.; Lerner, D.A. Synthesis of L-phenylalanine analogs by Rhodotorula glutinis. Bioconversion of cinnamic acids derivatives. Biotechnol. Lett. 1992, 14, 673–678. [Google Scholar] [CrossRef]
- Baedeker, M.; Schulz, G.E. Overexpression of a designed 2.2 kb gene of eukaryotic phenylalanine ammonia-lyase in Escherichia coli. FEBS Lett. 1999, 457, 57–60. [Google Scholar] [CrossRef]
- Moffitt, M.C.; Louie, G.V.; Bowman, M.E.; Pence, J.; Noel, J.P.; Moore, B.S. Discovery of two cyanobacterial phenylalanine ammonia lyases: Kinetic and structural characterization. Biochemistry 2007, 46, 1004–1012. [Google Scholar] [CrossRef] [PubMed]
- Paizs, C.; Katona, A.; Rétey, J. The interaction of heteroaryl-acrylates and alanines with phenylalanine ammonia-lyase from parsley. Chem. Eur. J. 2006, 12, 2739–2744. [Google Scholar] [CrossRef]
- Lovelock, S.L.; Turner, N.J. Bacterial Anabaena variabilis phenylalanine ammonia lyase: A biocatalyst with broad substrate specificity. Bioorg. Med. Chem. 2014, 22, 5555–5557. [Google Scholar] [CrossRef]
- Gloge, A.; Zoń, J.; Kövári, A.; Poppe, L.; Rétey, J. Phenylalanine ammonia-lyase: The use of its broad substrate specificity for mechanistic investigations and biocatalysis—Synthesis of L-arylalanines. Chem. Eur. J. 2000, 6, 3386–3390. [Google Scholar] [CrossRef]
- Parmeggiani, F.; Weise, N.J.; Ahmed, S.T.; Turner, N.J. Synthetic and therapeutic applications of ammonia-lyases and aminomutases. Chem. Rev. 2018, 118, 73–118. [Google Scholar] [CrossRef]
- Heberling, M.M.; Wu, B.; Bartsch, S.; Janssen, D.B. Priming ammonia lyases and aminomutases for industrial and therapeutic applications. Curr. Opin. Chem. Biol. 2013, 17, 250–260. [Google Scholar] [CrossRef]
- Liu, W. Synthesis of Optically Active Phenylalanine Analogs Using Rhodotorula graminis. U.S. Patent 5,981,239, 9 November 1999. [Google Scholar]
- Ahmed, S.T.; Parmeggiani, F.; Weise, N.J.; Flitsch, S.L.; Turner, N.J. Chemoenzymatic synthesis of optically pure L- and D-biarylalanines through biocatalytic asymmetric amination and palladium-catalyzed arylation. ACS Catal. 2015, 5, 5410–5413. [Google Scholar] [CrossRef]
- Bartsch, S.; Bornscheuer, U.T. Mutational analysis of phenylalanine ammonia lyase to improve reactions rates for various substrates. Protein Eng. Des. Sel. 2010, 23, 929–933. [Google Scholar] [CrossRef] [PubMed]
- Rowles, I.; Groenendaal, B.; Binay, B.; Malone, K.J.; Willies, S.C.; Turner, N.J. Engineering of phenylalanine ammonia lyase from Rhodotorula graminis for the enhanced synthesis of unnatural L-amino acids. Tetrahedron 2016, 72, 7343–7347. [Google Scholar] [CrossRef]
- Nagy, E.Z.A.; Tork, S.D.; Lang, P.A.; Filip, A.; Irimie, F.D.; Poppe, L.; Toşa, M.I.; Schofield, C.J.; Brem, J.; Paizs, C.; et al. Mapping the hydrophobic substrate binding site of phenylalanine ammonia-lyase from Petroselinum crispum. ACS Catal. 2019, 9, 8825–8834. [Google Scholar] [CrossRef]
- Bencze, L.C.; Filip, A.; Bánóczi, G.; Toşa, M.I.; Irimie, F.D.; Gellért, Á.; Poppe, L.; Paizs, C. Expanding the substrate scope of phenylalanine ammonia-lyase from Petroselinum crispum towards styrylalanines. Org. Biomol. Chem. 2017, 15, 3717–3727. [Google Scholar] [CrossRef]
- Filip, A.; Nagy, E.Z.A.; Tork, S.D.; Bánóczi, G.; Toşa, M.I.; Irimie, F.D.; Poppe, L.; Paizs, C.; Bencze, L.C. Tailored mutants of phenylalanine ammonia-lyase from Petroselinum crispum for the synthesis of bulky L- and D-arylalanines. ChemCatChem 2018, 10, 2627–2633. [Google Scholar] [CrossRef]
- Ahmed, S.T.; Parmeggiani, F.; Weise, N.J.; Flitsch, S.L.; Turner, N.J. Engineered ammonia lyases for the production of challenging electron-rich L-phenylalanines. ACS Catal. 2018, 8, 3129–3132. [Google Scholar] [CrossRef]
- Tork, S.D.; Nagy, E.Z.A.; Cserepes, L.; Bordea, D.M.; Nagy, B.; Toşa, M.I.; Paizs, C.; Bencze, L.C. The production of L- and D-phenylalanines using engineered phenylalanine ammonia lyases from Petroselinum crispum. Sci. Rep. 2019, 9, 1–11. [Google Scholar] [CrossRef]
- Wang, L.; Gamez, A.; Archer, H.; Abola, E.E.; Sarkissian, C.N.; Fitzpatrick, P.; Wendt, D.; Zhang, Y.; Vellard, M.; Bliesath, J.; et al. Structural and biochemical characterization of the therapeutic Anabaena variabilis phenylalanine ammonia lyase. J. Mol. Biol. 2008, 380, 623–635. [Google Scholar] [CrossRef]
- Zhang, F.; Huang, N.; Zhou, L.; Cui, W.; Liu, Z.; Zhu, L.; Liu, Y.; Zhou, Z. Modulating the pH activity profiles of phenylalanine ammonia lyase from Anabaena variabilis by modification of center-near surface residues. Appl. Biochem. Biotechnol. 2017, 183, 699–711. [Google Scholar] [CrossRef]
- Pedersen, J.N.; Zhou, Y.; Guo, Z.; Pérez, B. Genetic and chemical approaches for surface charge engineering of enzymes and their applicability in biocatalysis: A review. Biotechnol. Bioeng. 2019, 116, 1795–1812. [Google Scholar] [CrossRef]
- Liu, Q.; Xun, G.; Feng, Y. The state-of-the-art strategies of protein engineering for enzyme stabilization. Biotechnol. Adv. 2019, 37, 530–537. [Google Scholar] [CrossRef] [PubMed]
- Reetz, M.T. Biocatalysis in organic chemistry and biotechnology: Past, present, and future. J. Am. Chem. Soc. 2013, 135, 12480–12496. [Google Scholar] [CrossRef] [PubMed]
- Prasad, S.; Bocola, M.; Reetz, M.T. Revisiting the lipase from pseudomonas aeruginosa: Directed evolution of substrate acceptance and enantioselectivity using iterative saturation mutagenesis. ChemPhysChem 2011, 12, 1550–1557. [Google Scholar] [CrossRef] [PubMed]
- Reetz, M.T.; Bocola, M.; Carballeira, J.D.; Zha, D.; Vogel, A. Expanding the range of substrate acceptance of enzymes: Combinatorial active-site saturation test. Angew. Chem. Int. Ed. 2005, 44, 4192–4196. [Google Scholar] [CrossRef]
- Poppe, L.; Paizs, C.; Kovács, K.; Irimie, F.D.; Vértessy, B. Preparation of unnatural amino acids with ammonia-lyases and 2,3-aminomutases. In Unnatural Amino Acids: Methods and Protocols; Pollegioni, P., Servi, S., Eds.; Springer Science+Business Media: New York, NY, USA, 2012; Volume 794, pp. 3–19. [Google Scholar]
- Jendresen, C.B.; Stahlhut, S.G.; Li, M.J.; Gaspar, P.; Siedler, S.; Forster, J.; Maury, J.; Borodina, I.; Nielsen, A.T. Highly active and specific tyrosine ammonia-lyases from diverse origins enable enhanced production of aromatic compounds in bacteria and Saccharomyces cerevisiae. Appl. Environ. Microbiol. 2015, 81, 4458–4476. [Google Scholar] [CrossRef]
- Tian, Z.L.; Zhu, W.P.; Xu, Y.F.; Qian, X.H. An unnatural amino acid based fluorescent probe for phenylalanine ammonia lyase. Org. Biomol. Chem. 2014, 12, 5818–5821. [Google Scholar] [CrossRef]
- Yan, C.Y.; Parmeggiani, F.; Jones, E.A.; Claude, E.; Hussain, S.A.; Turner, N.J.; Flitsch, S.L.; Barran, P.E. Real-time screening of biocatalysts in live bacterial colonies. J. Am. Chem. Soc. 2017, 139, 1408–1411. [Google Scholar] [CrossRef]
- Reetz, M.T.; Carballeira, J.D. Iterative saturation mutagenesis (ISM) for rapid directed evolution of functional enzymes. Nat. Protoc. 2007, 2, 891–903. [Google Scholar] [CrossRef]
- Reetz, M.T.; Carballeira, J.D.; Vogel, A. Iterative saturation mutagenesis on the basis of B factors as a strategy for increasing protein thermostability. Angew. Chem. Int. Ed. 2006, 45, 7745–7751. [Google Scholar] [CrossRef]
- Clouthier, C.M.; Kayser, M.M.; Reetz, M.T. Designing new Baeyer-Villiger monooxygenases using restricted CASTing. J. Org. Chem. 2006, 71, 8431–8437. [Google Scholar] [CrossRef]
- Reetz, M.T.; Peyralans, J.J.P.; Maichele, A.; Fu, Y.; Maywald, M. Directed evolution of hybrid enzymes: Evolving enantioselectivity of an achiral Rh-complex anchored to a protein. Chem. Commun. 2006, 41, 4318–4320. [Google Scholar] [CrossRef] [PubMed]
- Parmeggiani, F.; Lovelock, S.L.; Weise, N.J.; Ahmed, S.T.; Turner, N.J. Synthesis of D- and L-phenylalanine derivatives by phenylalanine ammonia lyases: A multienzymatic cascade process. Angew. Chem. Int. Ed. 2015, 54, 4608–4611. [Google Scholar] [CrossRef] [PubMed]
- Dima, N.A.; Filip, A.; Bencze, L.C.; Oláh, M.; Sátorhelyi, P.; Vértessy, B.G.; Poppe, L.; Paizs, C. Expression and purification of recombinant phenylalanine ammonia-lyase from Petroselinum crispum. Studia Universitatis Babes-Bolyai Chemia 2016, 61, 21–34. [Google Scholar]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [PubMed]
- Jurrus, E.; Engel, D.; Star, K.; Monson, K.; Brandi, J.; Felberg, L.E.; Brookes, D.H.; Wilson, L.; Chen, J.H.; Liles, K.; et al. Improvements to the APBS biomolecular solvation software suite. Protein Sci. 2018, 27, 112–128. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Naismith, J.H. An efficient one-step site-directed deletion, insertion, single and multiple-site plasmid mutagenesis protocol. BMC Biotechnol. 2008, 8, 91. [Google Scholar] [CrossRef]
- Sanchis, J.; Fernández, L.; Carballeira, J.D.; Drone, J.; Gumulya, Y.; Höbenreich, H.; Kahakeaw, D.; Kille, S.; Lohmer, R.; Peyralans, J.J.P.; et al. Improved PCR method for the creation of saturation mutagenesis libraries in directed evolution: Application to difficult-to-amplify templates. Appl. Microbiol. Biotechnol. 2008, 81, 387–397. [Google Scholar] [CrossRef]
- Hoebenreich, S.; Zilly, F.E.; Acevedo-Rocha, C.G.; Zilly, M.; Reetz, M.T. Speeding up directed evolution: Combining the advantages of solid-phase combinatorial gene synthesis with statistically guided reduction of screening effort. ACS Synth. Biol. 2015, 4, 317–331. [Google Scholar] [CrossRef]
- Li, A.; Sun, Z.; Reetz, M.T. Solid-phase gene synthesis for mutant library construction: The future of directed evolution? ChemBioChem 2018, 19, 2023–2032. [Google Scholar] [CrossRef]
- Kováčik, J.; Klejdus, B. Tissue and method specificities of phenylalanine ammonia-lyase assay. J. Plant Physiol. 2012, 169, 1317–1320. [Google Scholar] [CrossRef]
- Paizs, C.; Toşa, M.I.; Bencze, L.C.; Brem, J.; Irimie, F.D.; Rétey, J. 2-amino-3-(5-phenylfuran-2-yl)propionic acids and 5-phenylfuran-2-ylacrylic acids are novel substrates of phenylalanine ammonia-lyase. Heterocycles 2011, 82, 1217–1228. [Google Scholar] [CrossRef]

| Compound | 4-MeO-Phe | l-Phe | ||||
|---|---|---|---|---|---|---|
| PcPAL | wt- | I460T | I460V | wt- | I460T | I460V |
| KM (μM) | 1741 ± 191.2 | 210.2 ± 27.9 | 256.2 ± 26.8 | 110.9 ± 12.3 | 127.1 ± 20.7 | 474.6 ± 39.8 |
| kcat (s−1) | 0.012 | 0.129 | 0.139 | 0.698 | 0.805 | 0.617 |
| vmax (μM/min) | 0.45 ± 0.03 | 4.8 ± 0.1 | 5.22 ± 0.1 | 26.2 ± 0.6 | 35.4 ± 1.1 | 5.2 ± 0.1 |
| kcat/ KM *10−3 (s−1xμM−1) | 0.007 | 0.617 | 0.543 | 6.297 | 6.334 | 1.300 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tomoiagă, R.B.; Tork, S.D.; Horváth, I.; Filip, A.; Nagy, L.C.; Bencze, L.C. Saturation Mutagenesis for Phenylalanine Ammonia Lyases of Enhanced Catalytic Properties. Biomolecules 2020, 10, 838. https://doi.org/10.3390/biom10060838
Tomoiagă RB, Tork SD, Horváth I, Filip A, Nagy LC, Bencze LC. Saturation Mutagenesis for Phenylalanine Ammonia Lyases of Enhanced Catalytic Properties. Biomolecules. 2020; 10(6):838. https://doi.org/10.3390/biom10060838
Chicago/Turabian StyleTomoiagă, Raluca Bianca, Souad Diana Tork, Ilka Horváth, Alina Filip, Levente Csaba Nagy, and László Csaba Bencze. 2020. "Saturation Mutagenesis for Phenylalanine Ammonia Lyases of Enhanced Catalytic Properties" Biomolecules 10, no. 6: 838. https://doi.org/10.3390/biom10060838
APA StyleTomoiagă, R. B., Tork, S. D., Horváth, I., Filip, A., Nagy, L. C., & Bencze, L. C. (2020). Saturation Mutagenesis for Phenylalanine Ammonia Lyases of Enhanced Catalytic Properties. Biomolecules, 10(6), 838. https://doi.org/10.3390/biom10060838





