Metallacarborane Complex Boosts the Rate of DNA Oligonucleotide Hydrolysis in the Reaction Catalyzed by Snake Venom Phosphodiesterase
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemistry
2.1.1. Automated Synthesis of Oligonucleotides
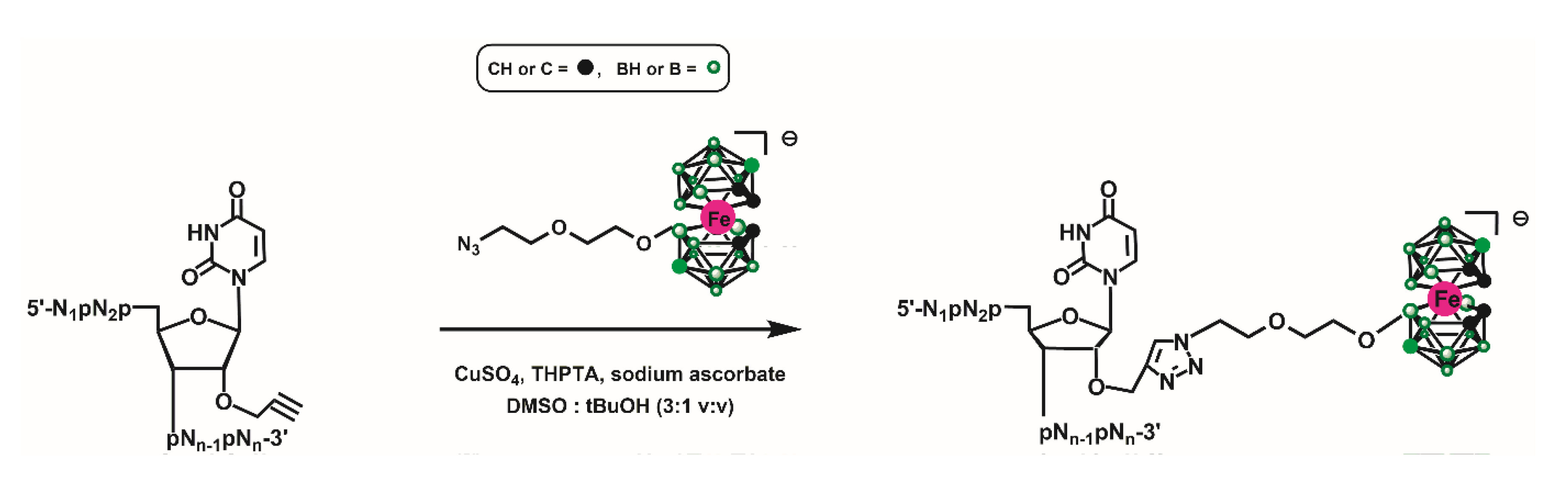
2.1.2. Synthesis of Oligonucleotides 1a, 1b, 1c and 2a, 2b Modified with Metallacarborane
2.2. Enzymatic Assays
2.2.1. 5’-Phosphorylation of Oligonucleotides
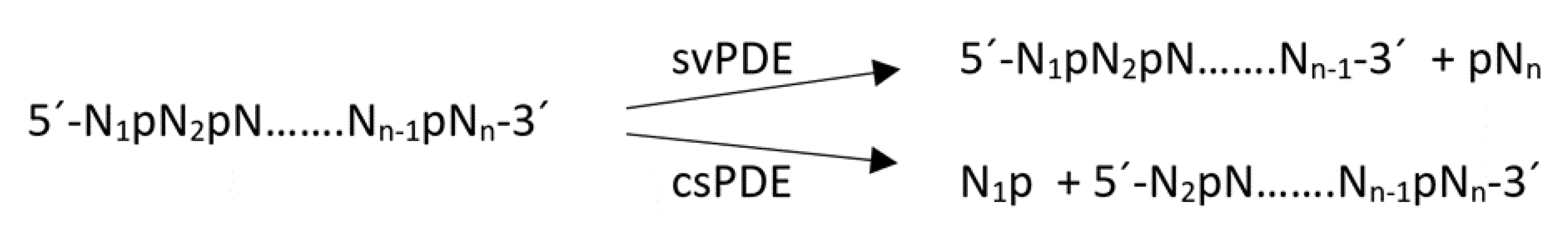
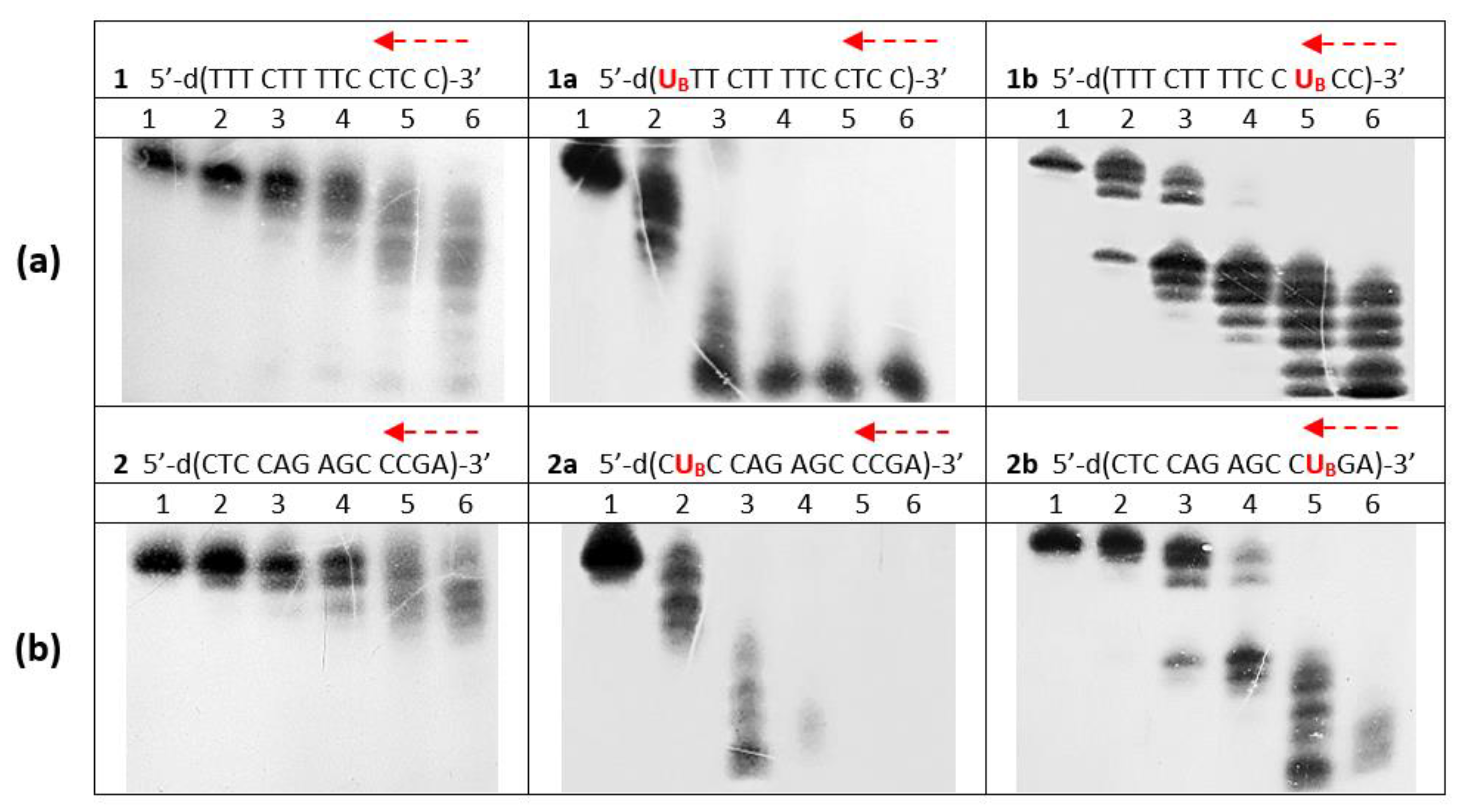
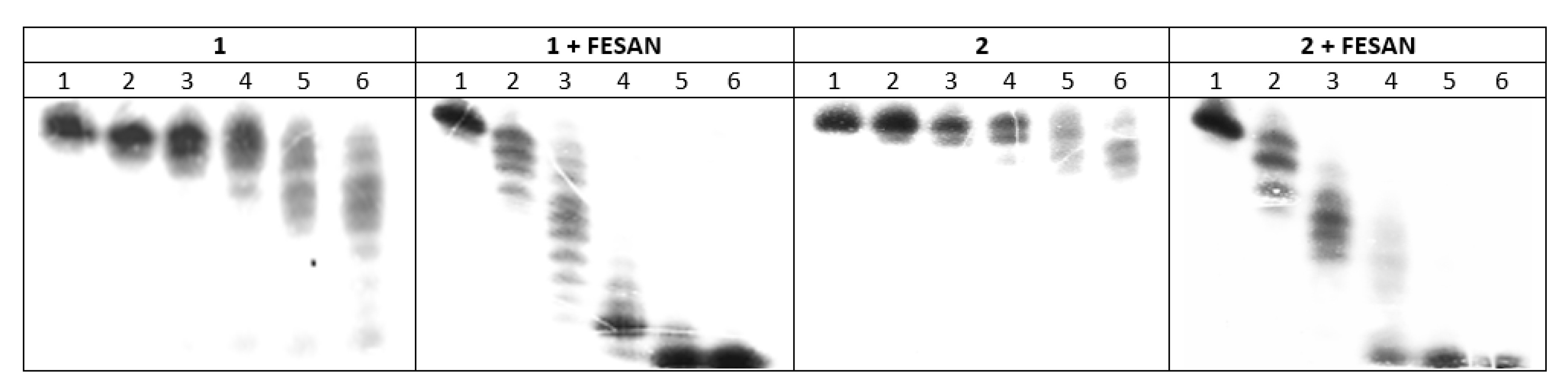
2.2.2. Hydrolysis of Oligonucleotides 1, 2, 1a, 1b, 1c, 2a and 2b in the Presence of svPDE Analyzed by PAGE
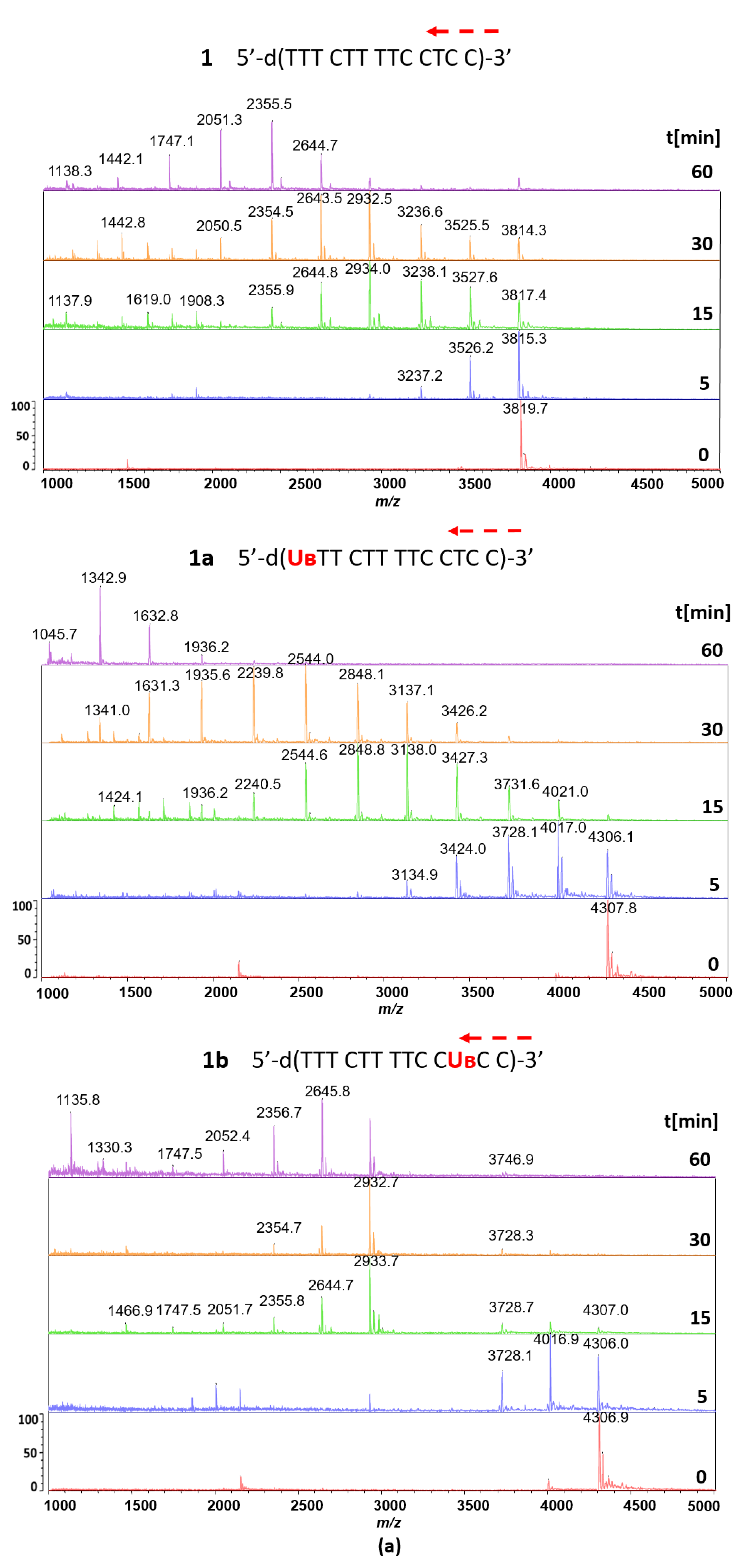
2.2.3. Hydrolysis of Oligonucleotides 1, 2, 1a, 1b, 1c, 2a and 2b in the Presence of svPDE Analyzed by MALDI-TOF MS
2.3. Affinity Measurements
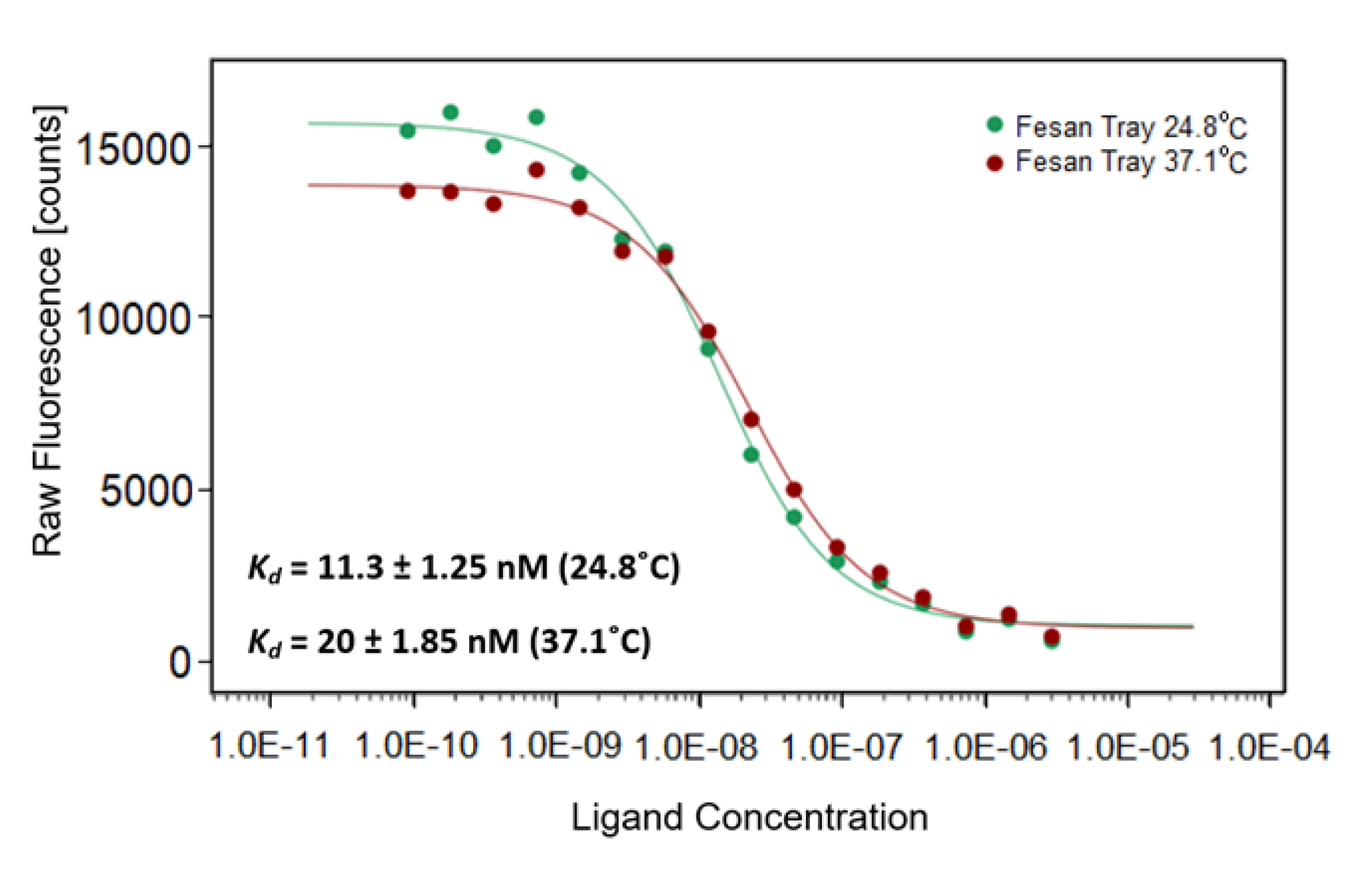
2.3.1. Affinity of Ferra(III) bis(dicarbollide) for Crude Crotalus Atrox Venom Analyzed by MST
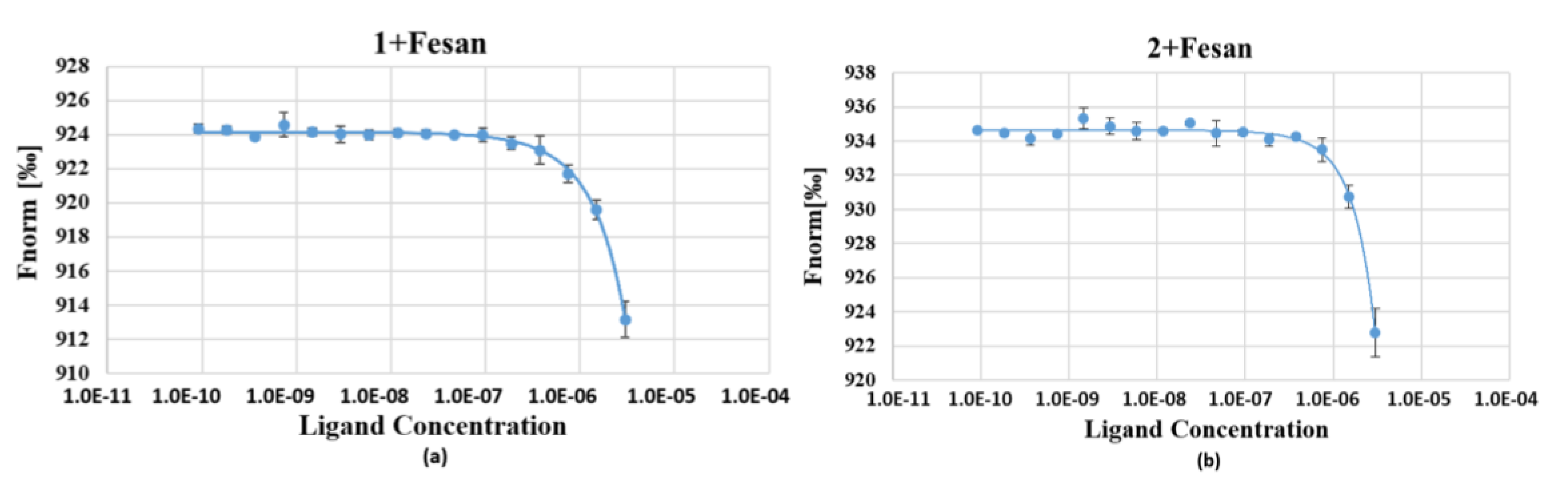
2.3.2. Affinity of Oligonucleotides FL-1 and FL-2 to Ferra(III) bis(dicarbollide) Analyzed by MST
3. Results
3.1. Chemical Synthesis of Metallacarborane-Conjugated ASO Models
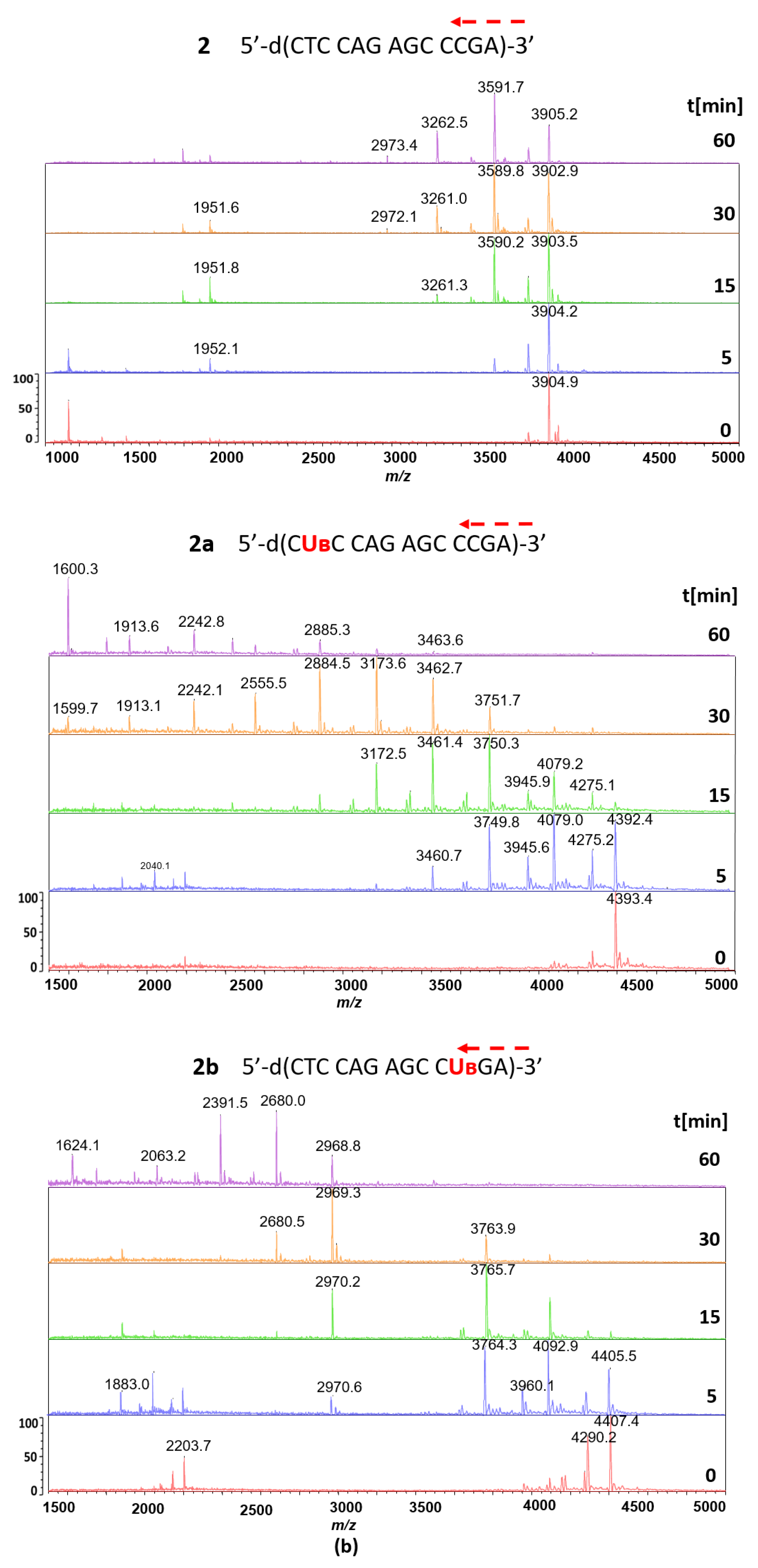
3.2. Hydrolysis of Reference Oligonucleotides 1 and 2 in the Presence of svPDE Monitored by MALDI-TOF Mass Spectrometry
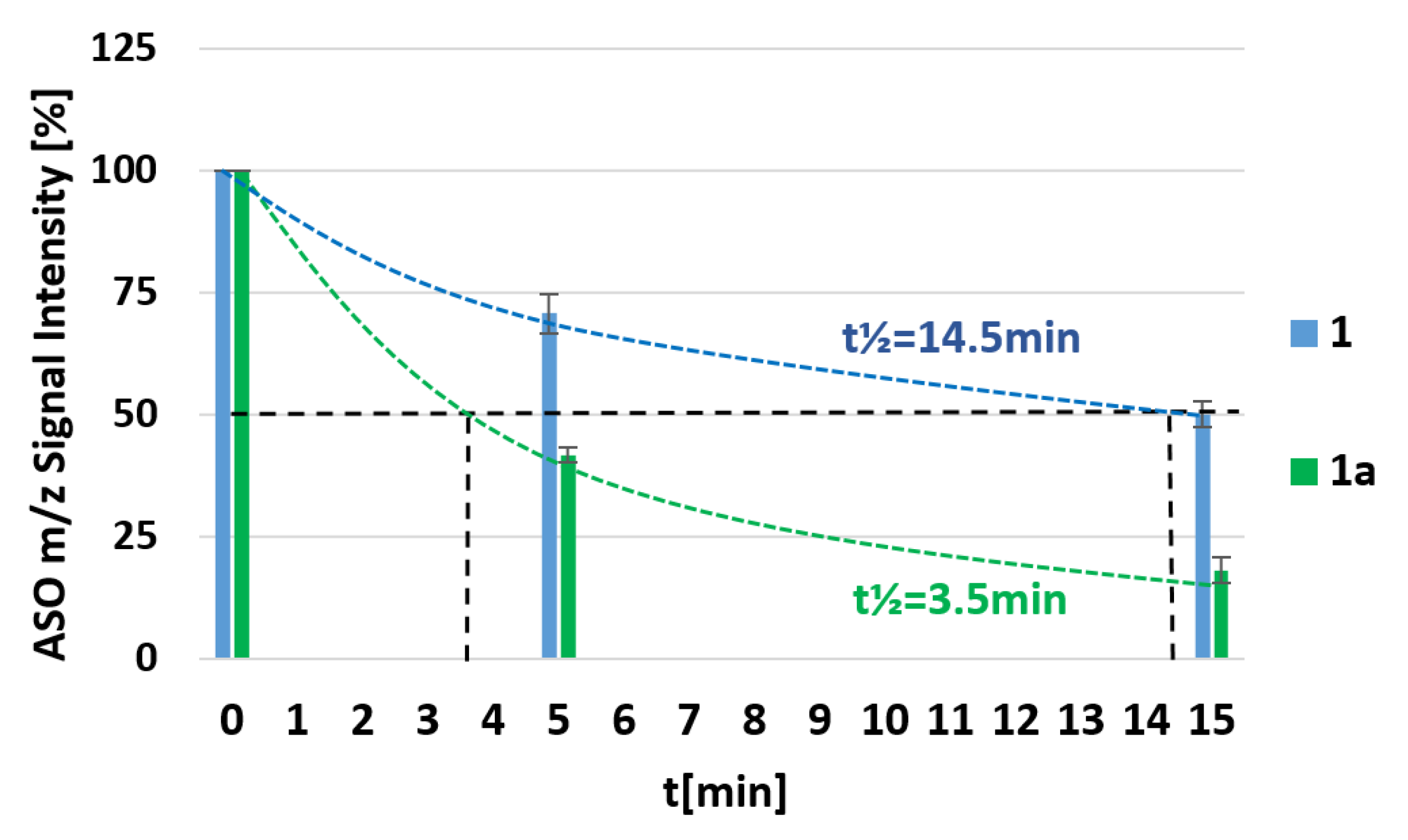
3.3. svPDE-Assisted Hydrolysis of Metallacarborane-Conjugated Oligonucleotides 1a, 1b, 1c, 2a, and 2b
3.4. svPDE-Catalyzed Cleavage of an Equimolar Mixture of 1 + 1a
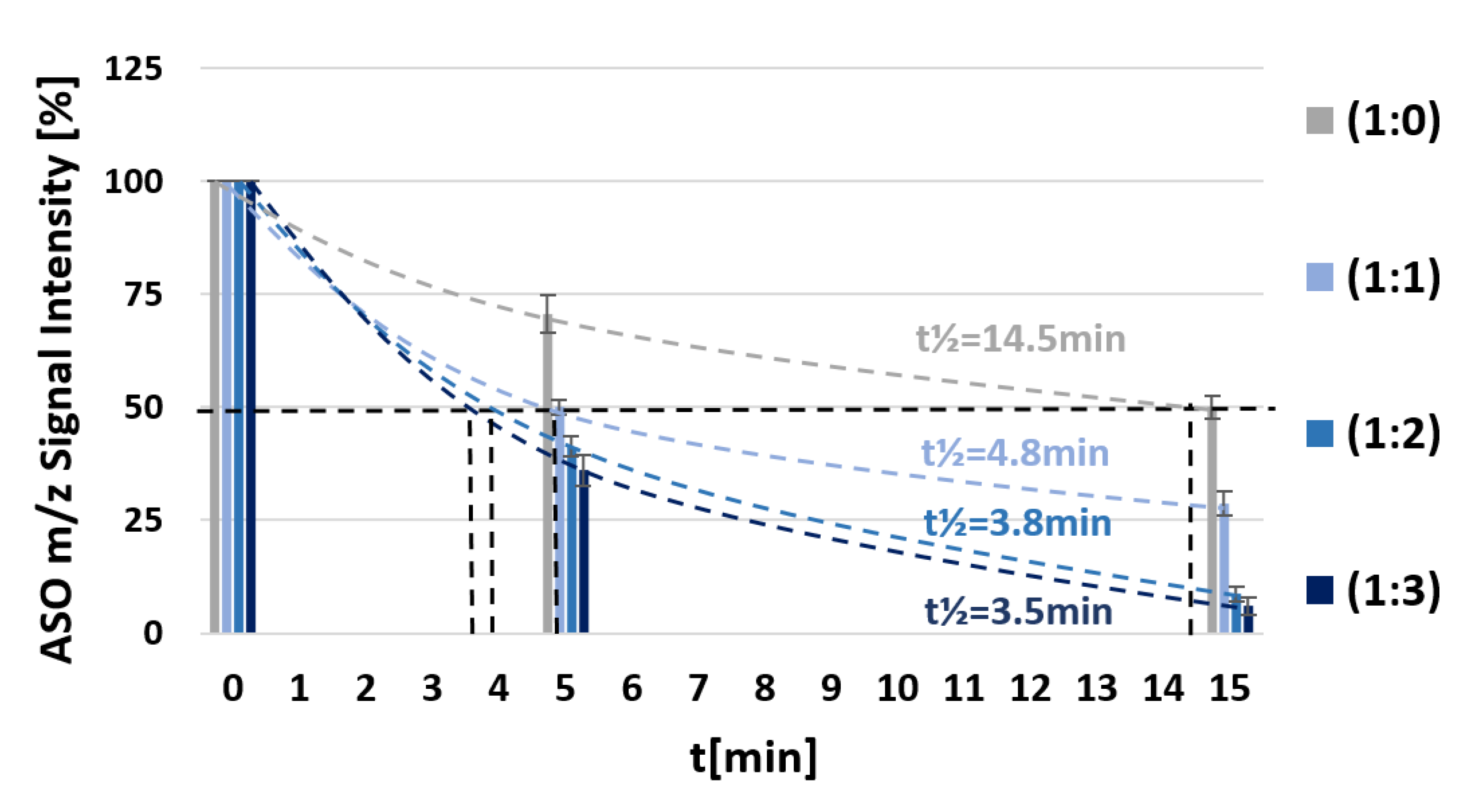
3.5. Enhanced 3’→5’ Exonucleolytic Activity of svPDE in the Presence of Free Metallacarborane
3.6. Affinity of Ferra(III) bis(dicarbollide) for Proteins Present in Crude Snake Venom Used as a Source of svPDE
3.7. Affinity of Ferra(III) bis(dicarbollide) to 5’-Fluorescently Labeled Oligonucleotides 1 and 2
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Crooke, S.T. Antisense Drug Technology: Principles, Strategies, and Applications, 2nd ed.; Edition CRC Press: Baco Raton, FL, USA, 2008. [Google Scholar] [CrossRef]
- Dias, N.; Stein, C.A. Antisense oligonucleotides: Basic concepts and mechanisms. Mol. Cancer Ther. 2002, 1, 347–355. [Google Scholar] [PubMed]
- Liao, W.; Dong, J.; Peh, H.Y.; Tan, L.H.; Lim, K.S.; Li, L.; Wong, W.F. Oligonucleotide Therapy for Obstructive and Restrictive Respiratory Diseases. Molecules 2017, 22, 139. [Google Scholar] [CrossRef] [PubMed]
- Di, C.; Syafrizayanti, Z.Q.; Chen, Y.; Wang, Y.; Zhang, X.; Liu, Y.; Sun, C.; Zhang, H.; Hoheisel, J.D. Function, clinical application, and strategies of Pre-mRNA splicing in cancer. Cell Death Differ. 2019, 26, 1181–1194. [Google Scholar] [CrossRef] [PubMed]
- Chery, J. RNA therapeutics: RNAi and antisense mechanisms and clinical applications. Postdoc J. 2016, 7, 35–50. [Google Scholar] [CrossRef]
- Singh, N.N.; Luo, D.; Singh, R.N. Pre-mRNA Splicing Modulation by Antisense Oligonucleotides. Methods Mol. Biol. 2018, 1828, 415–437. [Google Scholar] [CrossRef]
- Elkon, R.; Ugalde, A.P.; Agami, R. Alternative cleavage and polyadenylation: Extent, regulation and function. Nat. Rev. Genet. 2013, 14, 496–506. [Google Scholar] [CrossRef]
- Huang, L.; Aghajan, M.; Quesenberry, T.; Low, A.; Murray, S.F.; Monia, B.P.; Guo, S. Targeting Translation Termination Machinery with Antisense Oligonucleotides for Diseases Caused by Nonsense Mutations. Nucleic Acid Ther. 2019, 29, 175–186. [Google Scholar] [CrossRef]
- Svidritskiy, E.; Demo, G.; Korostelev, A.A. Mechanism of premature translation termination on a sense codon. J. Biol. Chem. 2018, 32, 12472–12479. [Google Scholar] [CrossRef]
- Yang, W. Nucleases: Diversity of structure, function and mechanism. Q. Rev. Biophys. 2011, 44, 1–93. [Google Scholar] [CrossRef]
- Cobb, A.J.A. Recent highlights in modified oligonucleotide chemistry. Org. Biomol. Chem. 2007, 5, 3260–3275. [Google Scholar] [CrossRef]
- Deleavey, G.F.; Damha, M.J. Designing chemically modified oligonucleotides for targeted gene silencing. Chem. Biol. 2012, 19, 937–954. [Google Scholar] [CrossRef]
- Chen, X.; Fleming, A.M.; Muller, J.G.; Burrows, C.J. Endonuclease and Exonuclease Activities on Oligodeoxynucleotides Containing Spiroiminodihydantoin Depend on the Sequence Context and the Lesion Stereochemistry. New J. Chem. 2013, 37, 3440–3449. [Google Scholar] [CrossRef]
- Bentzley, C.M.; Johnston, M.V.; Larsen, B.S. Base specificity of oligonucleotide digestion by calf spleen phosphodiesterase with matrix-assisted laser desorption ionization analysis. Anal. Biochem. 1998, 258, 31–37. [Google Scholar] [CrossRef]
- Dhananjaya, B.L.; D’souza, C.J.M. An overview on nucleases (DNase, RNase, and phosphodiesterase) in snake venoms. Biochemistry Moscow 2010, 75, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Scoles, D.R.; Minikel, E.V.; Pulst, S.M. Antisense oligonucleotides: A primer. Neurol. Genet. 2019, 5, 323–331. [Google Scholar] [CrossRef] [PubMed]
- DeVos, S.L.; Miller, T.M. Antisense oligonucleotides: Treating neurodegeneration at the level of RNA. Neurotherapeutics 2013, 10, 486–497. [Google Scholar] [CrossRef] [PubMed]
- Frieden, M.; Christensen, S.M.; Mikkelsen, N.D.; Rosenbohm, C.; Thrue, C.A.; Westergaard, M.; Hansen, H.F.; Ørum, H.; Koch, T. Expanding the design horizon of antisense oligonucleotides with alpha-l-LNA. Nucleic Acids Res. 2003, 31, 6365–6372. [Google Scholar] [CrossRef]
- Ebenryter-Olbińska, K.; Kaniowski, D.; Sobczak, M.; Wojtczak, B.A.; Janczak, S.; Wielgus, E.; Nawrot, B.; Leśnikowski, Z.J. Versatile Method for the Site-Specific Modification of DNA with Boron Clusters: Anti-Epidermal Growth Factor Receptor (EGFR) Antisense Oligonucleotide Case. Chem. Eur. J. 2017, 23, 16535–16546. [Google Scholar] [CrossRef]
- Kaniowski, D.; Ebenryter-Olbińska, K.; Sobczak, M.; Wojtczak, B.; Janczak, S.; Leśnikowski, Z.J.; Nawrot, B. High Boron-loaded DNA-Oligomers as Potential Boron Neutron Capture Therapy and Antisense Oligonucleotide Dual-Action Anticancer Agents. Molecules 2017, 22, 1393. [Google Scholar] [CrossRef]
- Kaniowski, D.; Ebenryter-Olbinska, K.; Kulik, K.; Janczak, S.; Maciaszek, A.; Bednarska-Szczepaniak, K.; Nawrot, B.; Lesnikowski, Z. Boron clusters as a platform for new materials: Composites of nucleic acids and oligofunctionalized carboranes (C(2)B(10)H(12)) and their assembly into functional nanoparticles. Nanoscale 2020, 12, 103–114. [Google Scholar] [CrossRef]
- Kwiatkowska, A.; Sobczak, M.; Mikolajczyk, B.; Janczak, S.; Olejniczak, A.B.; Sochacki, M.; Lesnikowski, Z.J.; Nawrot, B. siRNAs modified with boron cluster and their physicochemical and biological characterization. Bioconjug Chem. 2013, 24, 1017–1026. [Google Scholar] [CrossRef] [PubMed]
- Barth, R.F.; Mi, P.; Yang, W. Boron delivery agents for neutron capture therapy of cancer. Cancer Commun. (Lond.) 2018, 38, 35. [Google Scholar] [CrossRef] [PubMed]
- Olejniczak, A.B.; Koziołkiewicz, M.; Leśnikowski, Z.J. Carboranyl oligonucleotides. 4. Synthesis, and physicochemical studies of oligonucleotides containing 2’-O-(o-carboran-1-yl)methyl group. Antisense Nucl. Acid Drug Develop. 2002, 12, 79–94. [Google Scholar] [CrossRef] [PubMed]
- Fulcrand-El Kattan, G.; Lesnikowski, Z.J.; Yao, S.; Tanious, F.; Wilson, W.D.; Shinazi, R.F. Carboranyl Oligonucleotides. 2. Synthesis and Physicochemical Properties of Dodecathymidylate Containing 5-(o-Carboranyl-1-yl)-2’-O-Deoxyuridine. J. Am. Chem. Soc. 1994, 116, 7494–7501. [Google Scholar] [CrossRef]
- Lesnikowski, Z.J. Boron clusters—A new entity for DNA-oligonucleotide modification. Eur. J. Org. Chem. 2003, 23, 4489–4500. [Google Scholar] [CrossRef]
- Olejniczak, A.B.; Plesek, J.; Kriz, O.; Lesnikowski, Z.J. Nucleoside Conjugate Containing Metallacarborane Group and its Incorporation into DNA-oligonucleotide. Angew. Chem. Int. Ed. Engl. 2003, 42, 5740–5743. [Google Scholar] [CrossRef]
- Olejniczak, A. Metallacarboranes for the labelling of DNA—Synthesis of oligonucleotides bearing a 3,3’-iron-1,2,1’,2’-dicarbollide complex. Can. J. Chem. 2011, 89, 465–470. [Google Scholar] [CrossRef]
- Olejniczak, A.B.; Kierzek, R.; Wickstrom, E.; Leśnikowski, Z.J. Synthesis, physicochemical and biochemical studies of anti-IRS-1 oligonucleotides containing carborane and/or metallacarborane modification. J. Organomet. Chem. 2013, 747, 201–210. [Google Scholar] [CrossRef]
- Sanghvi, Y.S. A status update of modified oligonucleotides for chemotherapeutics applications. In Current Protocols in Nucleic Acid Chemistry; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2011; Chapter 4, Unit 4.1.1-4.1.22. [Google Scholar] [CrossRef]
- Glen Research. Available online: http:/glenresearch.com/GlenReports/GR25-24.html (accessed on 1 April 2020).
- Munawar, A.; Ali, S.A.; Akrem, A.; Betzel, C. Snake Venom Peptides: Tools of Biodiscovery. Toxins 2018, 10, 474. [Google Scholar] [CrossRef]
- Valério, A.A.; Corradini, A.C.; Panunto, P.C.; Mello, S.M.; Hyslop, S. Purification and characterization of a phosphodiesterase from Bothrops alternatus snake venom. J. Protein Chem. 2002, 21, 495–503. [Google Scholar] [CrossRef]
- Ullah, A.; Ullah, K.; Ali, H.; Betzel, C.; Ur Rehman, S. The Sequence and a Three-Dimensional Structural Analysis Reveal Substrate Specificity Among Snake Venom Phosphodiesterases. Toxins 2019, 11, 625. [Google Scholar] [CrossRef] [PubMed]
- Fayaz, A.; Narayan, S.H.; Yinghuai, Z. Boron Chemistry for Medical Applications. Molecules 2020, 25, 828. [Google Scholar] [CrossRef]
- Fuentes, I.; García-Mendiola, T.; Sato, S.; Pita, M.; Nakamura, H.; Lorenzo, E.; Teixidor, F.; Marques, F.; Viñas, C. Metallacarboranes on the Road to Anticancer Therapies: Cellular Uptake, DNA Interaction, and Biological Evaluation of Cobaltabisdicarbollide [COSAN](-). Chem. Eur. J. 2018, 24, 17239–17254. [Google Scholar] [CrossRef]
- Leśnikowski, Z.J.; Paradowska, E.; Olejniczak, A.B.; Studzińska, M.; Seekamp, P.; Schüßler, U.; Gabel, D.; Schinazi, R.F.; Plešek, J. Towards new boron carriers for boron neutron capture therapy: Metallacarboranes and their nucleoside conjugates. Bioorg. Med. Chem. 2005, 13, 4168–4175. [Google Scholar] [CrossRef]
- Goszczyński, T.M.; Fink, K.; Kowalski, K.; Leśnikowski, Z.J.; Boratyński, J. Interactions of Boron Clusters and their Derivatives with Serum Albumin. Sci. Rep. 2017, 7, 9800–9812. [Google Scholar] [CrossRef] [PubMed]
- Viñas, C.; Núñez, R.; Bennour, I.; Teixidor, F. Periphery Decorated and Core Initiated Neutral and Polyanionic Borane large molecules: Forthcoming and Promising properties for medicinal applications. Curr. Med. Chem. 2019, 26, 1–41. [Google Scholar] [CrossRef] [PubMed]
- Fink, K.; Kobak, K.; Kasztura, M.; Boratyński, J.; Goszczyński, T.M. Synthesis and Biological Activity of Thymosin β4-Anionic Boron Cluster Conjugates. Bioconjug. Chem. 2018, 29, 3509–3515. [Google Scholar] [CrossRef]
- Fink, K.; Boratyński, J.; Paprocka, M.; Goszczyński, T.M. Metallacarboranes as a tool for enhancing the activity of therapeutic peptides. Ann. N. Y. Acad. Sci. 2019, 1457, 128–141. [Google Scholar] [CrossRef]
- Agrawal, S.; Jiang, Z.; Zhao, Q.; Shaw, D.; Cai, Q.; Roskey, A.; Channavajjala, L.; Saxinger, C.; Zhang, R. Mixed-backbone oligonucleotides as second generation antisense oligonucleotides: In vitro and in vivo studies. PNAS U.S.A. 1997, 94, 2620–2625. [Google Scholar] [CrossRef]
- Fuentes, I.; Pujols, J.; Viñas, C.; Ventura, S.; Teixidor, F. Dual Binding Mode of Metallacarborane Produces a Robust Shield on Proteins. Chem. Eur. J. 2019, 25, 12820–12829. [Google Scholar] [CrossRef]
- Lesnikowski, Z.J.; Fulcrand, G.; Lloyd, R.M.; Juodawlkis, A.; Schinazi, R.F. Carboranyl Oligonucleotides. 3. Biochemical Properties of Oligonucleotides Containing 5-(o-carboranyl-1-yl)-2’-deoxyuridine. Biochemistry 1996, 35, 5741–5746. [Google Scholar] [CrossRef] [PubMed]



| No. | Sequences | MW Calc. (g/mol) | MALDI-TOF MS (m/z) | RP-HPLC (Rt, min) |
|---|---|---|---|---|
| 1 | 5’-d(TTT CTT TTC CTC C)-3’ | 3817.50 | 3817.98 1 | 13.04 |
| 2 | 5’-d(CTC CAG AGC CCG A)-3’ | 3904.56 | 3904.82 1 | 12.27 |
| P1a | 5’-d(UPrTT CTT TTC CTC C)-3’ | 3857.52 | 3857.98 1 | 13.13 |
| P1b | 5’-d(TTT CTT TTC CUPrC C)-3’ | 3857.52 | 3858.02 1 | 13.14 |
| P1c | 5’-d(UPrTT CTT TTC CTC UPr)-3’ | 3912.55 | 3910.80 | 13.51 |
| P2a | 5’-d(CUPrC CAG AGC CCGA)-3’ | 3944.58 | 3945.20 | 12.06 |
| P2b | 5’-d(CTC CAG AGC CUPrGA)-3’ | 3959.59 | 3958.40 | 12.15 |
| 1a | 5’-d(UB TT CTT TTC CTC C)-3’ | 4307.29 | 4306.20 | 25.20 |
| 1b | 5’-d(TTT CTT TTC C UBC C)-3’ | 4307.29 | 4305.90 | 23.67 |
| 1c | 5’-d(UBTT CTT TTC CTCUB)-3’ | 4812.11 | 4810.90 | 29.12 |
| 2a | 5’-d(CUBC CAG AGC CCGA)-3’ | 4394.36 | 4393.70 | 24.39 |
| 2b | 5’-d(CTC CAG AGC CUBGA)-3’ | 4409.37 | 4411.10 1 | 23.95 |
| FL-1 | 5’-FL-d(TTT CTT TTC CTC C)-3’ | 4354.95 | 4355.70 1 | 15.57 |
| FL-2 | 5’-FL-d(CTC CAG AGC CCG A)-3’ | 4442.02 | 4442.65 1 | 14.54 |
| ASO | Sequence 5’ → 3’ | T1/2 (min) |
|---|---|---|
| 1 | 5’-d(TTT CTT TTC CTC C)-3’ | 14.5 |
| 2 | 5’-d(CTC CAG AGC CCG A)-3’ | 25.0 |
| 1a | 5’-d(UB TT CTT TTC CTC C)-3’ | 3.5 |
| 1b | 5’-d(TTT CTT TTC C UBC C)-3’ | 4.0 |
| 1c | 5’-d(UBTT CTT TTC CTCUB)-3’ | 20.0 |
| 2a | 5’-d(CUBC CAG AGC CCGA)-3’ | 3.5 |
| 2b | 5’-d(CTC CAG AGC CUBGA)-3’ | 4.0 |
| 1 | 5’-d(TTT CTT TTC CTC C)-3’ + FESAN (1 equiv.) | 4.8 |
| 1 | 5’-d(TTT CTT TTC CTC C)-3’ + FESAN (2 equiv.) | 3.8 |
| 1 | 5’-d(TTT CTT TTC CTC C)-3’ + FESAN (3 equiv.) | 3.5 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kaniowski, D.; Kulik, K.; Ebenryter-Olbińska, K.; Wielgus, E.; Lesnikowski, Z.; Nawrot, B. Metallacarborane Complex Boosts the Rate of DNA Oligonucleotide Hydrolysis in the Reaction Catalyzed by Snake Venom Phosphodiesterase. Biomolecules 2020, 10, 718. https://doi.org/10.3390/biom10050718
Kaniowski D, Kulik K, Ebenryter-Olbińska K, Wielgus E, Lesnikowski Z, Nawrot B. Metallacarborane Complex Boosts the Rate of DNA Oligonucleotide Hydrolysis in the Reaction Catalyzed by Snake Venom Phosphodiesterase. Biomolecules. 2020; 10(5):718. https://doi.org/10.3390/biom10050718
Chicago/Turabian StyleKaniowski, Damian, Katarzyna Kulik, Katarzyna Ebenryter-Olbińska, Ewelina Wielgus, Zbigniew Lesnikowski, and Barbara Nawrot. 2020. "Metallacarborane Complex Boosts the Rate of DNA Oligonucleotide Hydrolysis in the Reaction Catalyzed by Snake Venom Phosphodiesterase" Biomolecules 10, no. 5: 718. https://doi.org/10.3390/biom10050718
APA StyleKaniowski, D., Kulik, K., Ebenryter-Olbińska, K., Wielgus, E., Lesnikowski, Z., & Nawrot, B. (2020). Metallacarborane Complex Boosts the Rate of DNA Oligonucleotide Hydrolysis in the Reaction Catalyzed by Snake Venom Phosphodiesterase. Biomolecules, 10(5), 718. https://doi.org/10.3390/biom10050718







