Anaerobic Degradation of Bicyclic Monoterpenes in Castellaniella defragrans
Abstract
1. Introduction
2. Results
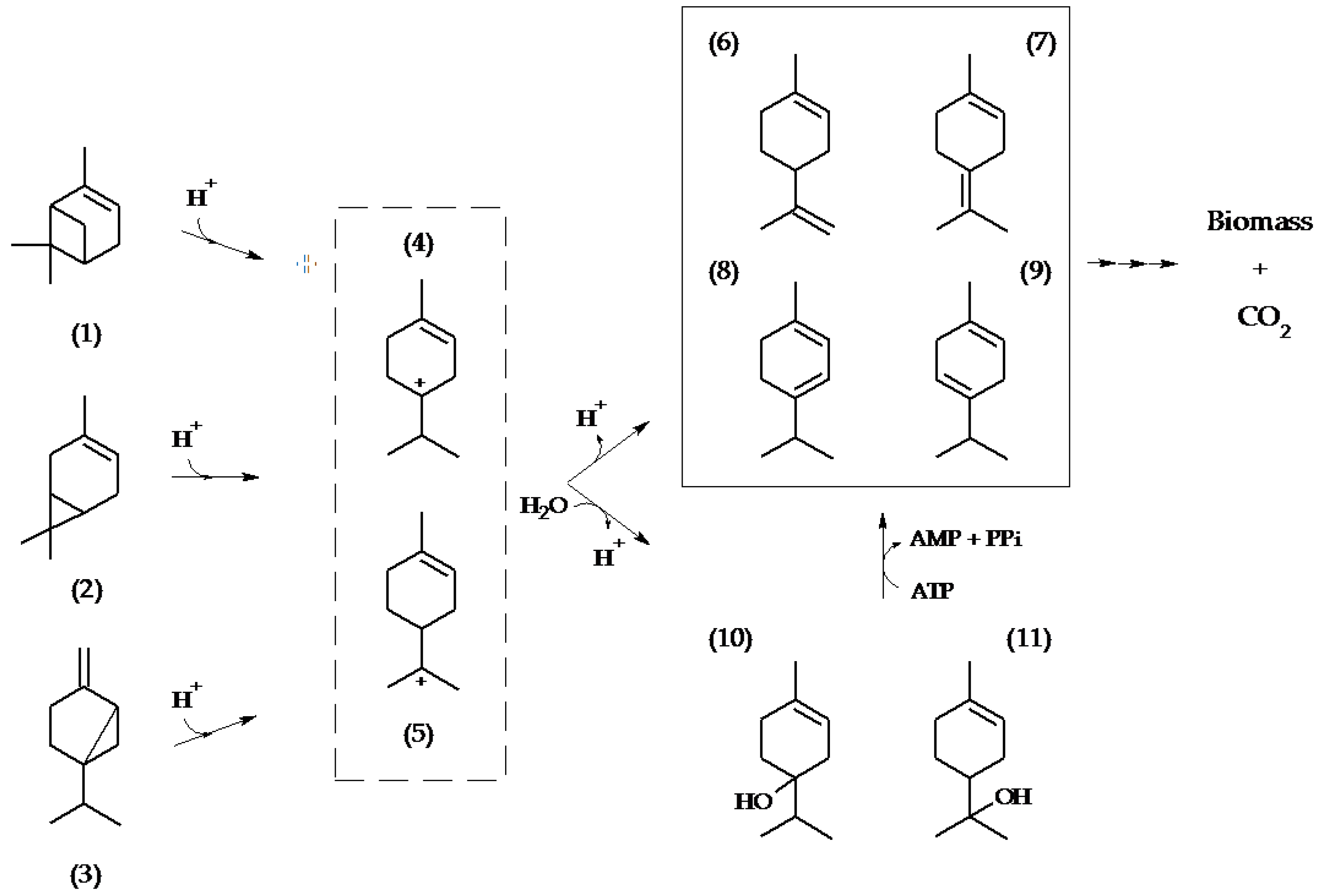
2.1. Metabolite Formation in Cultures
2.2. Metabolite Formation in Cell Lysates
2.3. Differential Proteomics
2.4. Transposon Conjugants Affected Growth on Monoterpenes
3. Discussion
4. Methods
4.1. Bacterial Cells
4.2. Culture Conditions
4.3. Metabolite Analysis
4.4. Enzyme Assays
4.5. Differential Proteomics: Preparation of Cell Lysates, Soluble Protein Fractionation and MALDI-ToF Analysis
4.6. Transposon Mutagenesis
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Kesselmeier, J.; Staudt, M. Biogenic volatile organic compounds (VOC): An overview on emission, physiology and ecology. J. Atmos. Chem. 1999, 33, 23–88. [Google Scholar] [CrossRef]
- Sharkey, T.D.; Wiberley, A.E.; Donohue, A.R. Isoprene emission from plants: Why and how. Ann. Bot. 2008, 101, 5–18. [Google Scholar] [CrossRef] [PubMed]
- Chizzola, R. Regular monoterpenes and sesquiterpenes (essential oils). In Natural Products: Phytochemistry, Botany and Metabolism of Alkaloids, Phenolics and Terpenes; Ramawat, K.G., Mérillon, J.-M., Eds.; Springer: Berlin/Heidelberg, Germany, 2013; pp. 2973–3008. [Google Scholar]
- Lathiere, J.; Hauglustaine, D.A.; Friend, A.D.; De Noblet-Ducoudre, N.; Viovy, N.; Folberth, G.A. Impact of climate variability and land use changes on global biogenic volatile organic compound emissions. Atmos. Chem. Phys. 2006, 6, 2129–2146. [Google Scholar] [CrossRef]
- Laothawornkitkul, J.; Taylor, J.E.; Paul, N.D.; Hewitt, C.N. Biogenic volatile organic compounds in the earth system. New Phytol. 2009, 183, 27–51. [Google Scholar] [CrossRef] [PubMed]
- Guenther, A.B.; Jiang, X.; Heald, C.L.; Sakulyanontvittaya, T.; Duhl, T.; Emmons, L.K.; Wang, X. The Model of Emissions of Gases and Aerosols from Nature version 2.1 (MEGAN2.1): An extended and updated framework for modeling biogenic emissions. Geosci. Model. Dev. 2012, 5, 1471–1492. [Google Scholar] [CrossRef]
- Muller, J.F.; Stavrakou, T.; Wallens, S.; De Smedt, I.; Van Roozendael, M.; Potosnak, M.J.; Rinne, J.; Munger, B.; Goldstein, A.; Guenther, A.B. Global isoprene emissions estimated using MEGAN, ECMWF analyses and a detailed canopy environment model. Atmos. Chem. Phys. 2008, 8, 1329–1341. [Google Scholar] [CrossRef]
- Hayward, S.; Muncey, R.J.; James, A.E.; Halsall, C.J.; Hewitt, C.N. Monoterpene emissions from soil in a sitka spruce forest. Atmos. Environ. 2001, 35, 4081–4087. [Google Scholar] [CrossRef]
- Owen, S.M.; Clark, S.; Pompe, M.; Semple, K.T. Biogenic volatile organic compounds as potential carbon sources for microbial communities in soil from the rhizosphere of Populus tremula. FEMS Microbiol. Lett. 2007, 268, 34–39. [Google Scholar] [CrossRef] [PubMed]
- Seubert, W. Degradation of isoprenoid compounds by micro-organisms. I. Isolation and characterization of an isoprenoid-degrading bacterium, Pseudomonas citronellolis n. sp. J. Bacteriol. 1960, 79, 426–434. [Google Scholar] [PubMed]
- Cantwell, S.G.; Lau, E.P.; Watt, D.S.; Fall, R.R. Biodegradation of acyclic isoprenoids by Pseudomonas species. J. Bacteriol. 1978, 135, 324–433. [Google Scholar] [PubMed]
- Förster-Fromme, K.; Jendrossek, D. Catabolism of citronellol and related acyclic terpenoids in pseudomonads. Appl. Microbiol. Biotechnol. 2010, 87, 859–869. [Google Scholar] [CrossRef] [PubMed]
- Harder, J.; Probian, C. Microbial degradation of monoterpenes in the absence of molecular oxygen. Appl. Environ. Microbiol. 1995, 61, 3804–3808. [Google Scholar] [PubMed]
- Foss, S.; Harder, J. Thauera linaloolentis sp. nov. and Thauera terpenica sp. nov., isolated on oxygen-containing monoterpenes (linalool, menthol, and eucalyptol) and nitrate. Syst. Appl. Microbiol. 1998, 21, 365–373. [Google Scholar] [CrossRef]
- Foss, S.; Heyen, U.; Harder, J. Alcaligenes defragrans sp. nov., description of four strains isolated on alkenoic monoterpenes ((+)-menthene, alpha-pinene, 2-carene, and alpha-phellandrene) and nitrate. Syst. Appl. Microbiol. 1998, 21, 237–244. [Google Scholar] [CrossRef]
- Kämpfer, P.; Denger, K.; Cook, A.M.; Lee, S.T.; Jackel, U.; Denner, E.B.M.; Busse, H.J. Castellaniella gen. nov., to accommodate the phylogenetic lineage of Alcaligenes defragrans, and proposal of Castellaniella defragrans gen. nov., comb. nov and Castellaniella denitrificans sp. nov. Int. J. Syst. Evol. Micr. 2006, 56, 815–819. [Google Scholar] [CrossRef] [PubMed]
- Marmulla, R.; Harder, J. Microbial monoterpene transformations—A review. Front. Microbiol. 2014, 5. [Google Scholar] [CrossRef] [PubMed]
- Brodkorb, D.; Gottschall, M.; Marmulla, R.; Lüddeke, F.; Harder, J. Linalool dehydratase-isomerase, a bifunctional enzyme in the anaerobic degradation of monoterpenes. J. Biol. Chem. 2010, 285, 30436–30442. [Google Scholar] [CrossRef] [PubMed]
- Petasch, J.; Disch, E.M.; Markert, S.; Becher, D.; Schweder, T.; Hüttel, B.; Reinhardt, R.; Harder, J. The oxygen-independent metabolism of cyclic monoterpenes in Castellaniella defragrans 65Phen. BMC Microbiol. 2014, 14, 164. [Google Scholar] [CrossRef] [PubMed]
- Lüddeke, F.; Wulfing, A.; Timke, M.; Germer, F.; Weber, J.; Dikfidan, A.; Rahnfeld, T.; Linder, D.; Meyerdierks, A.; Harder, J. Geraniol and geranial dehydrogenases induced in anaerobic monoterpene degradation by Castellaniella defragrans. Appl. Environ. Microbiol. 2012, 78, 2128–2136. [Google Scholar] [CrossRef] [PubMed]
- Harder, J.; Marmulla, R. Catabolic pathways and enzymes involved in the anaerobic degradation of terpenes. In Anaerobic Utilization of Hydrocarbons, Oils, and Lipids; Boll, M., Ed.; Springer International Publishing: Cham, Switzerland, 2017; pp. 1–15. [Google Scholar] [CrossRef]
- Weidenweber, S.; Marmulla, R.; Ermler, U.; Harder, J. X-ray structure of linalool dehydratase/isomerase from Castellaniella defragrans reveals enzymatic alkene synthesis. FEBS Lett. 2016, 590, 1375–1383. [Google Scholar] [CrossRef] [PubMed]
- Lüddeke, F.; Harder, J. Enantiospecific (S)-(+)-linalool formation from beta-myrcene by linalool dehydratase-isomerase. Z. Naturfr. C 2011, 66, 409–412. [Google Scholar] [CrossRef]
- Larsen, R.A.; Wilson, M.M.; Guss, A.M.; Metcalf, W.W. Genetic analysis of pigment biosynthesis in Xanthobacter autotrophicus Py2 using a new, highly efficient transposon mutagenesis system that is functional in a wide variety of bacteria. Arch. Microbiol. 2002, 178, 193–201. [Google Scholar] [CrossRef] [PubMed]
- Marmulla, R.; Cala, E.P.; Markert, S.; Schweder, T.; Harder, J. The anaerobic linalool metabolism in Thauera linaloolentis 47Lol. BMC Microbiol. 2016, 16, 76. [Google Scholar] [CrossRef] [PubMed]
- Yoo, S.K.; Day, D.F. Bacterial metabolism of α- and β-pinene and related monoterpenes by Pseudomonas sp. strain PIN. Process. Biochem. 2002, 37, 739–745. [Google Scholar] [CrossRef]
- Traas, P.C. Advances in the chemistry of some interesting cyclic monoterpene alcohols. In Fragrance Chemistry: The Science of the Sense of Smell; Heimer, E.T., Ed.; Academic Press: San Diego, CA, USA, 1982; pp. 165–219. [Google Scholar]
- Shukal, O.P.; Bhattacharyya, P.K. Microbiological transformations of terpenes: Part XI—Pathways of degradation of α-& β-pinenes in a soil pseudomonad (PL-strain). Indian J. Biochem. 1968, 5, 92–101. [Google Scholar]
- Cheng, Z.W.; Sun, P.F.; Jiang, Y.F.; Zhang, L.L.; Chen, J.M. Kinetic analysis and bacterium metabolization of alpha-pinene by a novel identified Pseudomonas sp. strain. J. Environ. Sci. (China) 2012, 24, 1806–1815. [Google Scholar] [CrossRef]
- Narushima, H.; Omori, T.; Minoda, Y. Microbial transformation of alpha-pinene. Eur. J. Appl. Microbiol. 1982, 16, 174–178. [Google Scholar] [CrossRef]
- Lüddeke, F.; Dikfidan, A.; Harder, J. Physiology of deletion mutants in the anaerobic beta-myrcene degradation pathway in Castellaniella defragrans. BMC Microbiol. 2012, 12, 192. [Google Scholar] [CrossRef] [PubMed]
- Metcalf, W.W.; Jiang, W.; Daniels, L.L.; Kim, S.K.; Haldimann, A.; Wanner, B.L. Conditionally replicative and conjugative plasmids carrying lacZα for cloning, mutagenesis, and allele replacement in bacteria. Plasmid 1996, 35, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef] [PubMed]
- Martin-Platero, A.M.; Valdivia, E.; Maqueda, M.; Martinez-Bueno, M. Fast, convenient, and economical method for isolating genomic DNA from lactic acid bacteria using a modification of the protein “salting-out” procedure. Anal. Biochem. 2007, 366, 102–104. [Google Scholar] [CrossRef] [PubMed]
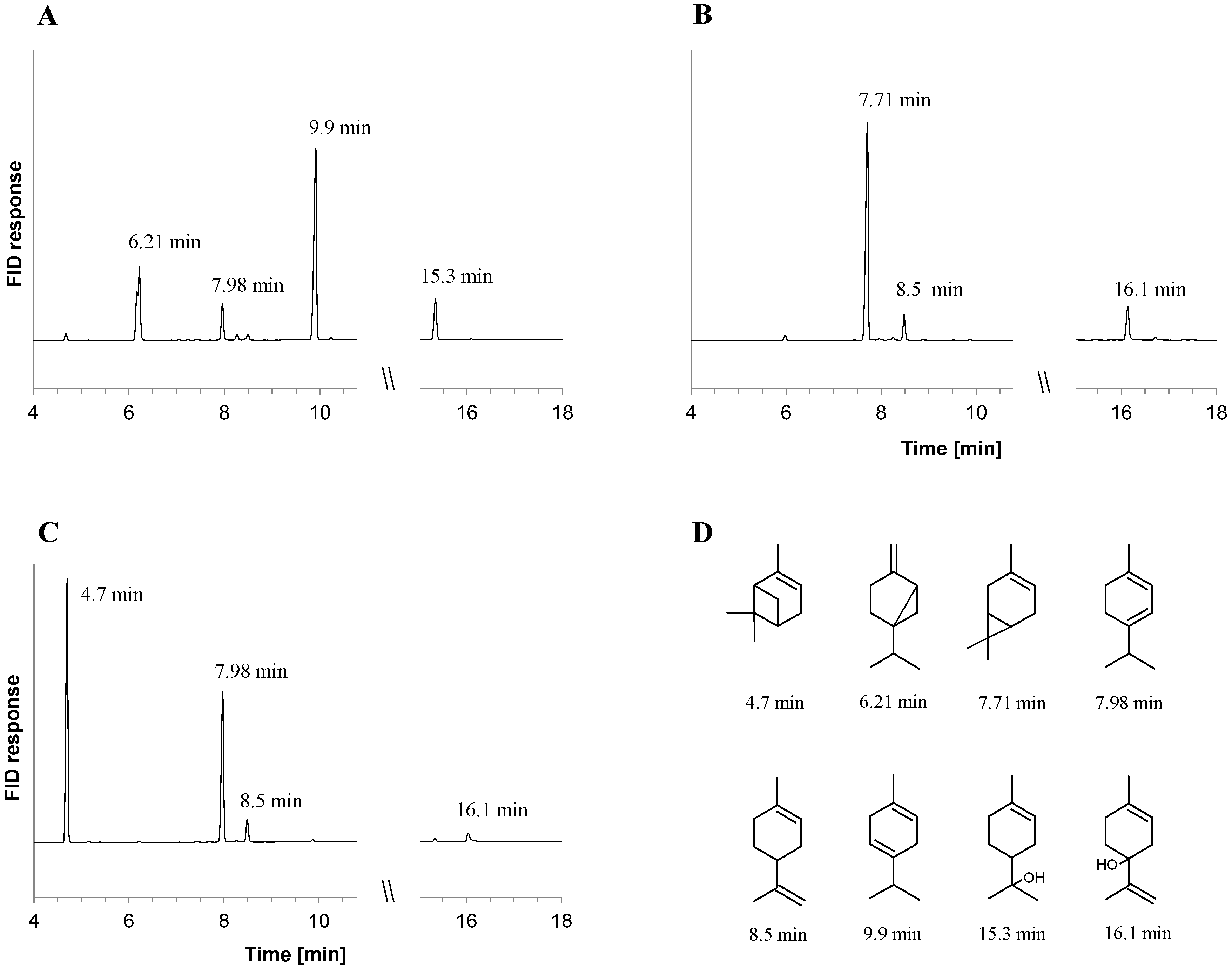
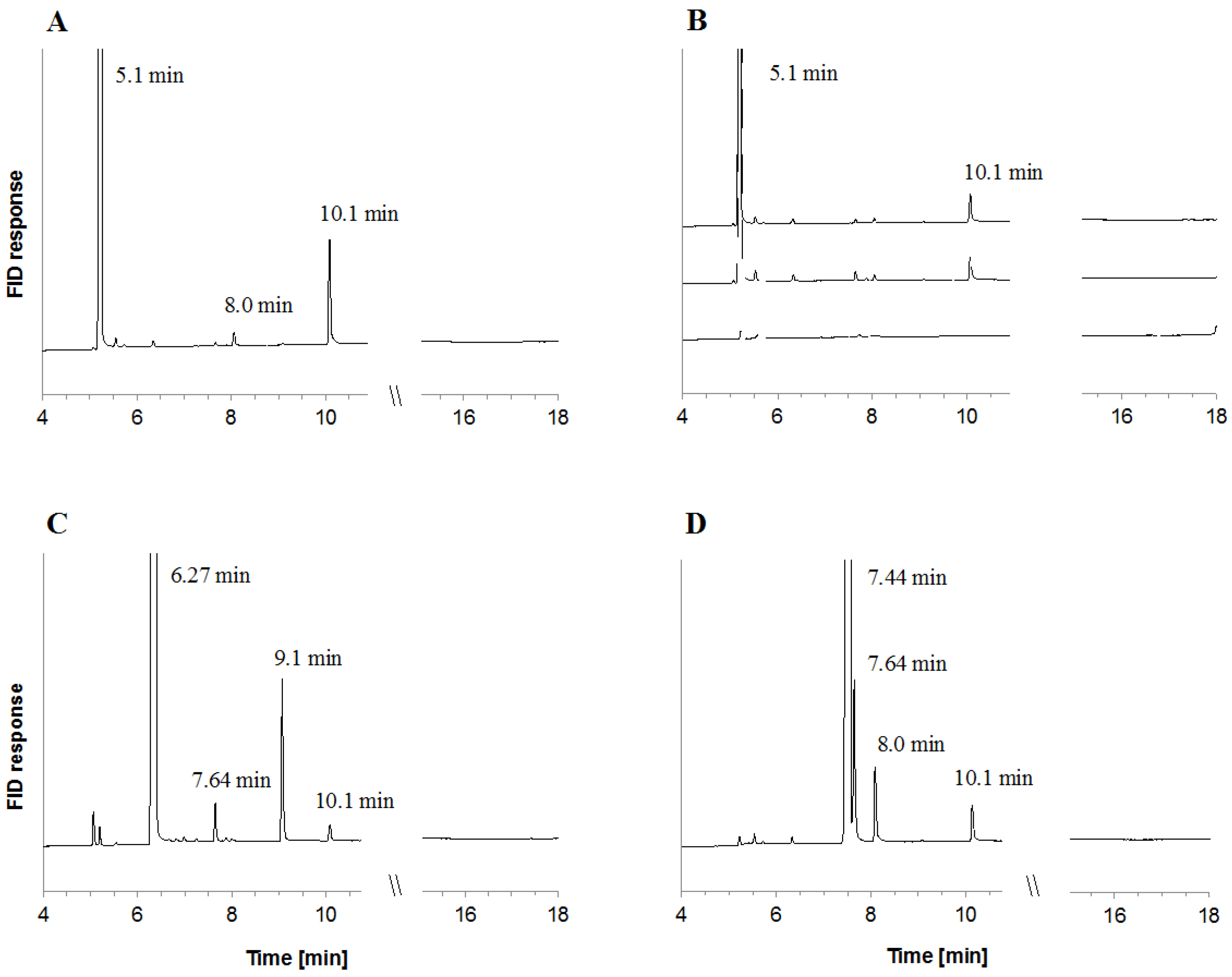
| Monoterpene Co-Substrate | Monoterpene (mM), (Monoterpene Consumed (%)) | |
|---|---|---|
| Wild-Type | ΔctmAB | |
| sabinene | 1.6 ± 0.6 (47%) | 1.44 ± 0.11 (52%) |
| 3-carene | 2.65 ± 0.15 (12%) | 2.68 ± 0.02 (11%) |
| α-pinene | 2.95 ± 0.03 (2%) | 2.69 ± 0.06 (10%) |
| Accession Number | Annotation | Growth Substrate | ||
|---|---|---|---|---|
| L | S | P | ||
| CDM22609 | Translation elongation factor Tu | + | - | - |
| CDM22610 | Translation elongation factor G | + | + | + |
| CDM22641 | Translation elongation factor Tu | + | - | - |
| CDM22907 | Copper-containing nitrite reductase | + | + | + |
| CDM23001 | Aconitate hydratase | + | + | + |
| CDM23572 | Methylmalonate-semialdehyde dehydrogenase | + | + | + |
| CDM23679 | Heat shock protein 60 family chaperone GroEL | + | + | + |
| CDM23795 | Chaperone protein DnaK | + | + | + |
| CDM23915 | 3-methylmercaptopropionyl-CoA dehydrogenase DmdC | + | + | + |
| CDM24415 | Hypothetical protein | + | + | + |
| CDM24550 | Glutamine synthetase type I | + | + | + |
| CDM24733 | Isoleucyl-tRNA synthetase | + | + | + |
| CDM24892 | Membrane alanine aminopeptidase N | - | + | + |
| CDM24998 | Transcription termination protein NusA | + | - | + |
| CDM25009 | Acetoacetyl-CoA reductase | + | + | + |
| CDM25072 | Citrate synthase | + | + | + |
| CDM25085 | Aconitate hydratase 2 | + | + | + |
| CDM25210 | Polyribonucleotide nucleotidyltransferase | (+) | + | + |
| CDM25241 | Acyl-CoA dehydrogenase | + | + | + |
| CDM25246 | 3-hydroxyacyl-CoA dehydrogenase | (+) | + | + |
| CDM25251 | 4-isopropenyl-2-oxo-cyclohexane-1-carboxyl-CoA hydrolase MrcE | (+) | + | + |
| CDM25253 | 2,4-enoyl-CoA reductase MrcC | + | + | + |
| CDM25259 | RND efflux transporter component | + | + | + |
| CDM25260 | RND efflux transporter component | + | + | + |
| CDM25267 | Geraniol dehydrogenase GeoA | (+) | + | + |
| CDM25281 | Geranial dehydrogenase GeoB | - | + | + |
| CDM25285 | NADH:ferredoxin oxidoreductase CtmF | + | + | + |
| CDM25289 | Limonene dehydrogenase CtmB | + | + | + |
| CDM25290 | Limonene dehydrogenase CtmA | + | + | + |
| CDM25340 | Branched-chain amino acid aminotransferase | - | + | + |
| CDM25770 | Protein export cytoplasm protein SecA | - | - | + |
| CDM25844 | Heat shock protein 60 family chaperone GroEL | + | + | + |
| CDM26013 | Glutamate aspartate periplasmic binding protein precursor GltI | + | + | + |
| Affected Protein (acc. no.) | Gen Length [bp] | Insertion Position [bp] | Annotation | Growth on Solid Media | Growth in Liquid Media | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| L | S | P | C | L | S | P | C | ||||
| Wild-Type | - | - | - | + | + | + | + | + | + | + | + |
| CDM22783 | 1217 | 1023 | Cystathionine beta-lyase | - | - | - | - | - | - | - | - |
| CDM23032 | 702 | 793 | Glutamate ABC transporter permease (periplasmic component) | - | - | - | - | + | - | - | - |
| CDM23105 | 930 | 627 | High-affinity branched-chain amino acid transport system (permease protein) LivH | - | - | - | - | - | - | - | - |
| CDM23105 | 930 | 161 | High-affinity branched-chain amino acid transport system (permease protein) LivH | (+) | (+) | - | - | + | - | - | - |
| CDM23279 | 2793 | 411 | Inositol phosphate phosphatase | (+) | - | - | - | - | (+) | (+) | - |
| CDM23452 | 1230 | 150 | Fused spore maturation proteins A and B | (+) | - | - | (+) | (+) | (+) | - | (+) |
| CDM23960 | 1230 | 765 | Glycolate dehydrogenase (iron-sulfur subunit) | - | - | - | - | + | - | - | (+) |
| CDM24063 | 279 | 243 | HigB toxin protein | - | - | - | (+) | (+) | - | - | (+) |
| CDM24591 | 3630 | 889 | O-antigen biosynthesis protein | (+) | - | - | - | (+) | - | - | - |
| CDM24600 | 1029 | 1012 | Glycosyl transferase, family 2 | - | - | - | (+) | + | + | - | - |
| CDM24629 | 1116 | 11 | ABC transport system (permease component) YbhR | - | - | - | - | - | - | - | (+) |
| CDM24629 | 1116 | 1104 | ABC transport system (permease component) YbhR | - | - | - | - | - | - | - | - |
| CDM24678 | 897 | 388 | Permease of the drug/metabolite transporter superfamily | - | - | (+) | (+) | - | - | (+) | (+) |
| CDM24706 | 3387 | 2695 | Trehalose synthase | - | - | - | - | - | - | - | - |
| CDM24919 | 3840 | 1904 | Putative ATP-dependent helicase | - | - | - | - | - | - | - | - |
| CDM24922 | 1356 | 1253 | N-acetylglutamate synthase | - | - | (+) | (+) | (+) | + | - | - |
| CDM25080 | 900 | 88 | Methylisocitrate lyase | (+) | - | - | - | (+) | + | + | - |
| CDM25154 | 525 | 334 | Hypothetical protein | (+) | - | - | - | + | + | - | + |
| CDM25252 | 771 | 497 | 2-hydroxy-4-isopropenyl-cyclohexane-1-carboxyl-CoA dehydrogenase MrcD | (+) | - | - | - | - | - | - | - |
| CDM25285 | 1230 | 492 | NADH:ferredoxin oxidoreductase CtmF | - | - | - | - | - | - | - | - |
| CDM25322 | 999 | 468 | Hypothetical protein | - | (+) | - | - | + | (+) | (+) | - |
| CDM25752 | 849 | 330 | Glucose-1-phosphate thymidylyltransferase | (+) | + | - | - | + | + | - | - |
| CDM25923 | 777 | 223 | Enoyl-CoA hydratase | - | - | - | - | - | - | - | - |
| CDM26084 | 1514 | 1428 | Putative transposase | - | - | - | (+) | (+) | - | (+) | + |
| Non-coding region | - | −500 | Upstream of CDM22657: single-stranded DNA-binding protein | - | - | - | (+) | + | + | (+) | - |
| Non-coding region | - | −10 | Upstream of CDM22986: phenylacetate-CoA ligase | (+) | - | - | - | + | + | - | - |
| Non-coding region | - | 26 | Downstream of CDM23018: auxin efflux transporter | - | - | - | (+) | - | - | - | + |
| Non-coding region | - | −387 | Upstream of CDM23059: hypothetical protein | - | - | - | - | - | - | - | - |
| Non-coding region | - | −97 | Upstream of CDM23110: glutamate ABC transporter (periplasmic component) | - | - | - | - | + | - | - | - |
| Non-coding region | - | −281 | Upstream of CDM23992: hypothetical transcriptional regulator | - | (+) | - | - | (+) | (+) | - | - |
| Non-coding region | - | −559 | Upstream of CDM23992: putative transposase | (+) | - | - | (+) | + | - | - | - |
| Non-coding region | - | −270 | Upstream of CDM23993: putative transcriptional regulator | (+) | - | - | - | + | + | - | - |
| Non-coding region | - | −78 | Usptream of CDM25290: limonene dehydrogenase CtmA | (+) | (+) | (+) | - | - | (+) | - | - |
| Non-coding region | - | −40 | Upstream of CDM25994: thiamin-phosphate pyrophosphatase | - | - | - | - | - | - | - | - |
| Non-coding region | - | −295 | Upstream of CDM26087: putative transposase | (+) | - | - | - | + | - | - | - |
| Strain | Genotype | Source |
|---|---|---|
| C. defragrans 65Phen | Rifampicin-resistant (RaR) | [31] |
| C. defragrans 65Phen ΔctmAB | 65Phen, RaR, ΔctmAB | This work |
| E. coli BW20767 | RP4-2-tet::Mu-1, kan::Tn7 integrant, leu-63::IS10, recA1, creC510, hsdR17, endA1, zbf-5, uidA, (ΔMlul)::pir+ thi | [32] |
| Plasmid | ||
| pRL27 | Tn5 with KmR, R6K ori, oriT, RP4, tnpA | [24] |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Puentes-Cala, E.; Liebeke, M.; Markert, S.; Harder, J. Anaerobic Degradation of Bicyclic Monoterpenes in Castellaniella defragrans. Metabolites 2018, 8, 12. https://doi.org/10.3390/metabo8010012
Puentes-Cala E, Liebeke M, Markert S, Harder J. Anaerobic Degradation of Bicyclic Monoterpenes in Castellaniella defragrans. Metabolites. 2018; 8(1):12. https://doi.org/10.3390/metabo8010012
Chicago/Turabian StylePuentes-Cala, Edinson, Manuel Liebeke, Stephanie Markert, and Jens Harder. 2018. "Anaerobic Degradation of Bicyclic Monoterpenes in Castellaniella defragrans" Metabolites 8, no. 1: 12. https://doi.org/10.3390/metabo8010012
APA StylePuentes-Cala, E., Liebeke, M., Markert, S., & Harder, J. (2018). Anaerobic Degradation of Bicyclic Monoterpenes in Castellaniella defragrans. Metabolites, 8(1), 12. https://doi.org/10.3390/metabo8010012






