The Redox Status of Cancer Cells Supports Mechanisms behind the Warburg Effect
Abstract
:1. Introduction
2. Results
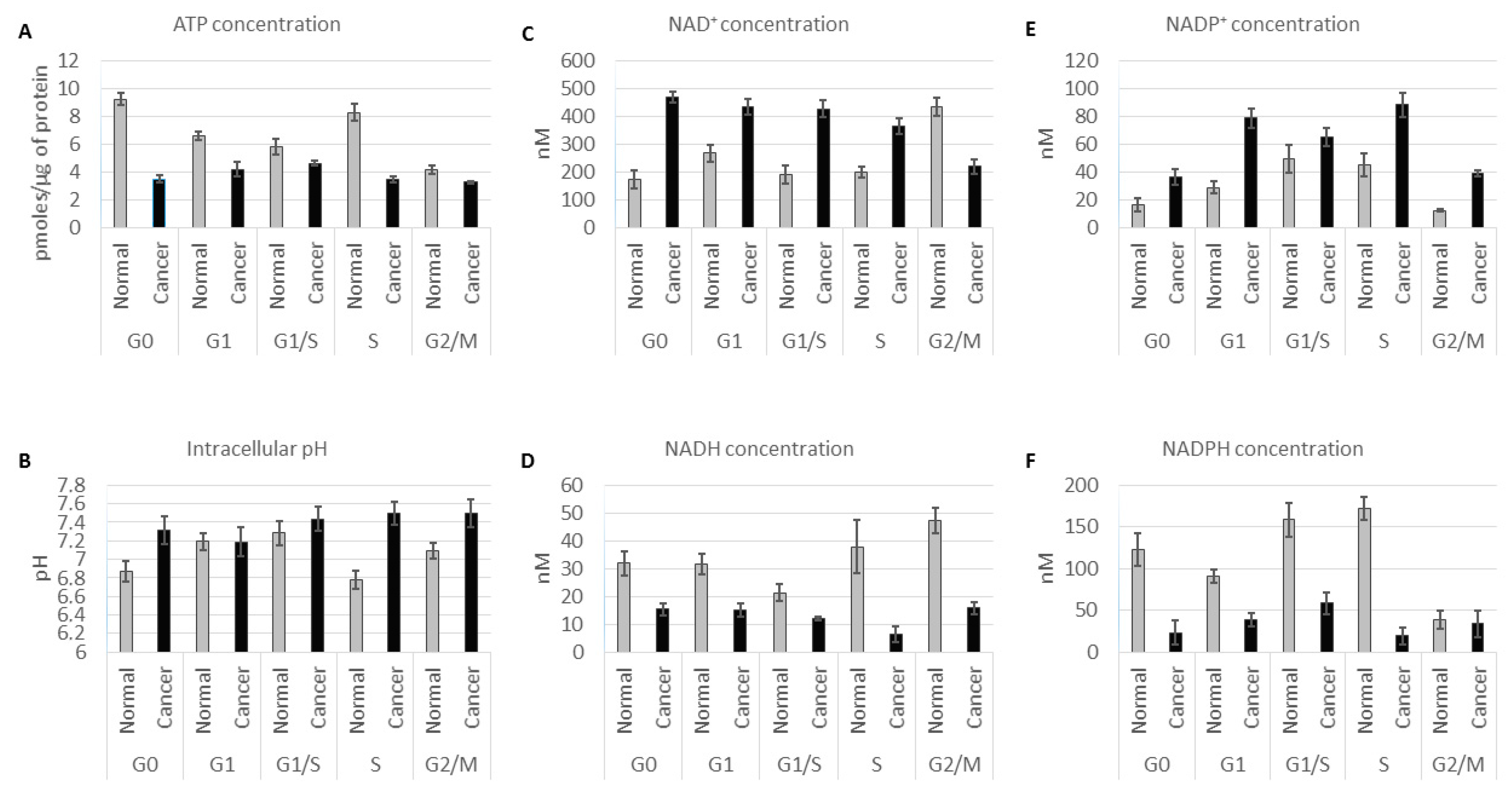
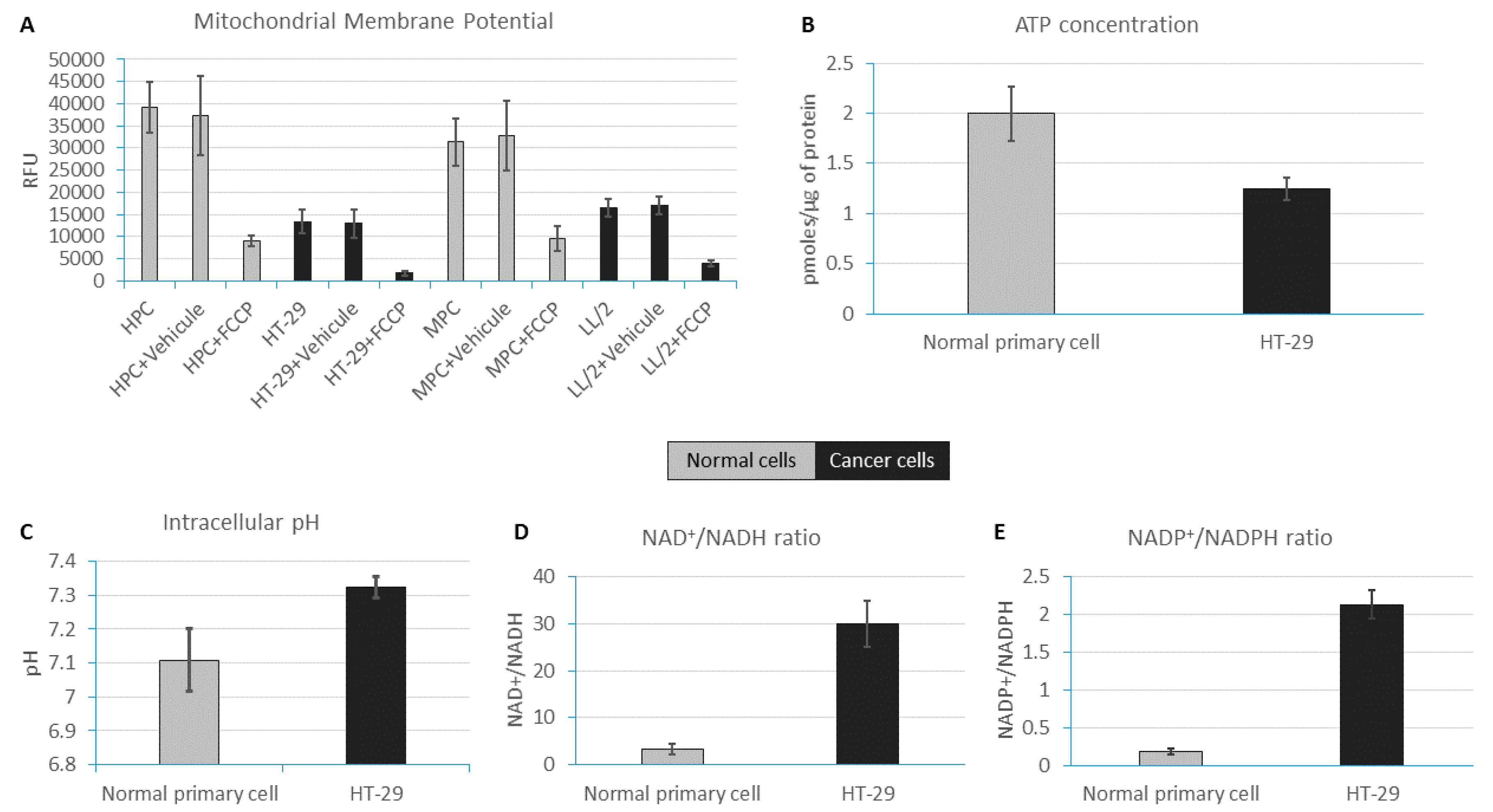
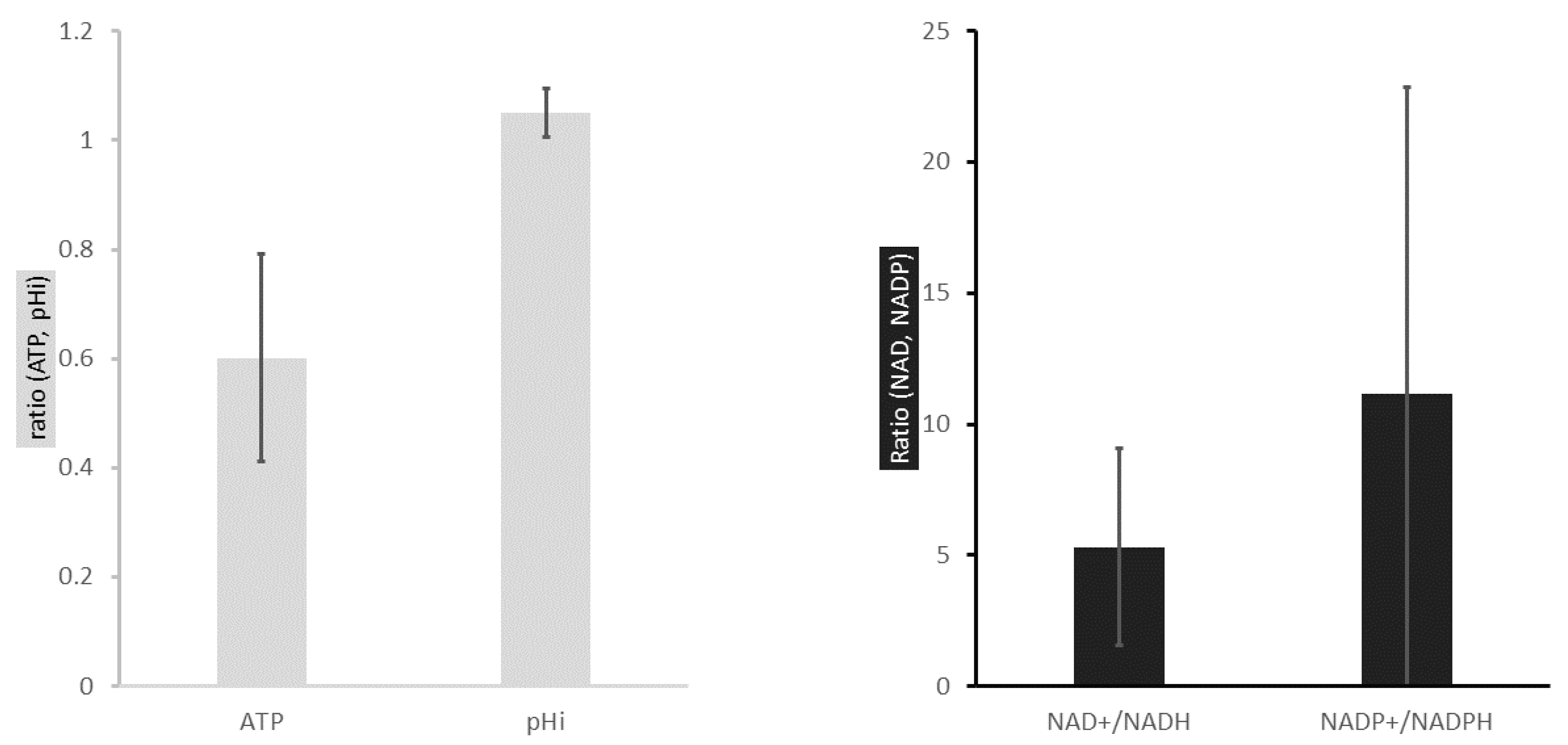
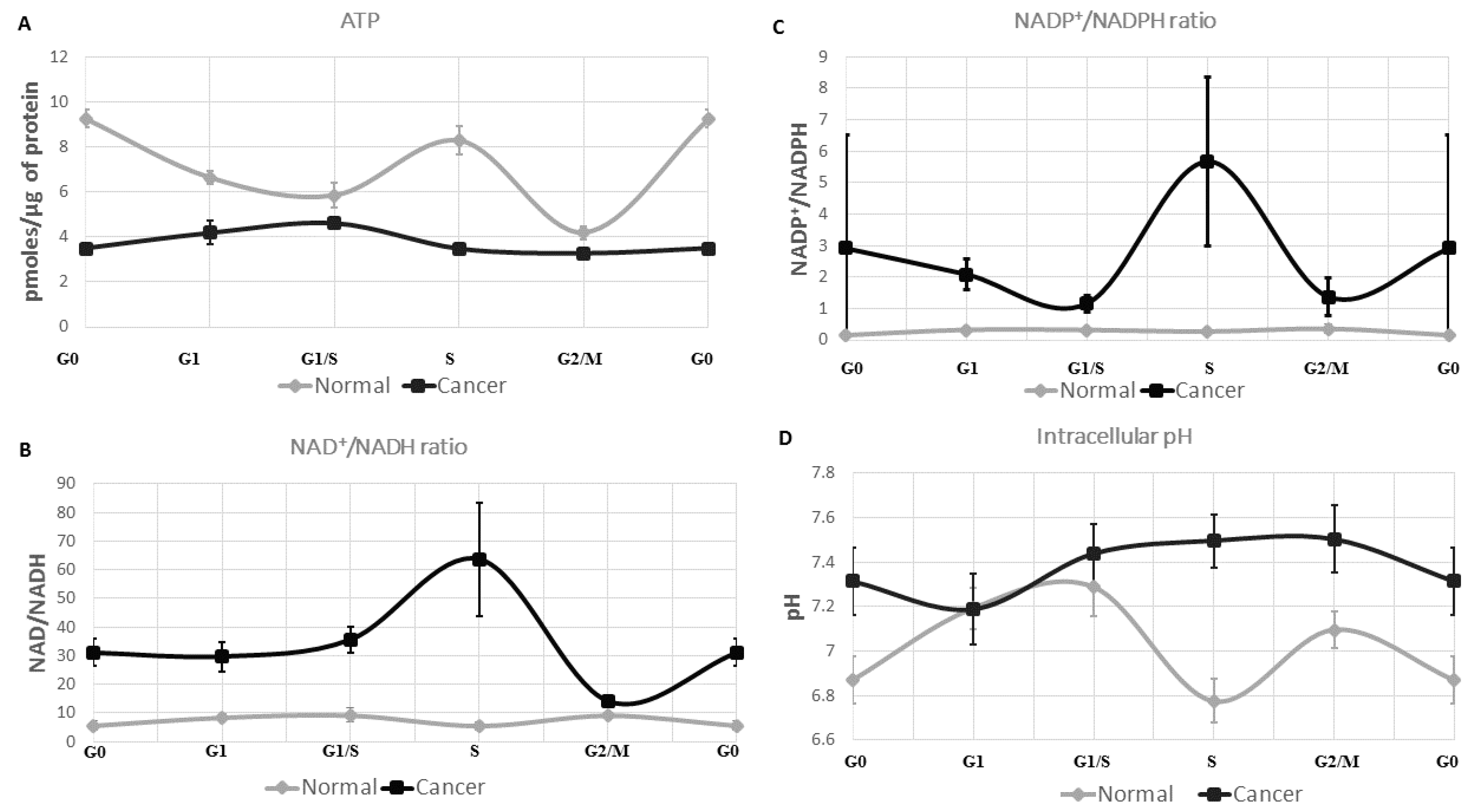
2.1. ATP Concentration Is Reduced in Colon Cancer Cells
2.2. Colon Cancer Cells Have a Reductive Energetic Status
2.3. The Intracellular pH of Cancer Cells Is Alkaline
2.4. Reduced Mitochondrial Membrane Potential in Immortal Cancer Cell Lines
3. Discussion
4. Materials and Methods
4.1. Cell Suspension Preparation and Centrifugal Elutriation
4.2. Fluorescence and Luminescence Measurements
4.3. pHi Measurement
4.4. Reduced Mitochondrial Membrane Potential in Immortal Cancer Cell Lines
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
Appendix
References
- Noor, E.; Eden, E.; Milo, R.; Alon, U. Central carbon metabolism as a minimal biochemical walk between precursors for biomass and energy. Mol. Cell 2010, 39, 809–820. [Google Scholar] [CrossRef] [PubMed]
- Da Veiga Moreira, J.; Peres, S.; Steyaert, J.-M.; Bigan, E.; Paulevé, L.; Nogueira, M.L.; Schwartz, L. Cell cycle progression is regulated by intertwined redox oscillators. Theor. Biol. Med. Model. 2015, 12, 10. [Google Scholar] [CrossRef] [PubMed]
- Tyson, J.J. Biochemical oscillations. In Computational Cell Biology; Springer: Berlin/Heidelberg, Germany, 2002; pp. 230–260. [Google Scholar]
- Yu, F.X.; Dai, R.P.; Goh, S.R.; Zheng, L.; Luo, Y. Logic of a mammalian metabolic cycle: An oscillated NAD+/NADH redox signaling regulates coordinated histone expression and S-phase progression. Cell Cycle 2009, 8, 773–779. [Google Scholar] [CrossRef] [PubMed]
- Diaz-Moralli, S.; Tarrado-Castellarnau, M.; Miranda, A.; Cascante, M. Targeting cell cycle regulation in cancer therapy. Pharmacol. Ther. 2013, 138, 255–271. [Google Scholar] [CrossRef] [PubMed]
- Norbury, C.; Nurse, P. Animal cell cycles and their control. Annu. Rev. Biochem. 1992, 6, 441–468. [Google Scholar] [CrossRef] [PubMed]
- Nurse, P. A long twentieth century of the cell cycle and beyond. Cell 2000, 100, 71–78. [Google Scholar] [CrossRef]
- Burhans, W.C.; Heintz, N.H. The cell cycle is a redox cycle: Linking phase-specific targets to cell fate. Free Radic. Biol. Med. 2009, 4, 1282–1293. [Google Scholar] [CrossRef] [PubMed]
- Chiu, J.; Dawes, W. Redox control of cell proliferation. Trends Cell Biol. 2012, 20, 592–601. [Google Scholar] [CrossRef] [PubMed]
- Menon, S.G.; Goswami, P.C. A redox cycle within the cell cycle: Ring in the old with the new. Oncogene 2007, 26, 1101–1109. [Google Scholar] [CrossRef] [PubMed]
- Sarsour, E.H.; Kumar, M.G.; Chaudhuri, L.; Kalen, A.L.; Goswami, P.C. Redox control of the cell cycle in health and disease. Antioxid. Redox Signal. 2009, 11, 2985–3011. [Google Scholar] [CrossRef] [PubMed]
- DeBerardinis, R.J.; Lum, J.J.; Hatzivassiliou, G.; Thompson, C.B. The biology of cancer: Metabolic reprogramming fuels cell growth and proliferation. Cell Metab. 2008, 7, 11–20. [Google Scholar] [CrossRef] [PubMed]
- Warburg, O. The metabolism of carcinoma cells. J. Cancer Res. 1925, 9, 148–163. [Google Scholar] [CrossRef]
- Warburg, O. On the origin of cancer cells. Science 1956, 123, 309–314. [Google Scholar] [CrossRef] [PubMed]
- Fabregat, I.; Vitorica, J.; Satrustegui, J.; Machado, A. The pentose phosphate cycle is regulated by NADPH/NADP ratio in rat liver. Arch. Biochem. Biophys. 1985, 236, 110–118. [Google Scholar] [CrossRef]
- Fabregat, I.; Revilla, E.; Machado, A. Short-term control of the pentose phosphate cycle by insulin could be modulated by the NADPH/NADP ratio in rat adipocytes and hepatocytes. Biochem. Biophy. Res. Commun. 1987, 146, 920–925. [Google Scholar] [CrossRef]
- Revilla, E.; Fabregat, I.; Santa-María, C.; Machado, A. The NADPH-producing pathways (pentose phosphate and malic enzyme) are regulated by the NADPH consumption in rat mammary gland. Biochem. Int. 1987, 14, 957–962. [Google Scholar] [PubMed]
- McBride, H.M.; Neuspiel, M.; Wasiak, S. Mitochondria: More than just a powerhouse. Curr. Biol. 2006, 16, 551–560. [Google Scholar] [CrossRef] [PubMed]
- Scalettar, B.A.; Abney, J.R.; Hackenbrock, C.R. Dynamics, structure, and function are coupled in the mitochondrial matrix. Proc. Natl. Acad. Sci. USA 1991, 88, 8057–8061. [Google Scholar] [CrossRef] [PubMed]
- Hackenbrock, C.R. Ultrastructural bases for metabolically linked mechanical activity in mitochondria II. Electron transport-linked ultrastructural transformations in mitochondria. J. Cell Biol. 1968, 37, 345–369. [Google Scholar] [CrossRef] [PubMed]
- Mitra, K.; Wunder, C.; Roysam, B.; Lin, G.; Lippincott-Schwartz, J. A hyperfused mitochondrial state achieved at G1–S regulates cyclin E buildup and entry into S phase. Proc. Natl. Acad. Sci. USA 2009, 106, 11960–11965. [Google Scholar] [CrossRef] [PubMed]
- Christen, R.; Schackmann, R.W.; Shapiro, B.M. Metabolism of sea urchin sperm. Interrelationships between intracellular pH, ATPase activity, and mitochondrial respiration. J. Biol. Chem. 1983, 258, 5392–5399. [Google Scholar] [PubMed]
- Jones, R.G.; Plas, D.R.; Kubek, S.; Buzzai, M.; Mu, J.; Xu, Y.; Birnbaum, M.J.; Thompson, C.B. AMP-activated protein kinase induces a p53-dependent metabolic checkpoint. Mol. Cell 2005, 18, 283–293. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, L.; Buhler, L.; Icard, P.; Lincet, H.; Steyaert, J.M. Metabolic treatment of cancer: Intermediate results of a prospective case series. Anticancer Res. 2014, 34, 973–980. [Google Scholar] [PubMed]
- Pouyssegur, J.; Franchi, A.; L’allemain, G.; Paris, S. Cytoplasmic pH, a key determinant of growth factor-induced DNA synthesis in quiescent fibroblasts. FEBS Lett. 1985, 190, 115–119. [Google Scholar] [CrossRef]
- Aerts, R.J.; Durston, A.J.; Moolenaar, W.H. Cytoplasmic pH and the regulation of the Dictyostelium cell cycle. Cell 1985, 43, 653–657. [Google Scholar] [CrossRef]
- Finkel, T.; Hwang, P.M. The Krebs cycle meets the cell cycle: Mitochondria and the G1–S transition. Proc. Natl. Acad. Sci. USA 2009, 106, 11825–11826. [Google Scholar] [CrossRef] [PubMed]
- Westrate, L.M.; Sayfie, A.D.; Burgenske, D.M.; MacKeigan, J.P. Persistent mitochondrial hyperfusion promotes G2/M accumulation and caspase-dependent cell death. PLoS ONE 2014, 9, e91911. [Google Scholar] [CrossRef] [PubMed]
- Toyama, E.Q.; Herzig, S.; Courchet, J.; Lewis, T.L., Jr.; Losón, O.C.; Hellberg, K.; Young, N.P.; Chen, H.; Polleux, F.; Chan, D.C.; et al. AMP-activated protein kinase mediates mitochondrial fission in response to energy stress. Science 2016, 351, 275–281. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Yang, C.; Yang, Z.; Zhang, D.; Ma, X.; Mills, G.; Liu, Z. Homeostasis of redox status derived from glucose metabolic pathway could be the key to understanding the Warburg effect. Am. J. Cancer Res. 2015, 5, 928–944. [Google Scholar] [PubMed]
- Israël, M.; Schwartz, L. The metabolic advantage of tumor cells. Mol. Cancer 2011, 10, 1–12. [Google Scholar] [CrossRef] [PubMed]
- McBrian, M.A.; Behbahan, I.S.; Ferrari, R.; Su, T.; Huang, T.W.; Li, K.; Hong, C.S.; Christofk, H.R.; Vogelauer, M.; Seligson, D.B.; et al. Histone acetylation regulates intracellular pH. Mol. Cell 2013, 49, 310–321. [Google Scholar] [CrossRef] [PubMed]
- Kurdistani, S.K. Chromatin: A capacitor of acetate for integrated regulation of gene expression and cell physiology. Curr. Opin. Genet. Dev. 2014, 26, 53–58. [Google Scholar] [CrossRef] [PubMed]
- Kurdistani, S.K.; Grunstein, M. Histone acetylation and deacetylation in yeast. Nat. Rev. Mol. Cell Biol. 2003, 4, 276–284. [Google Scholar] [CrossRef] [PubMed]
- Imai, S.I.; Armstrong, C.M.; Kaeberlein, M.; Guarente, L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature 2000, 403, 795–800. [Google Scholar] [PubMed]
- Busa, W.B.; Nuccitelli, R. Metabolic regulation via intracellular pH. Am. J. Physiol. 1984, 246, 409–438. [Google Scholar]
- Abolhassani, M.; Aloulou, N.; Chaumette, M.T.; Aparicio, T.; Martin-Garcia, N.; Mansour, H.; Le Gouvello, S.; Delchier, J.C.; Sobhani, I. Leptin receptor–related immune response in colorectal tumors: The role of colonocytes and interleukin-8. Cancer Res. 2008, 68, 9423–9432. [Google Scholar] [CrossRef] [PubMed]
- Michelakis, E.D.; Abdulkarim, B. Metabolic modulation of glioblastoma with dichloroacetate. Sci. Transl. Med. 2010, 2. [Google Scholar] [CrossRef] [PubMed]
- Kinnaird, A.; Michelakis, E. 297 Dichloroacetate is a novel therapy for renal cell carcinoma. J. Urol. 2012, 187, e120–e121. [Google Scholar] [CrossRef]
- Kafara, P.; Icard, P.; Guillamin, M.; Schwartz, L.; Lincet, H. Lipoic acid decreases Mcl-1, Bcl-x L and up regulates Bim on ovarian carcinoma cells leading to cell death. J. Ovarian Res. 2015, 8, 36. [Google Scholar] [CrossRef] [PubMed]

© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Moreira, J.D.V.; Hamraz, M.; Abolhassani, M.; Bigan, E.; Pérès, S.; Paulevé, L.; Nogueira, M.L.; Steyaert, J.-M.; Schwartz, L. The Redox Status of Cancer Cells Supports Mechanisms behind the Warburg Effect. Metabolites 2016, 6, 33. https://doi.org/10.3390/metabo6040033
Moreira JDV, Hamraz M, Abolhassani M, Bigan E, Pérès S, Paulevé L, Nogueira ML, Steyaert J-M, Schwartz L. The Redox Status of Cancer Cells Supports Mechanisms behind the Warburg Effect. Metabolites. 2016; 6(4):33. https://doi.org/10.3390/metabo6040033
Chicago/Turabian StyleMoreira, Jorgelindo Da Veiga, Minoo Hamraz, Mohammad Abolhassani, Erwan Bigan, Sabine Pérès, Loïc Paulevé, Marcel Levy Nogueira, Jean-Marc Steyaert, and Laurent Schwartz. 2016. "The Redox Status of Cancer Cells Supports Mechanisms behind the Warburg Effect" Metabolites 6, no. 4: 33. https://doi.org/10.3390/metabo6040033
APA StyleMoreira, J. D. V., Hamraz, M., Abolhassani, M., Bigan, E., Pérès, S., Paulevé, L., Nogueira, M. L., Steyaert, J.-M., & Schwartz, L. (2016). The Redox Status of Cancer Cells Supports Mechanisms behind the Warburg Effect. Metabolites, 6(4), 33. https://doi.org/10.3390/metabo6040033





