Lipidomics of Glycosphingolipids
Abstract
:Abbreviations
| GSL nomenclature: | [1] |
| Ac: | Acetyl |
| APCI: | Atmospheric Pressure Chemical Ionization |
| APPI: | Atmospheric Pressure Photo-Ionization |
| Cer: | Ceramide (N-Acylsphingosine) |
| CID: | Collision induced dissociation |
| DESI: | Desorption electrospray ionization |
| ESI: | Electrospray ionization |
| Forssman-GSL: | GalNAcα1,3GalNAcβ1,3Galα1,4Galβ1,4Glcβ1,1′Cer |
| FT-ICR: | Fourier-Transform-Ion-Cyclotron |
| GA1: | Galβ1,3GalNAcβ1,4Galβ1,4Glcβ1,1′Cer |
| GA2: | GalNAcβ1,4Galβ1,4Glcβ1,1′Cer |
| Gal: | D-Galactose |
| GalCer: | β-Galactosylceramide |
| GalNAc: | 2-Deoxy-2-N-Acetyl-D-Galactosamine |
| Gb3Cer: | Galα1,4Galβ1,4Glcβ1,1′Cer |
| Gb4Cer: | GalNAcβ1,3Galα1,4Galβ1,4Glcβ1,1′Cer |
| GD1a: | Neu5Acα2,3Galβ1,3GalNAcβ1,4( Neu5Acα2,3)Galβ1,4Glcβ1,1′Cer |
| GD1b: | Galβ1,3GalNAcβ1,4( Neu5Acα2,8 Neu5Acα2,3)Galβ1,4Glcβ1,1′Cer |
| GD2: | GalNAcβ1,4( Neu5Acα2,8 Neu5Acα2,3)Galβ1,4Glcβ1,1′Cer |
| GD3: | Neu5Acα2,8 Neu5Acα2,3Galβ1,4Glcβ1,1′Cer |
| Glc: | D-Glucose |
| GlcNAc: | 2-Deoxy-2-N-Acetyl-D-Glucosamine |
| GM1(a): | Galβ1,3GalNAcβ1,4(NeuAcα2,3)Galβ1,4Glcβ1,1′Cer |
| GM1b: | Neu5Acα2,3Galβ1,3GalNAcβ1,4Galβ1,4Glcβ1,1′Cer |
| GM3: | Neu5Acα2,3Galβ1,4Glcβ1,1′Cer |
| GQ1b: | Neu5Acα2,8Neu5Acα2,3Galβ1,3GalNAcβ1,4(Neu5Acα2,8Neu5Acα2,3)Galβ1,4Glcβ1,1′Cer |
| GSL: | Glycosphingolipid |
| GT1a: | Neu5Acα2,8 Neu5Acα2,3Galβ1,3GalNAcβ1,4( Neu5Acα2,3)Galβ1,4Glcβ1,1′Cer |
| GT1b: | Neu5Acα2,3Galβ1,3GalNAcβ1,4( Neu5Acα2,8 Neu5Acα2,3)Galβ1,4Glcβ1,1′Cer |
| HDL: | High Density Lipoprotein |
| HexCer: | Hexosylceramide (GlcCer or GalCer) |
| HexNAc: | N-Acetylhexosamine (usually GlcNAc or GalNAc) |
| HILIC: | Hydrophilic interaction liquid chromatography |
| IR: | Infrared |
| LacCer: | Lactosylceramide |
| LC: | Liquid chromatography |
| LDL: | Low Density Lipoprotein |
| LTQ: | Linear trap quadrupole |
| lysoGSL: | glycosylated sphingoid bases |
| MDCK: | Madin Darby canine kidney |
| MS/MS: | Tandem mass spectrometry |
| MRM: | Multiple reaction monitoring |
| MS: | Mass spectrometry |
| m/z: | Mass per charge ratio |
| Neu5Ac: | N-Acetylneuraminic Acid |
| Neu5Gc: | N-Glycolylneuraminic Acid |
| QTOF: | Quadrupole Time-of-Flight |
| RP: | Reversed phase |
| Sulfatide: | GalCer-3-sulfate |
| MALDI: | Matrix-Assisted Laser Desorption/Ionization |
| TLC: | thin layer chromatography |
| TOF: | Time-of-Flight |
| VLDL: | Very Low Density Lipoprotein |
1. Introduction
| Series | Core structure |
|---|---|
| Gala | Galα1,4Galβ1,1′Cer |
| Ganglio | Galβ1,3GalNAcβ1,4Galβ1,4Glcβ1,1′Cer |
| Globo | GalNAcβ1,3Galα1,4Galβ1,4Glcβ1,1′Cer |
| Isoglobo | GalNAcβ1,3Galα1,3Galβ1,4Glcβ1,1′Cer |
| Lacto | Galβ1,3GlcNAcβ1,3Galβ1,4Glcβ1,1′Cer |
| Neolacto | Galβ1,4GlcNAcβ1,3Galβ1,4Glcβ1,1′Cer |
| Muco | Galβ1,3Galβ1,4Galβ1,4Glcβ1,1′Cer |
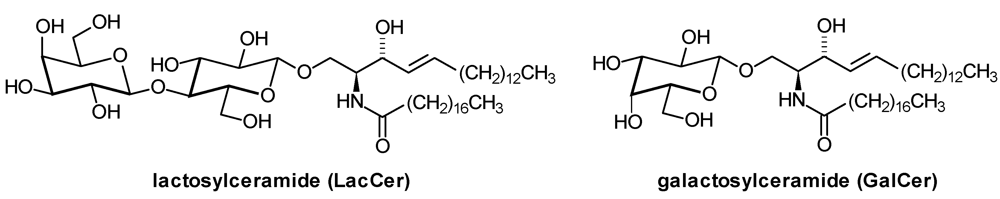

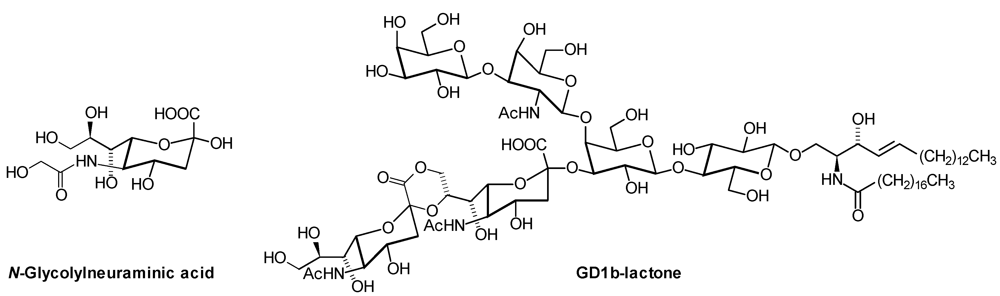
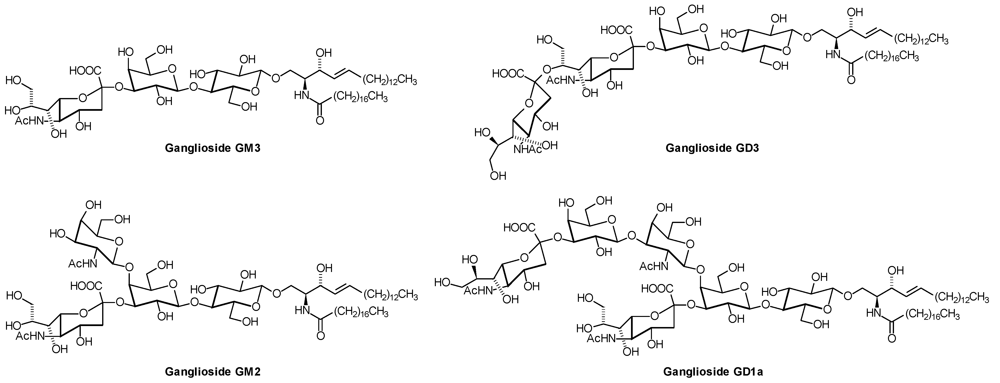

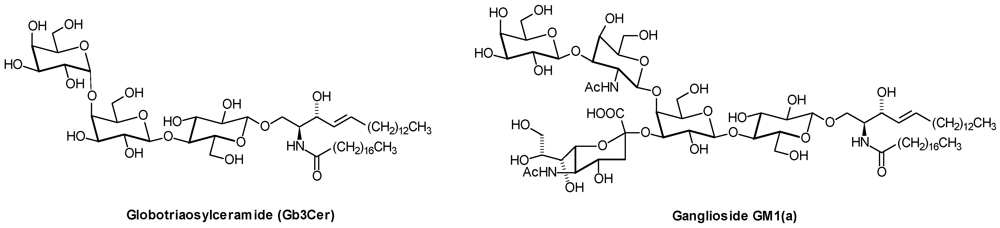
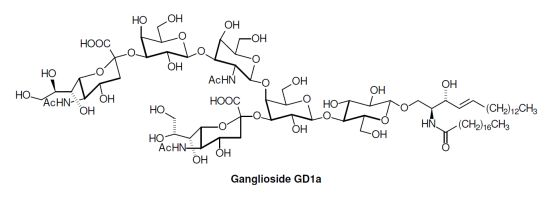
2. Glycosphingolipid Structure Elucidation and Analysis
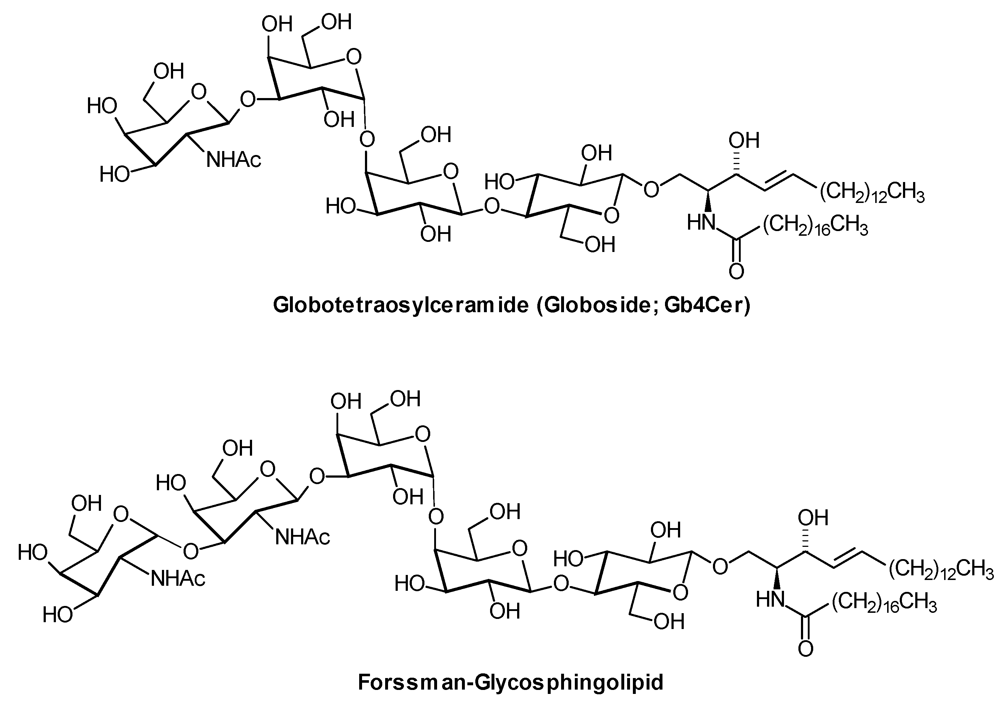
2.1. Sample Preparation and Glycosphingolipid Extraction
2.2. Glycosphingolipid Standards
2.3. Separation
2.4. Shotgun Lipidomics
2.5. Electrospray Ionization (ESI)
2.6. Matrix-Assisted Laser Desorption/Ionization (MALDI)
2.7. Other Ionization Techniques
2.8. Tandem Mass Spectrometry
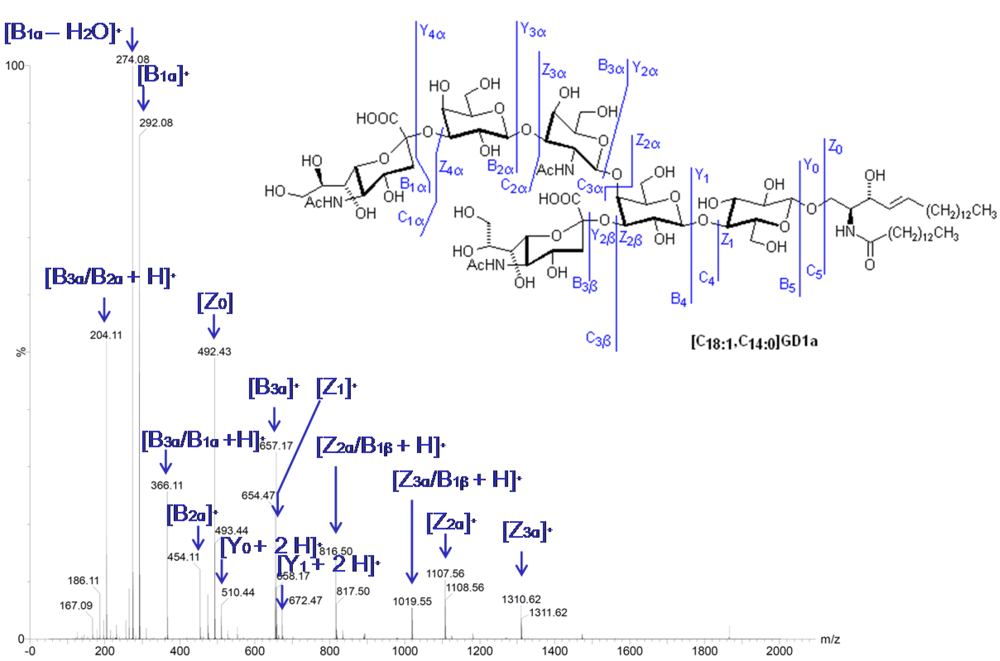
2.9. Imaging Mass Spectrometry (IMS)
2.10. Indirect Methods
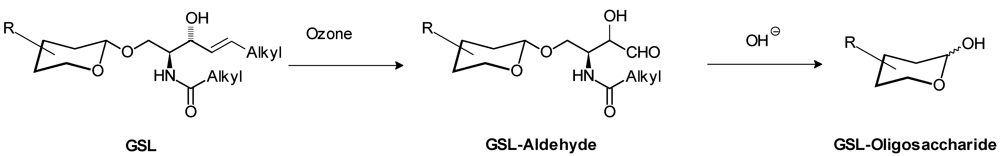
3. Identification of Interacting Molecules
4. Applications
4.1. Analysis of Lipidomes
4.2. Mutants and Other Functional Studies
4.3. Inherited Diseases

4.4. Acquired Diseases
4.4.1. Folding Diseases
4.4.2. Diabetes
4.4.3. Cancer
5. Outlook
Acknowledgments
Conflict of Interest
References and Notes
- Chester, M.A. Nomenclature of glycolipids. Pure Appl. Chem. 1997, 69, 2475–2487. [Google Scholar] [CrossRef]
- Merrill, A.H. Sphingolipid and glycosphingolipid metabolic pathways in the era of sphingolipidomics. Chem. Rev. 2011, 111, 6387–6422. [Google Scholar] [CrossRef]
- Fahy, E.; Subramaniam, S.; Brown, H.A.; Glass, C.K.; Merrill, A.H., Jr.; Murphy, R.C.; Raetz, C.R.; Russell, D.W.; Seyama, Y.; Shaw, W.; et al. A comprehensive classification system for lipids. J. Lipid Res. 2005, 46, 839–861. [Google Scholar] [CrossRef]
- Wenk, M.R. Lipidomics: New tools and applications. Cell 2010, 143, 888–895. [Google Scholar] [CrossRef]
- Jung, H.R.; Sylvanne, T.; Koistinen, K.M.; Tarasov, K.; Kauhanen, D.; Ekroos, K. High throughput quantitative molecular lipidomics. Biochim. Biophys. Acta 2011, 1811, 925–934. [Google Scholar] [CrossRef]
- Hart, G.W.; Copeland, R.J. Glycomics hits the big time. Cell 2010, 143, 672–676. [Google Scholar] [CrossRef]
- Rakus, J.F.; Mahal, L.K. New technologies for glycomic analysis: Toward a systematic understanding of the glycome. Annu. Rev. Anal. Chem. (Palo Alto Calif) 2011, 4, 367–392. [Google Scholar] [CrossRef]
- Gupta, G.; Surolia, A.; Sampathkumar, S.G. Lectin microarrays for glycomic analysis. OMICS 2010, 14, 419–436. [Google Scholar]
- Rillahan, C.D.; Paulson, J.C. Glycan microarrays for decoding the glycome. Annu. Rev. Biochem. 2011, 80, 797–823. [Google Scholar] [CrossRef]
- Zaia, J. Mass spectrometry and glycomics. OMICS 2010, 14, 401–418. [Google Scholar] [CrossRef]
- Kolter, T.; Proia, R.L.; Sandhoff, K. Combinatorial ganglioside biosynthesis. J. Biol. Chem. 2002, 277, 25859–25862. [Google Scholar]
- Levery, S.B. Glycosphingolipid structural analysis and glycosphingolipidomics. Mass Spectrom. Modified Proteins Glycoconjugates 2005, 405, 300–369. [Google Scholar] [CrossRef]
- Teufel, A.; Maass, T.; Galle, P.R.; Malik, N. The longevity assurance homologue of yeast lag1 (Lass) gene family (review). Int. J. Mol. Med. 2009, 23, 135–140. [Google Scholar]
- Sandhoff, R.; Geyer, R.; Jennemann, R.; Paret, C.; Kiss, E.; Yamashita, T.; Gorgas, K.; Sijmonsma, T.P.; Iwamori, M.; Finaz, C.; et al. Novel class of glycosphingolipids involved in male fertility. J. Biol. Chem. 2005, 280, 27310–27318. [Google Scholar]
- Schauer, R. Sialic acids as regulators of molecular and cellular interactions. Curr. Opin. Struct. Biol. 2009, 19, 507–514. [Google Scholar] [CrossRef]
- Riboni, L.; Sonnino, S.; Acquotti, D.; Malesci, A.; Ghidoni, R.; Egge, H.; Mingrino, S.; Tettamanti, G. Natural occurrence of ganglioside lactones. Isolation and characterization of GD1b inner ester from adult human brain. J. Biol. Chem. 1986, 261, 8514–8519. [Google Scholar]
- Jiang, X.; Cheng, H.; Yang, K.; Gross, R.W.; Han, X. Alkaline methanolysis of lipid extracts extends shotgun lipidomics analyses to the low-abundance regime of cellular sphingolipids. Anal. Biochem. 2007, 371, 135–145. [Google Scholar]
- He, H.; Conrad, C.A.; Nilsson, C.L.; Ji, Y.; Schaub, T.M.; Marshall, A.G.; Emmett, M.R. Method for lipidomic analysis: p53 expression modulation of sulfatide, ganglioside, and phospholipid composition of U87 MG glioblastoma cell. Anal. Chem. 2007, 79, 8423–8430. [Google Scholar]
- Vukelic, Z.; Kalanj-Bognar, S.; Froesch, M.; Bindila, L.; Radic, B.; Allen, M.; Peter-Katalinic, J.; Zamfir, A.D. Human gliosarcoma-associated ganglioside composition is complex and distinctive as evidenced by high-performance mass spectrometric determination and structural characterization. Glycobiology 2007, 17, 504–515. [Google Scholar]
- Kohla, G.; Stockfleth, E.; Schauer, R. Gangliosides with O-acetylated sialic acids in tumors of neuroectodermal origin. Neurochem. Res. 2002, 27, 583–592. [Google Scholar] [CrossRef]
- Herrler, G.; Schwegmann-Wessels, C. Sialic acids as receptor determinants for coronaviruses. Glycoconjugate J. 2006, 23, 51–58. [Google Scholar] [CrossRef]
- Zarei, M.; Muthing, J.; Peter-Katalinic, J.; Bindila, L. Separation and identification of GM1b pathway Neu5Ac- and Neu5Gc gangliosides by on-line nanoHPLC-QToF MS and tandem MS: Toward glycolipidomics screening of animal cell lines. Glycobiology 2010, 20, 118–126. [Google Scholar] [CrossRef]
- Svennerholm, L. Chromatographic separation of human brain gangliosides. J. Neurochem. 1963, 10, 613–623. [Google Scholar] [CrossRef]
- Posse de Chaves, E.; Sipione, S. Sphingolipids and gangliosides of the nervous system in membrane function and dysfunction. FEBS Lett. 2010, 584, 1748–1759. [Google Scholar] [CrossRef]
- Yu, R.K.; Nakatani, Y.; Yanagisawa, M. The role of glycosphingolipid metabolism in the developing brain. J. Lipid Res. 2009, 50, S440–S445. [Google Scholar]
- Hakomori, S.I. Structure and function of glycosphingolipids and sphingolipids: Recollections and future trends. Biochim. Biophys. Acta 1780, 325–346. [Google Scholar]
- Schnaar, R.L.; Mehta, N.R.; Nguyen, T.; Griffin, J.W. Ganglioside engagement by myelin-associated glycoprotein (MAG) protects axons from acute toxic insults. J. Neurochem. 2010, 113, 26. [Google Scholar]
- Schengrund, C.L. Lipid rafts: Keys to neurodegeneration. Brain Res. Bull. 2010, 82, 7–17. [Google Scholar] [CrossRef]
- Yamashita, T.; Hashiramoto, A.; Haluzik, M.; Mizukami, H.; Beck, S.; Norton, A.; Kono, M.; Tsuji, S.; Daniotti, J.L.; Werth, N.; et al. Enhanced insulin sensitivity in mice lacking ganglioside GM3. Proc. Natl. Acad. Sci. USA 2003, 100, 3445–3449. [Google Scholar]
- Eggeling, C.; Ringemann, C.; Medda, R.; Schwarzmann, G.; Sandhoff, K.; Polyakova, S.; Belov, V.N.; Hein, B.; von Middendorff, C.; Schonle, A.; et al. Direct observation of the nanoscale dynamics of membrane lipids in a living cell. Nature 2009, 457, 1159–1162. [Google Scholar]
- Sandhoff, K.; Gallala, H.D. Principles of microdomain formation in biological membranes-Are there lipid liquid ordered domains in living cellular membranes? Trends Glycosci. Glycotechnol. 2008, 20, 277–295. [Google Scholar] [CrossRef]
- Malorni, W.; Garofaloa, T.; Tinari, A.; Matarrese, P.; Giammarioli, A.M.; Manganelli, V.; Ciarlo, L.; Misasi, R.; Sorice, M. Do mitochondria act as "Cargo boats" in the journey of GD3 to the nucleus during apoptosis? FEBS Lett. 2007, 581, 3899–3903. [Google Scholar]
- Hanada, K. Sphingolipids in infectious diseases. Jpn. J. Infect. Dis. 2005, 58, 131–148. [Google Scholar]
- Holgersson, J.; Stromberg, N.; Breimer, M.E. Glycolipids of human large intestine: Difference in glycolipid expression related to anatomical localization, epithelial/non-epithelial tissue and the ABO, Le and Se phenotypes of the donors. Biochimie 1988, 70, 1565–1574. [Google Scholar] [CrossRef]
- Zumbrun, S.D.; Hanson, L.; Sinclair, J.F.; Freedy, J.; Melton-Celsa, A.R.; Rodriguez-Canales, J.; Hanson, J.C.; O'Brien, A.D. Human intestinal tissue and cultured colonic cells contain globotriaosylceramide synthase mRNA and the alternate Shiga toxin receptor globotetraosylceramide. Infect. Immun. 2010, 78, 4488–4499. [Google Scholar]
- Schweppe, C.H.; Hoffmann, P.; Nofer, J.R.; Pohlentz, G.; Mormann, M.; Karch, H.; Friedrich, A.W.; Muthing, J. Neutral glycosphingolipids in human blood: A precise mass spectrometry analysis with special reference to lipoprotein-associated Shiga toxin receptors. J. Lipid Res. 2010, 51, 2282–2294. [Google Scholar] [CrossRef]
- Senn, H.J.; Orth, M.; Fitzke, E.; Wieland, H.; Gerok, W. Gangliosides in normal human serum. Concentration, pattern and transport by lipoproteins. Eur. J. Biochem. 1989, 181, 657–662. [Google Scholar] [CrossRef]
- Kusunoki, S.; Kaida, K. Antibodies against ganglioside complexes in Guillain-Barre syndrome and related disorders. J. Neurochem. 2011, 116, 828–832. [Google Scholar] [CrossRef]
- Haynes, C.A.; Allegood, J.C.; Park, H.; Sullards, M.C. Sphingolipidomics: Methods for the comprehensive analysis of sphingolipids. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2009, 877, 2696–2708. [Google Scholar] [CrossRef]
- Egge, H.; Peter-Katalinic, J.; Reuter, G.; Schauer, R.; Ghidoni, R.; Sonnino, S.; Tettamanti, G. Analysis of gangliosides using fast atom bombardment mass spectrometry. Chem. Phys. Lipids 1985, 37, 127–141. [Google Scholar] [CrossRef]
- Chen, Y.F.; Liu, Y.; Sullards, M.C.; Merrill, A.H. An Introduction to Sphingolipid Metabolism and Analysis by New Technologies. Neuromol. Med. 2010, 12, 306–319. [Google Scholar] [CrossRef]
- Muthing, J.; Distler, U. Advances on the compositional analysis of glycosphingolipids combining thin-layer chromatography with mass spectrometry. Mass Spectrom Rev. 2010, 29, 425–479. [Google Scholar]
- Farwanah, H.; Kolter, T.; Sandhoff, K. Mass spectrometric analysis of neutral sphingolipids: Methods, applications, and limitation. Biochim. Biophys. Acta 1811, 854–860. [Google Scholar]
- Hsu, F.F.; Turk, J. Structural determination of glycosphingolipids as lithiated adducts by electrospray ionization mass spectrometry using low-energy collisional-activated dissociation on a triple stage quadrupole instrument. J. Am. Soc. Mass Spectrom. 2001, 12, 61–79. [Google Scholar] [CrossRef]
- Tanaka, K.; Yamada, M.; Tamiya-Koizumi, K.; Kannagi, R.; Aoyama, T.; Hara, A.; Kyogashima, M. Systematic analyses of free ceramide species and ceramide species comprising neutral glycosphingolipids by MALDI-TOF MS with high-energy CID. Glycoconjugate J. 2011, 28, 67–87. [Google Scholar] [CrossRef]
- Zamfir, A.D.; Vukelic, Z.; Schneider, A.; Sisu, E.; Dinca, N.; Ingendoh, A. A novel approach for ganglioside structural analysis based on electrospray multiple-stage mass spectrometry. J. Biomol. Tech. 2007, 18, 188–193. [Google Scholar]
- Han, X.; Cheng, H. Characterization and direct quantitation of cerebroside molecular species from lipid extracts by shotgun lipidomics. J. Lipid Res. 2005, 46, 163–175. [Google Scholar]
- Merrill, A.H., Jr.; Sullards, M.C.; Allegood, J.C.; Kelly, S.; Wang, E. Sphingolipidomics: High-throughput, structure-specific, and quantitative analysis of sphingolipids by liquid chromatography tandem mass spectrometry. Methods 2005, 36, 207–224. [Google Scholar] [CrossRef]
- Holgersson, J.; Jovall, P.A.; Samuelsson, B.E.; Breimer, M.E. Blood group type glycosphingolipids of human kidneys. Structural characterization of extended globo-series compounds. Glycoconjugate J. 1991, 8, 424–433. [Google Scholar] [CrossRef]
- Simons, K.; Sampaio, J.L. Membrane organization and lipid rafts. Cold Spring Harb. Perspect. Biol. 2011, 3, a004697. [Google Scholar] [CrossRef]
- Lacomba, R.; Salcedo, J.; Alegria, A.; Jesus Lagarda, M.; Barbera, R.; Matencio, E. Determination of sialic acid and gangliosides in biological samples and dairy products: A review. J. Pharm. Biomed. Anal. 2010, 51, 346–357. [Google Scholar]
- Svennerholm, L.; Fredman, P. A procedure for the quantitative isolation of brain gangliosides. Biochim. Biophys. Acta 1980, 617, 97–109. [Google Scholar] [CrossRef]
- Byrne, M.C.; Sbaschnig-Agler, M.; Aquino, D.A.; Sclafani, J.R.; Ledeen, R.W. Procedure for isolation of gangliosides in high yield and purity: Simultaneous isolation of neutral glycosphingolipids. Anal. Biochem. 1985, 148, 163–173. [Google Scholar]
- Wang, W.Q.; Gustafson, A. Ganglioside extraction from erythrocytes: A comparison study. Acta Chem. Scand. 1995, 49, 929–936. [Google Scholar] [CrossRef]
- Folch, J.; Lees, M.; Sloane Stanley, G.H. A simple method for the isolation and purification of total lipids from animal tissues. J. Biol. Chem. 1957, 226, 497–509. [Google Scholar]
- Bodennec, J.; Pelled, D.; Futerman, A.H. Aminopropyl solid phase extraction and 2 D TLC of neutral glycosphingolipids and neutral lysoglycosphingolipids. J. Lipid Res. 2003, 44, 218–226. [Google Scholar] [CrossRef]
- Manzi, A.E.; Hayes, B.K. HPLC methods for the fractionation and analysis of negatively charged oligosaccharides and gangliosides. Curr. Protoc. Mol. Biol. 2001. Chapter 17, Unit17 21A. [Google Scholar]
- Yu, R.K.; Ledeen, R.W. Gangliosides of human, bovine, and rabbit plasma. J. Lipid Res. 1972, 13, 680–686. [Google Scholar]
- Dass, C. Fundamentals of Contemporary Mass Spectrometry; Wiley-Interscience: Hoboken, NJ, USA, 2007. [Google Scholar]
- Fong, B.; Norris, C.; Lowe, E.; McJarrow, P. Liquid chromatography-high-resolution mass spectrometry for quantitative analysis of gangliosides. Lipids 2009, 44, 867–874. [Google Scholar]
- Moore, J.D.; Caufield, W.V.; Shaw, W.A. Quantitation and standardization of lipid internal standards for mass spectroscopy. Meth. Enzymology 2007, 432, 351–367. [Google Scholar]
- Quehenberger, O.; Armando, A.M.; Brown, A.H.; Milne, S.B.; Myers, D.S.; Merrill, A.H.; Bandyopadhyay, S.; Jones, K.N.; Kelly, S.; Shaner, R.L.; et al. Lipidomics reveals a remarkable diversity of lipids in human plasma. J. Lipid Res. 2010, 51, 3299–3305. [Google Scholar] [CrossRef]
- Mills, K.; Johnson, A.; Winchester, B. Synthesis of novel internal standards for the quantitative determination of plasma ceramide trihexoside in Fabry disease by tandem mass spectrometry. FEBS Lett. 2002, 515, 171–176. [Google Scholar] [CrossRef]
- Mills, K.; Eaton, S.; Ledger, V.; Young, E.; Winchester, B. The synthesis of internal standards for the quantitative determination of sphingolipids by tandem mass spectrometry. Rapid Commun. Mass Spectrom. 2005, 19, 1739–1748. [Google Scholar]
- Sandhoff, R.; Hepbildikler, S.T.; Jennemann, R.; Geyer, R.; Gieselmann, V.; Proia, R.L.; Wiegandt, H.; Grone, H.J. Kidney sulfatides in mouse models of inherited glycosphingolipid disorders: Determination by nano-electrospray ionization tandem mass spectrometry. J. Biol. Chem. 2002, 277, 20386–20398. [Google Scholar]
- Ejsing, C.S.; Sampaio, J.L.; Surendranath, V.; Duchoslav, E.; Ekroos, K.; Klemm, R.W.; Simons, K.; Shevchenko, A. Global analysis of the yeast lipidome by quantitative shotgun mass spectrometry. Proc. Natl. Acad. Sci. USA 2009, 106, 2136–2141. [Google Scholar]
- Fuchs, B.; Suss, R.; Teuber, K.; Eibisch, M.; Schiller, J. Lipid analysis by thin-layer chromatography--a review of the current state. J. Chromatogr. A 1218, 2754–2774. [Google Scholar]
- Fuchs, B.; Suss, R.; Schiller, J. An update of MALDI-TOF mass spectrometry in lipid research. Prog. Lipid Res. 2010, 49, 450–475. [Google Scholar] [CrossRef]
- Rohlfing, A.; Muthing, J.; Pohlentz, G.; Distler, U.; Peter-Katalinic, J.; Berkenkamp, S.; Dreisewerd, K. IR-MALDI-MS analysis of HPTLC-separated phospholipid mixtures directly from the TLC plate. Anal. Chem. 2007, 79, 5793–5808. [Google Scholar]
- Meisen, I.; Mormann, M.; Muthing, J. Thin-layer chromatography, overlay technique and mass spectrometry: A versatile triad advancing glycosphingolipidomics. Biochim. Biophys. Acta 2011, 1811, 875–896. [Google Scholar] [CrossRef]
- Suzuki, A.; Miyazaki, M.; Matsuda, J.; Yoneshige, A. High-performance thin-layer chromatography/mass spectrometry for the analysis of neutral glycosphingolipids. Biochim. Biophys. Acta 2011, 1811, 861–874. [Google Scholar] [CrossRef]
- Kirsch, S.; Muthing, J.; Peter-Katalinic, J.; Bindila, L. On-line nano-HPLC/ESI QTOF MS monitoring of alpha2-3 and alpha2-6 sialylation in granulocyte glycosphingolipidome. Biol. Chem. 2009, 390, 657–672. [Google Scholar] [CrossRef]
- Meisen, I.; Peter-Katalinic, J.; Muthing, J. Discrimination of neolacto-series gangliosides with alpha2-3- and alpha2-6-linked N-acetylneuraminic acid by nanoelectrospray ionization low-energy collision-induced dissociation tandem quadrupole TOF MS. Anal. Chem. 2003, 75, 5719–5725. [Google Scholar]
- Alpert, A.J. Hydrophilic-interaction chromatography for the separation of peptides, nucleic acids and other polar compounds. J. Chromatogr. 1990, 499, 177–196. [Google Scholar]
- Scherer, M.; Leuthauser-Jaschinski, K.; Ecker, J.; Schmitz, G.; Liebisch, G. A rapid and quantitative LC-MS/MS method to profile sphingolipids. J. Lipid Res. 2010, 51, 2001–2011. [Google Scholar] [CrossRef]
- Zamfir, A.; Vukelic, Z.; Peter-Katalinic, J. A capillary electrophoresis and off-line capillary electrophoresis/electrospray ionization-quadrupole time of flight-tandem mass spectrometry approach for ganglioside analysis. Electrophoresis 2002, 23, 2894–2903. [Google Scholar]
- Han, X.; Gross, R.W. Shotgun lipidomics: Electrospray ionization mass spectrometric analysis and quantitation of cellular lipidomes directly from crude extracts of biological samples. Mass Spectrom. Rev. 2005, 24, 367–412. [Google Scholar]
- Bou Khalil, M.; Hou, W.; Zhou, H.; Elisma, F.; Swayne, L.A.; Blanchard, A.P.; Yao, Z.; Bennett, S.A.; Figeys, D. Lipidomics era: Accomplishments and challenges. Mass Spectrom. Rev. 2010, 29, 877–929. [Google Scholar] [CrossRef]
- Farwanah, H.; Kolter, T. Lipidomics. In Wiley Encyclopedia of Chemical Biology; John Wiley & Sons, Inc.: New Jersey, NJ, USA, 2007. [Google Scholar]
- Ikeda, K.; Shimizu, T.; Taguchi, R. Targeted analysis of ganglioside and sulfatide molecular species by LC/ESI-MS/MS with theoretically expanded multiple reaction monitoring. J. Lipid Res. 2008, 49, 2678–2689. [Google Scholar]
- Hammad, S.M.; Pierce, J.S.; Soodavar, F.; Smith, K.J.; Al Gadban, M.M.; Rembiesa, B.; Klein, R.L.; Hannun, Y.A.; Bielawski, J.; Bielawska, A. Blood sphingolipidomics in healthy humans: Impact of sample collection methodology. J. Lipid Res. 2010, 51, 3074–3087. [Google Scholar]
- Liebisch, G.; Scherer, M.; Bottcher, A.; Schmitz, G. Sphingolipid profiling of human plasma and FPLC-separated lipoprotein fractions by hydrophilic interaction chromatography tandem mass spectrometry. Biochimica Et Biophysica Acta-Mol. Cell Biol. Lipids 2011, 1811, 68–75. [Google Scholar]
- Scherer, M.; Bottcher, A.; Liebisch, G. Lipid profiling of lipoproteins by electrospray ionization tandem mass spectrometry. Biochim. Biophys. Acta 2011, 1811, 918–924. [Google Scholar]
- Kirsch, S.; Zarei, M.; Cindric, M.; Muthing, J.; Bindila, L.; Peter-Katalinic, J. On-line nano-HPLC/ESI QTOF MS and tandem MS for separation, detection, and structural elucidation of human erythrocytes neutral glycosphingolipid mixture. Anal. Chem. 2008, 80, 4711–4722. [Google Scholar]
- Farwanah, H.; Wirtz, J.; Kolter, T.; Raith, K.; Neubert, R.H.; Sandhoff, K. Normal phase liquid chromatography coupled to quadrupole time of flight atmospheric pressure chemical ionization mass spectrometry for separation, detection and mass spectrometric profiling of neutral sphingolipids and cholesterol. J. Chromatogr. B Analyt. Technol. Biomed. life Sci. 2009, 877, 2976–2982. [Google Scholar] [CrossRef]
- Ivleva, V.B.; Elkin, Y.N.; Budnik, B.A.; Moyer, S.C.; O'Connor, P.B.; Costello, C.E. Coupling thin-layer chromatography with vibrational cooling matrix-assisted laser desorption/ionization Fourier transform mass spectrometry for the analysis of ganglioside mixtures. Anal. Chem. 2004, 76, 6484–6491. [Google Scholar]
- Zarei, M.; Bindila, L.; Souady, J.; Dreisewerd, K.; Berkenkamp, S.; Muthing, J.; Peter-Katalinic, J. A sialylation study of mouse brain gangliosides by MALDI a-TOF and o-TOF mass spectrometry. J. Mass Spectrom. 2008, 43, 716–725. [Google Scholar]
- Zarei, M.; Kirsch, S.; Muthing, J.; Bindila, L.; Peter-Katalinic, J. Automated normal phase nano high performance liquid chromatography/matrix assisted laser desorption/ionization mass spectrometry for analysis of neutral and acidic glycosphingolipids. Anal. Bioanal. Chem. 2008, 391, 289–297. [Google Scholar]
- Kliman, M.; May, J.C.; McLean, J.A. Lipid analysis and lipidomics by structurally selective ion mobility-mass spectrometry. Biochim. Biophys. Acta 2011, 1811, 935–945. [Google Scholar]
- Jackson, S.N.; Ugarov, M.; Egan, T.; Post, J.D.; Langlais, D.; Albert Schultz, J.; Woods, A.S. MALDI-ion mobility-TOFMS imaging of lipids in rat brain tissue. J. Mass Spectrom. 2007, 42, 1093–1098. [Google Scholar]
- Jackson, S.N.; Colsch, B.; Egan, T.; Lewis, E.K.; Schultz, J.A.; Woods, A.S. Gangliosides' analysis by MALDI-ion mobility MS. Analyst 2011, 136, 463–466. [Google Scholar]
- Kushi, Y.; Rokukawa, C.; Numajir, Y.; Kato, Y.; Handa, S. Analysis of underivatized glycosphingolipids by high-performance liquid chromatography/atmospheric pressure ionization mass spectrometry. Anal. Biochem. 1989, 182, 405–410. [Google Scholar]
- Roy, S.; Delobel, A.; Gaudin, K.; Touboul, D.; Germain, D.P.; Baillet, A.; Prognon, P.; Laprevote, O.; Chaminade, P. Liquid chromatography on porous graphitic carbon with atmospheric pressure photoionization mass spectrometry and tandem mass spectrometry for the analysis of glycosphingolipids. J. Chromatogr. A 2006, 1117, 154–162. [Google Scholar] [CrossRef]
- Tsui, Z.C.; Chen, Q.R.; Thomas, M.J.; Samuel, M.; Cui, Z. A method for profiling gangliosides in animal tissues using electrospray ionization-tandem mass spectrometry. Anal. Biochem. 2005, 341, 251–258. [Google Scholar]
- Sorensen, L.K. A liquid chromatography/tandem mass spectrometric approach for the determination of gangliosides GD3 and GM3 in bovine milk and infant formulae. Rapid Commun. Mass Spectrom. 2006, 20, 3625–3633. [Google Scholar] [CrossRef]
- Hsu, F.F.; Turk, J. Studies on sulfatides by quadrupole ion-trap mass spectrometry with electrospray ionization: Structural characterization and the fragmentation processes that include an unusual internal galactose residue loss and the classical charge-remote fragmentation. J. Am. Soc. Mass Spectrom. 2004, 15, 536–546. [Google Scholar]
- Adams, J.; Ann, Q.H. Structure Determination of Sphingolipids by Mass-Spectrometry. Mass Spectrom. Rev. 1993, 12, 51–85. [Google Scholar]
- Vukelic, Z.; Zamfir, A.D.; Bindila, L.; Froesch, M.; Peter-Katalinic, J.; Usuki, S.; Yu, R.K. Screening and sequencing of complex sialylated and sulfated glycosphingolipid mixtures by negative ion electrospray Fourier transform ion cyclotron resonance mass spectrometry. J. Am. Soc. Mass Spectrom. 2005, 16, 571–580. [Google Scholar]
- Serb, A.; Schiopu, C.; Flangea, C.; Sisu, E.; Zamfir, A.D. Top-down glycolipidomics: Fragmentation analysis of ganglioside oligosaccharide core and ceramide moiety by chip-nanoelectrospray collision-induced dissociation MS2-MS6. J. Mass Spectrom. 2009, 44, 1434–1442. [Google Scholar]
- Goto-Inoue, N.; Hayasaka, T.; Zaima, N.; Setou, M. Imaging mass spectrometry for lipidomics. Biochim. Biophys. Acta 2011, 1811, 961–969. [Google Scholar]
- Fernandez, J.A.; Ochoa, B.; Fresnedo, O.; Giralt, M.T.; Rodriguez-Puertas, R. Matrix-assisted laser desorption ionization imaging mass spectrometry in lipidomics. Anal. Bioanal. Chem. 2011, 401, 29–51. [Google Scholar] [CrossRef]
- Passarelli, M.K.; Winograd, N. Lipid imaging with time-of-flight secondary ion mass spectrometry (ToF-SIMS). Biochim. Biophys. Acta 2011, 1811, 976–990. [Google Scholar] [CrossRef]
- Eberlin, L.S.; Ferreira, C.R.; Dill, A.L.; Ifa, D.R.; Cooks, R.G. Desorption electrospray ionization mass spectrometry for lipid characterization and biological tissue imaging. Biochim. Biophys. Acta 2011, 1811, 946–960. [Google Scholar]
- Sjovall, P.; Lausmaa, J.; Johansson, B. Mass spectrometric imaging of lipids in brain tissue. Anal. Chem. 2004, 76, 4271–4278. [Google Scholar]
- Borner, K.; Nygren, H.; Hagenhoff, B.; Malmberg, P.; Tallarek, E.; Mansson, J.E. Distribution of cholesterol and galactosylceramide in rat cerebellar white matter. Biochim. Biophys. Acta 1761, 335–344. [Google Scholar]
- Nygren, H.; Borner, K.; Hagenhoff, B.; Malmberg, P.; Mansson, J.E. Localization of cholesterol, phosphocholine and galactosylceramide in rat cerebellar cortex with imaging TOF-SIMS equipped with a bismuth cluster ion source. Biochim. Biophys. Acta 2005, 1737, 102–110. [Google Scholar]
- Pernber, Z.; Richter, K.; Mansson, J.E.; Nygren, H. Sulfatide with different fatty acids has unique distributions in cerebellum as imaged by time-of-flight secondary ion mass spectrometry (TOF-SIMS). Biochim. Biophys. Acta 2007, 1771, 202–209. [Google Scholar]
- Manicke, N.E.; Nefliu, M.; Wu, C.; Woods, J.W.; Reiser, V.; Hendrickson, R.C.; Cooks, R.G. Imaging of lipids in atheroma by desorption electrospray ionization mass spectrometry. Anal. Chem. 2009, 81, 8702–8707. [Google Scholar]
- Sugiura, Y.; Shimma, S.; Konishi, Y.; Yamada, M.K.; Setou, M. Imaging mass spectrometry technology and application on ganglioside study; visualization of age-dependent accumulation of C20-ganglioside molecular species in the mouse hippocampus. PLoS One 2008, 3, e3232. [Google Scholar]
- Chen, Y.; Allegood, J.; Liu, Y.; Wang, E.; Cachon-Gonzalez, B.; Cox, T.M.; Merrill, A.H., Jr.; Sullards, M.C. Imaging MALDI mass spectrometry using an oscillating capillary nebulizer matrix coating system and its application to analysis of lipids in brain from a mouse model of Tay-Sachs/Sandhoff disease. Anal. Chem. 2008, 80, 2780–2788. [Google Scholar]
- Colsch, B.; Woods, A.S. Localization and imaging of sialylated glycosphingolipids in brain tissue sections by MALDI mass spectrometry. Glycobiology 2010, 20, 661–667. [Google Scholar] [CrossRef]
- Marsching, C.; Eckhardt, M.; Grone, H.J.; Sandhoff, R.; Hopf, C. Imaging of complex sulfatides SM3 and SB1a in mouse kidney using MALDI-TOF/TOF mass spectrometry. Anal. Bioanal. Chem. 2011, 401, 53–64. [Google Scholar]
- Ageta, H.; Asai, S.; Sugiura, Y.; Goto-Inoue, N.; Zaima, N.; Setou, M. Layer-specific sulfatide localization in rat hippocampus middle molecular layer is revealed by nanoparticle-assisted laser desorption/ionization imaging mass spectrometry. Med. Mol. Morphol. 2009, 42, 16–23. [Google Scholar] [CrossRef]
- Vanderschaeghe, D.; Festjens, N.; Delanghe, J.; Callewaert, N. Glycome profiling using modern glycomics technology: Technical aspects and applications. Biol. Chem. 2010, 391, 149–161. [Google Scholar]
- Hakomori, S.I. Release of carbohydrates from sphingoglycolipid by osmium-catalyzed periodate oxidation followed by treatment with mild alkali. J. Lipid Res. 1966, 7, 789–792. [Google Scholar]
- Wiegandt, H.; Bucking, H.W. Carbohydrate components of extraneuronal gangliosides from bovine and human spleen, and bovine kidney. Eur. J. Biochem. 1970, 15, 287–292. [Google Scholar] [CrossRef]
- Nagahori, N.; Abe, M.; Nishimura, S. Structural and functional glycosphingolipidomics by glycoblotting with an aminooxy-functionalized gold nanoparticle. Biochemistry 2009, 48, 583–594. [Google Scholar]
- Li, Y.T.; Chou, C.W.; Li, S.C.; Kobayashi, U.; Ishibashi, Y.H.; Ito, M. Preparation of homogenous oligosaccharide chains from glycosphingolipids. Glycoconjugate J. 2009, 26, 929–933. [Google Scholar]
- Song, X.; Lasanajak, Y.; Xia, B.; Heimburg-Molinaro, J.; Rhea, J.M.; Ju, H.; Zhao, C.; Molinaro, R.J.; Cummings, R.D.; Smith, D.F. Shotgun glycomics: A microarray strategy for functional glycomics. Nat. Methods 2011, 8, 85–90. [Google Scholar]
- Wenk, M.R. The emerging field of lipidomics. Nat. Rev. Drug Discov. 2005, 4, 594–610. [Google Scholar] [CrossRef]
- Magnani, J.L.; Smith, D.F.; Ginsburg, V. Detection of gangliosides that bind cholera toxin: Direct binding of 125I-labeled toxin to thin-layer chromatograms. Anal. Biochem. 1980, 109, 399–402. [Google Scholar]
- Muthing, J.; Schweppe, C.H.; Karch, H.; Friedrich, A.W. Shiga toxins, glycosphingolipid diversity, and endothelial cell injurY. Thromb. Haemost. 2009, 101, 252–264. [Google Scholar]
- Musken, A.; Souady, J.; Dreisewerd, K.; Zhang, W.; Distler, U.; Peter-Katalinic, J.; Miller-Podraza, H.; Karch, H.; Muthing, J. Application of thin-layer chromatography/infrared matrix-assisted laser desorption/ionization orthogonal time-of-flight mass spectrometry to structural analysis of bacteria-binding glycosphingolipids selected by affinity detection. Rapid Commun. Mass Spectrom. 2010, 24, 1032–1038. [Google Scholar]
- Meisen, I.; Friedrich, A.W.; Karch, H.; Witting, U.; Peter-Katalinic, J.; Muthing, J. Application of combined high-performance thin-layer chromatography immunostaining and nanoelectrospray ionization quadrupole time-of-flight tandem mass spectrometry to the structural characterization of high- and low-affinity binding ligands of Shiga toxin 1. Rapid Commun. Mass Spectrom. 2005, 19, 3659–3665. [Google Scholar]
- Lopez, P.H.; Schnaar, R.L. Determination of glycolipid-protein interaction specificity. Meth. Enzymology 2006, 417, 205–220. [Google Scholar]
- Sampaio, J.L.; Gerl, M.J.; Klose, C.; Ejsing, C.S.; Beug, H.; Simons, K.; Shevchenko, A. Membrane lipidome of an epithelial cell line. Proc. Natl. Acad. Sci. USA 2011, 108, 1903–1907. [Google Scholar]
- Dennis, E.A.; Deems, R.A.; Harkewicz, R.; Quehenberger, O.; Brown, H.A.; Milne, S.B.; Myers, D.S.; Glass, C.K.; Hardiman, G.; Reichart, D.; et al. A mouse macrophage lipidome. J. Biol. Chem. 2010, 285, 39976–39985. [Google Scholar]
- Kalvodova, L.; Sampaio, J.L.; Cordo, S.; Ejsing, C.S.; Shevchenko, A.; Simons, K. The lipidomes of vesicular stomatitis virus, semliki forest virus, and the host plasma membrane analyzed by quantitative shotgun mass spectrometry. J. Virol. 2009, 83, 7996–8003. [Google Scholar]
- Imgrund, S.; Hartmann, D.; Farwanah, H.; Eckhardt, M.; Sandhoff, R.; Degen, J.; Gieselmann, V.; Sandhoff, K.; Willecke, K. Adult ceramide synthase 2 (CERS2)-deficient mice exhibit myelin sheath defects, cerebellar degeneration, and hepatocarcinomas. J. Biol. Chem. 2009, 284, 33549–33560. [Google Scholar]
- Iwabuchi, K.; Prinetti, A.; Sonnino, S.; Mauri, L.; Kobayashi, T.; Ishii, K.; Kaga, N.; Murayama, K.; Kurihara, H.; Nakayama, H.; et al. Involvement of very long fatty acid-containing lactosylceramide in lactosylceramide-mediated superoxide generation and migration in neutrophils. Glycoconjugate J. 2008, 25, 357–374. [Google Scholar]
- Xu, Y.H.; Barnes, S.; Sun, Y.; Grabowski, G.A. Multi-system disorders of glycosphingolipid and ganglioside metabolism. J. Lipid Res. 2010, 51, 1643–1675. [Google Scholar]
- Wolf, C.; Quinn, P.J. Lipidomics in diagnosis of lipidoses. Subcell. Biochem. 2008, 49, 567–588. [Google Scholar]
- Fauler, G.; Rechberger, G.N.; Devrnja, D.; Erwa, W.; Plecko, B.; Kotanko, P.; Breunig, F.; Paschke, E. Rapid determination of urinary globotriaosylceramide isoform profiles by electrospray ionization mass spectrometry using stearoyl-d35-globotriaosylceramide as internal standard. Rapid Commun. Mass Spectrom. 2005, 19, 1499–1506. [Google Scholar]
- Nelson, B.C.; Roddy, T.; Araghi, S.; Wilkens, D.; Thomas, J.J.; Zhang, K.; Sung, C.C.; Richards, S.M. Globotriaosylceramide isoform profiles in human plasma by liquid chromatography-tandem mass spectrometry. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2004, 805, 127–134. [Google Scholar]
- Roddy, T.P.; Nelson, B.C.; Sung, C.C.; Araghi, S.; Wilkens, D.; Zhang, X.K.; Thomas, J.J.; Richards, S.M. Liquid chromatography-tandem mass spectrometry quantification of globotriaosylceramide in plasma for long-term monitoring of Fabry patients treated with enzyme replacement therapy. Clin. Chem. 2005, 51, 237–240. [Google Scholar]
- Kitagawa, T.; Ishige, N.; Suzuki, K.; Owada, M.; Ohashi, T.; Kobayashi, M.; Eto, Y.; Tanaka, A.; Mills, K.; Winchester, B.; et al. Non-invasive screening method for Fabry disease by measuring globotriaosylceramide in whole urine samples using tandem mass spectrometry. Mol. Genet. Metab. 2005, 85, 196–202. [Google Scholar]
- Kruger, R.; Bruns, K.; Grunhage, S.; Rossmann, H.; Reinke, J.; Beck, M.; Lackner, K.J. Determination of globotriaosylceramide in plasma and urine by mass spectrometry. Clin. Chem. Lab. Med. 2010, 48, 189–198. [Google Scholar]
- Whitfield, P.D.; Sharp, P.C.; Johnson, D.W.; Nelson, P.; Meikle, P.J. Characterization of urinary sulfatides in metachromatic leukodystrophy using electrospray ionization-tandem mass spectrometry. Mol. Genet. Metab. 2001, 73, 30–37. [Google Scholar]
- Fujiwaki, T.; Yamaguchi, S.; Tasaka, M.; Takayanagi, M.; Isobe, M.; Taketomi, T. Evaluation of sphingolipids in vitreous bodies from a patient with Gaucher disease, using delayed extraction matrix-assisted laser desorption ionization time-of-flight mass spectrometry. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2004, 806, 47–51. [Google Scholar] [CrossRef]
- Fujiwaki, T.; Tasaka, M.; Yamaguchi, S. Quantitative evaluation of sphingomyelin and glucosylceramide using matrix-assisted laser desorption ionization time-of-flight mass spectrometry with sphingosylphosphorylcholine as an internal standard. Practical application to tissues from patients with Niemann-Pick disease types A and C, and Gaucher disease. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2008, 870, 170–176. [Google Scholar] [CrossRef]
- Fuller, M.; Rozaklis, T.; Lovejoy, M.; Zarrinkalam, K.; Hopwood, J.J.; Meikle, P.J. Glucosylceramide accumulation is not confined to the lysosome in fibroblasts from patients with Gaucher disease. Mol. Genet. Metab. 2008, 93, 437–443. [Google Scholar]
- Kolter, T.; Sandhoff, K. Sphingolipid metabolism diseases. Biochimica et Biophysica Acta 2006, 1758, 2057–2079. [Google Scholar] [CrossRef]
- Auray-Blais, C.; Ntwari, A.; Clarke, J.T.R.; Warnock, D.G.; Oliveira, J.P.; Young, S.P.; Millington, D.S.; Bichet, D.G.; Sirrs, S.; West, M.L.; et al. How well does urinary lyso-Gb(3) function as a biomarker in Fabry disease? Clin. Chim. Acta 2010, 411, 1906–1914. [Google Scholar]
- Sakuraba, H.; Togawa, T.; Kawashima, I.; Kodama, T.; Tsukimura, T.; Suzuki, T.; Fukushige, T.; Kanekura, T. Tissue and plasma globotriaosylsphingosine could be a biomarker for assessing enzyme replacement therapy for Fabry disease. Biochem. Biophys. Res. Commun. 2010, 399, 716–720. [Google Scholar]
- Schiller, J.; Zschornig, O.; Petkovic, M.; Muller, M.; Arnhold, J.; Arnold, K. Lipid analysis of human HDL and LDL by MALDI-TOF mass spectrometry and (31)P-NMR. J. Lipid Res. 2001, 42, 1501–1508. [Google Scholar]
- Kolter, T. A view on sphingolipids and disease. Chem. Phys. Lipids 2011, 164, 590–606. [Google Scholar] [CrossRef]
- Hernandez-Corbacho, M.J.; Jenkins, R.W.; Clarke, C.J.; Hannun, Y.A.; Obeid, L.M.; Snider, A.J.; Siskind, L.J. Accumulation of long-chain glycosphingolipids during aging is prevented by caloric restriction. PLoS One 2011, 6, e20411. [Google Scholar]
- Ariga, T.; McDonald, M.P.; Yu, R.K. Role of ganglioside metabolism in the pathogenesis of Alzheimer's disease--a review. J. Lipid Res. 2008, 49, 1157–1175. [Google Scholar]
- Yuyama, K.; Yanagisawa, K. Late endocytic dysfunction as a putative cause of amyloid fibril formation in Alzheimer's disease. J. Neurochem. 2009, 109, 1250–1260. [Google Scholar]
- Han, X. Multi-dimensional mass spectrometry-based shotgun lipidomics and the altered lipids at the mild cognitive impairment stage of Alzheimer's disease. Biochim. Biophys. Acta 2010, 1801, 774–783. [Google Scholar]
- Kabayama, K.; Sato, T.; Saito, K.; Loberto, N.; Prinetti, A.; Sonnino, S.; Kinjo, M.; Igarashi, Y.; Inokuchi, J. Dissociation of the insulin receptor and caveolin-1 complex by ganglioside GM3 in the state of insulin resistance. Proc. Natl. Acad. Sci. USA 2007, 104, 13678–13683. [Google Scholar]
- Sato, T.; Nihei, Y.; Nagafuku, M.; Tagami, S.; Chin, R.; Kawamura, M.; Miyazaki, S.; Suzuki, M.; Sugahara, S.I.; Takahashi, Y.; et al. Circulating levels of ganglioside GM3 in metabolic syndrome: A pilot study. Obes. Res. Clin. Pract. 2008, 2, 231–238. [Google Scholar]
- Inokuchi, J. Physiopathological function of hematoside (GM3 ganglioside). Proc. Jpn. Acad. Ser. B 2011, 87, 179–198. [Google Scholar] [CrossRef]
- Jennemann, R.; Rothermel, U.; Wang, S.; Sandhoff, R.; Kaden, S.; Out, R.; van Berkel, T.J.; Aerts, J.M.; Ghauharali, K.; Sticht, C.; et al. Hepatic glycosphingolipid deficiency and liver function in mice. Hepatology 2010, 51, 1799–1809. [Google Scholar]
- Fox, T.E.; Bewley, M.C.; Unrath, K.A.; Pedersen, M.M.; Anderson, R.E.; Jung, D.Y.; Jefferson, L.S.; Kim, J.K.; Bronson, S.K.; Flanagan, J.M.; et al. Circulating sphingolipid biomarkers in models of type 1 diabetes. J. Lipid Res. 2011, 52, 509–517. [Google Scholar]
- Ryland, L.K.; Fox, T.E.; Liu, X.; Loughran, T.P.; Kester, M. Dysregulation of sphingolipid metabolism in cancer. Cancer Biol. Ther. 2011, 11, 138–149. [Google Scholar] [CrossRef]
- Patwardhan, G.A.; Liu, Y.Y. Sphingolipids and expression regulation of genes in cancer. Prog. Lipid Res. 2011, 50, 104–114. [Google Scholar] [CrossRef]
- Fernandez, L.E.; Gabri, M.R.; Guthmann, M.D.; Gomez, R.E.; Gold, S.; Fainboim, L.; Gomez, D.E.; Alonso, D.F. NGcGM3 ganglioside: A privileged target for cancer vaccines. Clin. Dev. Immunol. 2010, 814397. [Google Scholar]
- Miyagi, T. Aberrant expression of sialidase and cancer progression. Proc. Jpn. Acad. Ser. B 2008, 84, 407–418. [Google Scholar] [CrossRef]
- Bernstein, M.L. Targeted therapy in pediatric and adolescent oncology. Cancer 2011, 117, 2268–2274. [Google Scholar] [CrossRef]
- Yu, A.L.; Gilman, A.L.; Ozkaynak, M.F.; London, W.B.; Kreissman, S.G.; Chen, H.X.; Smith, M.; Anderson, B.; Villablanca, J.G.; Matthay, K.K.; et al. Anti-GD2 antibody with GM-CSF, interleukin-2, and isotretinoin for neuroblastoma. N. Engl. J. Med. 2010, 363, 1324–1334. [Google Scholar]
- Distler, U.; Souady, J.; Hulsewig, M.; Drmic-Hofman, I.; Haier, J.; Friedrich, A.W.; Karch, H.; Senninger, N.; Dreisewerd, K.; Berkenkamp, S.; et al. Shiga toxin receptor Gb3Cer/CD77: Tumor-association and promising therapeutic target in pancreas and colon cancer. PLoS One 2009, 4, e6813. [Google Scholar]
- Gehrmann, M.; Liebisch, G.; Schmitz, G.; Anderson, R.; Steinem, C.; De Maio, A.; Pockley, G.; Multhoff, G. Tumor-specific Hsp70 plasma membrane localization is enabled by the glycosphingolipid Gb3. PLoS One 2008, 3, e1925. [Google Scholar]
- Zamfir, A.D.; Serb, A.; Vukeli, Z.; Flangea, C.; Schiopu, C.; Fabris, D.; Kalanj-Bognar, S.; Capitan, F.; Sisu, E. Assessment of the Molecular Expression and Structure of Gangliosides in Brain Metastasis of Lung Adenocarcinoma by an Advanced Approach Based on Fully Automated Chip-Nanoelectrospray Mass Spectrometry. J. Am. Soc. Mass Spectrom. 2011, 22, 2145–2159. [Google Scholar]
- Momin, A.A.; Park, H.; Portz, B.J.; Haynes, C.A.; Shaner, R.L.; Kelly, S.L.; Jordan, I.K.; Merrill, A.H., Jr. A method for visualization of "omic" datasets for sphingolipid metabolism to predict potentially interesting differences. J. Lipid Res. 2011, 52, 1073–1083. [Google Scholar]
- Liu, Y.; Chen, Y.; Momin, A.; Shaner, R.; Wang, E.; Bowen, N.J.; Matyunina, L.V.; Walker, L.D.; McDonald, J.F.; Sullards, M.C.; et al. Elevation of sulfatides in ovarian cancer: An integrated transcriptomic and lipidomic analysis including tissue-imaging mass spectrometry. Mol. Cancer 2010, 9, 186. [Google Scholar]
© 2012 by the authors; licensee MDPI, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Farwanah, H.; Kolter, T. Lipidomics of Glycosphingolipids. Metabolites 2012, 2, 134-164. https://doi.org/10.3390/metabo2010134
Farwanah H, Kolter T. Lipidomics of Glycosphingolipids. Metabolites. 2012; 2(1):134-164. https://doi.org/10.3390/metabo2010134
Chicago/Turabian StyleFarwanah, Hany, and Thomas Kolter. 2012. "Lipidomics of Glycosphingolipids" Metabolites 2, no. 1: 134-164. https://doi.org/10.3390/metabo2010134
APA StyleFarwanah, H., & Kolter, T. (2012). Lipidomics of Glycosphingolipids. Metabolites, 2(1), 134-164. https://doi.org/10.3390/metabo2010134