Exploring 6-aza-2-Thiothymine as a MALDI-MSI Matrix for Spatial Lipidomics of Formalin-Fixed Paraffin-Embedded Clinical Samples
Abstract
1. Introduction
2. Materials and Methods
2.1. Specimen Selection
2.2. Chemicals and Reagents
2.3. Sample Preparation and Analysis
2.3.1. MALDI-MS Profiling
2.3.2. MALDI-MSI Analysis
2.4. Data Analysis
2.4.1. MALDI-MS Profiling
2.4.2. MALDI-MSI Analysis
3. Results and Discussion
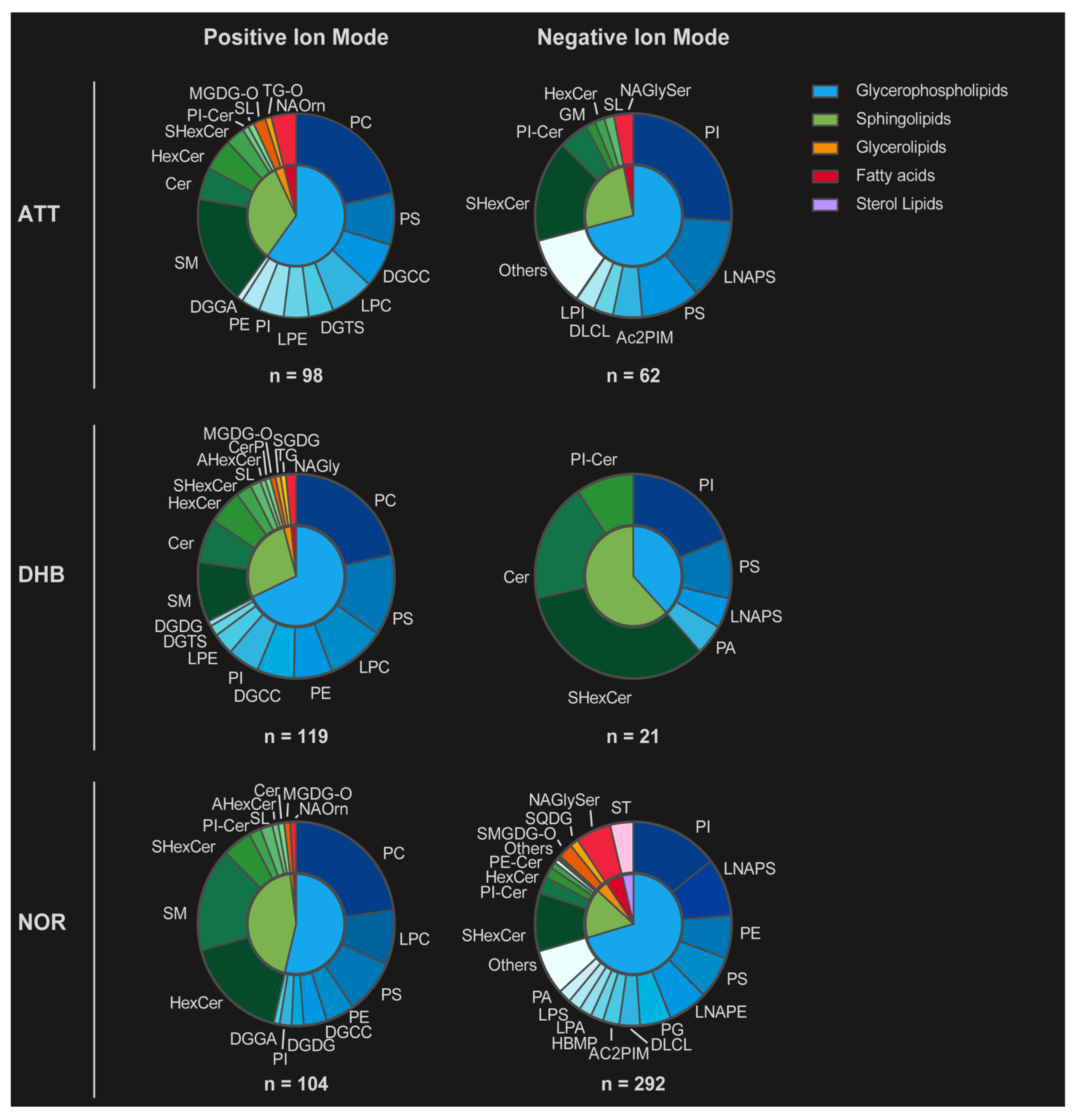
3.1. Feasibility of ATT as a MALDI Matrix for Lipid Detection
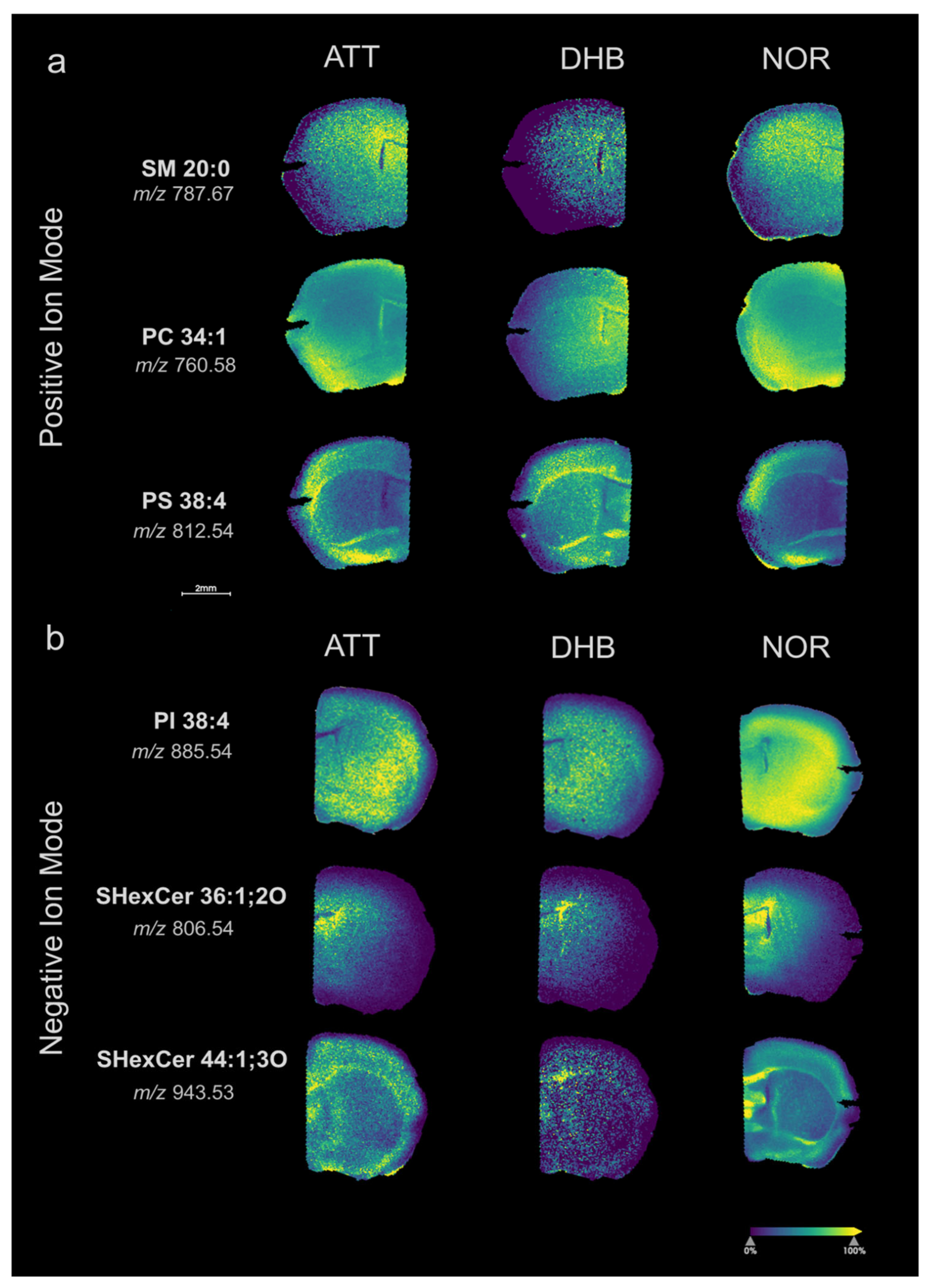
3.2. ATT for Lipid Detection in Mouse Brain Tissue
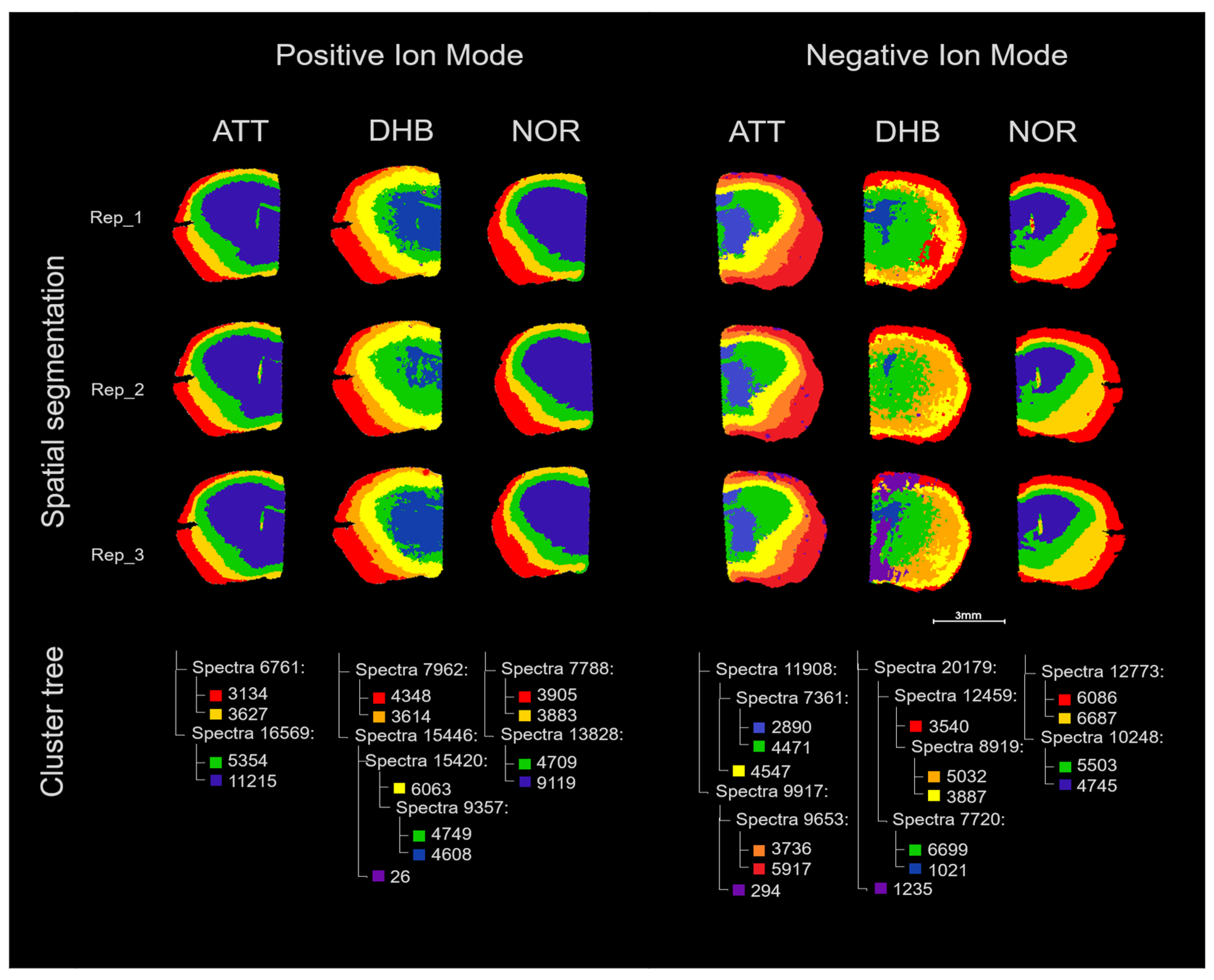
3.3. Feasibility of ATT Matrix for Spatial Lipidomics
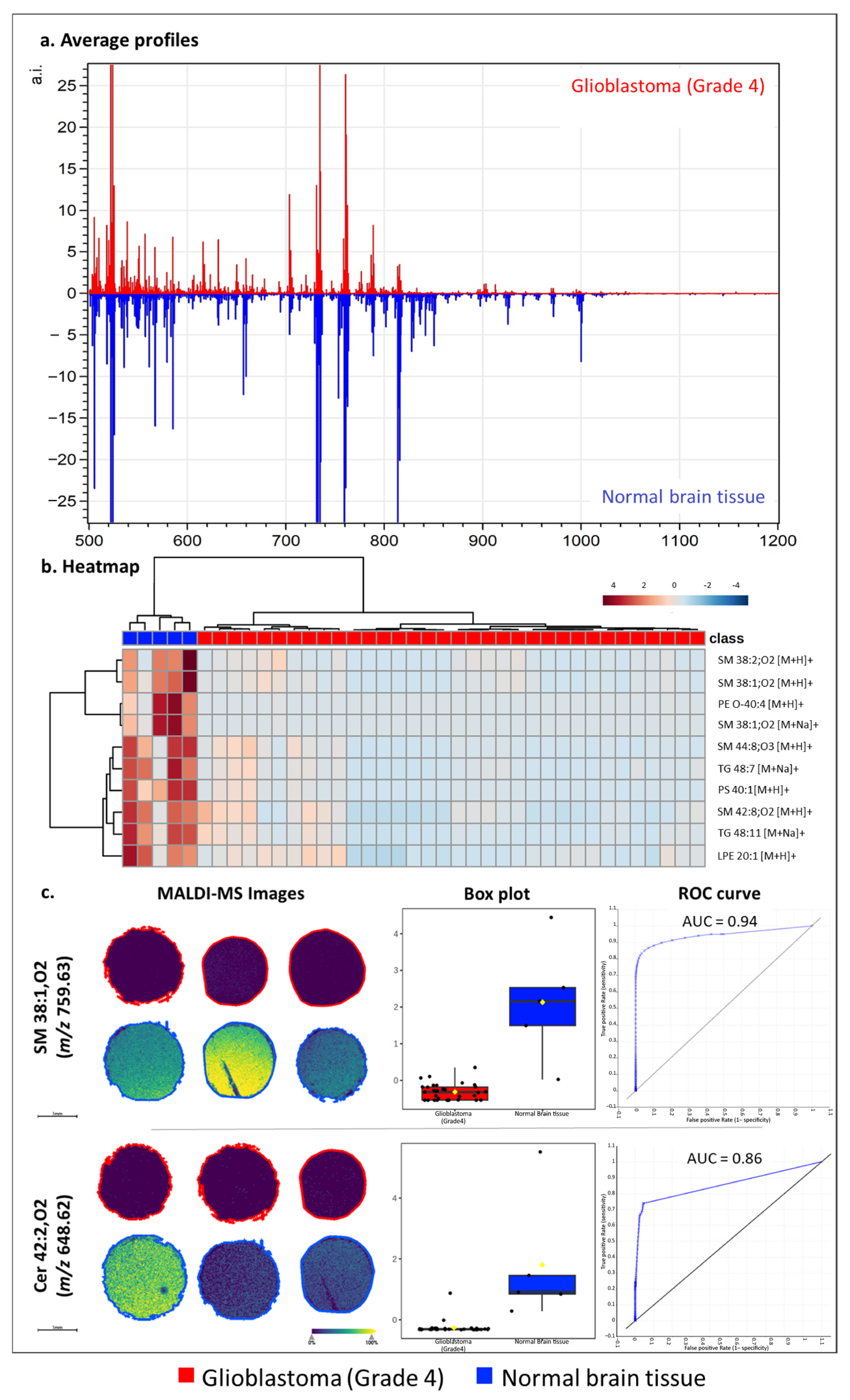
3.4. Application of ATT to Human Glioblastoma Tissue
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| MALDI-MSI | Matrix-Assisted Laser Desorption/Ionization-MS Imaging |
| FFPE | Formalin fixation and paraffin embedding |
| DHB | 2′,5′-dihydroxybenzoic acid |
| NOR | Norharmane |
| ATT | 6-aza-2-thiothymine |
| CHCA | α-cyano-4-hydroxycinnamic acid |
| DHAP | 2′,5′-dihydroxyacetophenone |
| GB | glioblastoma |
| TMAs | tissue microarrays |
| HPLC | high-performance liquid chromatography |
| MeOH | methanol |
| TFA | trifluoroacetic-acid |
| GUA | guanidinium chloride |
| DAHC | diammonium hydrogen citrate |
| ITO | Indium-Tin-Oxide-coated |
| EtOH | ethanol |
| MS | mass spectrometry |
| TIMS | trapped ion mobility spectrometry |
| PC | phosphocholines |
| SM | sphingomyelins |
| 9-AA | 9-Aminoacridine |
| Ac2PIM | diacylated phosphatidylinositol monomannosides |
| AHexCer | acylhexosylceramide |
| Cer | ceramides |
| CerP | ceramide phosphates |
| DGCC | diacylglyceryl-carboxyhydroxymethylcholines |
| DGDG | digalactosyldiacylglycerols |
| DGGA | diacylglyceryl glucuronides |
| DGTS | Diacylglyceryl trimethylhomoserines |
| DLCL | Dilysocardiolipins |
| GM | Gangliosides |
| HBMP | Hemibismonoacylglycerophosphates |
| HexCer | Hexosylceramides |
| LNAPE | N-acyl-lysophosphatidylethanolamines |
| LNAPS | N-acyl-lysophosphatidylserine |
| LPA | Lysophosphatidic acids |
| LPC | Lysophosphatidylcholines |
| LPE | Lysophosphatidylethanolamines |
| LPG | Lysophosphatidylglycerols |
| LPI | Lysophosphatidylinositols |
| LPS | Lysophosphatidylserines |
| MGDG-O | Ether-linked monogalactosyldiacylglycerols |
| MLCL | Lysocardiolipins |
| NAGly | N-acyl glycine |
| NAGlySer | N-acyl glycyl serine |
| NAOrn | N-acyl ornithine |
| PA | Phosphatidic acids |
| PE | Phosphoethanolamines |
| PE-Cer | Ceramide phosphoethanolamines |
| PEtOH | Phosphatidylethanol |
| PG | Phosphoglycerols |
| PI | Phosphoinositols |
| PI-Cer | Ceramide phosphoinositols |
| PMeOH | Phosphatidylmethanol |
| PS | Phosphoserines |
| SGDG | Sulfogalactosyl diacylglycerols |
| SHexCer | Sulfatides |
| SL | Sulfonolipid |
| SM | Sphingomielins |
| SMGDG-O | Semino lipid |
| ST | Sulfoquinovosyl diacylglycerol SQDG Cholic acids |
| TG | Triacylglycerols |
| TG-O | Ether-linked triacylglycerols |
Appendix A
Appendix A.1
Appendix A.2
Appendix A.3
References
- Kwon, Y.W.; Jo, H.-S.; Bae, S.; Seo, Y.; Song, P.; Song, M.; Yoon, J.H. Application of Proteomics in Cancer: Recent Trends and Approaches for Biomarkers Discovery. Front. Med. 2021, 8, 747333. [Google Scholar] [CrossRef] [PubMed]
- Muro, E.; Atilla-Gokcumen, G.E.; Eggert, U.S. Lipids in Cell Biology: How Can We Understand Them Better? MBoC 2014, 25, 1819–1823. [Google Scholar] [CrossRef] [PubMed]
- Snaebjornsson, M.T.; Janaki-Raman, S.; Schulze, A. Greasing the Wheels of the Cancer Machine: The Role of Lipid Metabolism in Cancer. Cell Metab. 2020, 31, 62–76. [Google Scholar] [CrossRef] [PubMed]
- Butler, L.M.; Perone, Y.; Dehairs, J.; Lupien, L.E.; De Laat, V.; Talebi, A.; Loda, M.; Kinlaw, W.B.; Swinnen, J.V. Lipids and Cancer: Emerging Roles in Pathogenesis, Diagnosis and Therapeutic Intervention. Adv. Drug Deliv. Rev. 2020, 159, 245–293. [Google Scholar] [CrossRef]
- Maan, M.; Peters, J.M.; Dutta, M.; Patterson, A.D. Lipid Metabolism and Lipophagy in Cancer. Biochem. Biophys. Res. Commun. 2018, 504, 582–589. [Google Scholar] [CrossRef]
- Wang, R.; Li, B.; Lam, S.M.; Shui, G. Integration of Lipidomics and Metabolomics for In-Depth Understanding of Cellular Mechanism and Disease Progression. J. Genet. Genom. 2020, 47, 69–83. [Google Scholar] [CrossRef]
- Salihovic, S.; Lamichane, S.; Hyötyläinen, T.; Orešič, M. Recent Advances towards Mass Spectrometry-Based Clinical Lipidomics. Curr. Opin. Chem. Biol. 2023, 76, 102370. [Google Scholar] [CrossRef]
- Anh, N.K.; Thu, N.Q.; Tien, N.T.N.; Long, N.P.; Nguyen, H.T. Advancements in Mass Spectrometry-Based Targeted Metabolomics and Lipidomics: Implications for Clinical Research. Molecules 2024, 29, 5934. [Google Scholar] [CrossRef]
- Duong, V.-A.; Lee, H. Bottom-Up Proteomics: Advancements in Sample Preparation. Int. J. Mol. Sci. 2023, 24, 5350. [Google Scholar] [CrossRef]
- Ma, X.; Fernández, F.M. Advances in Mass Spectrometry Imaging for Spatial Cancer Metabolomics. Mass Spectrom. Rev. 2024, 43, 235–268. [Google Scholar] [CrossRef]
- Planque, M.; Igelmann, S.; Ferreira Campos, A.M.; Fendt, S.-M. Spatial Metabolomics Principles and Application to Cancer Research. Curr. Opin. Chem. Biol. 2023, 76, 102362. [Google Scholar] [CrossRef]
- Dannhorn, A.; Swales, J.G.; Hamm, G.; Strittmatter, N.; Kudo, H.; Maglennon, G.; Goodwin, R.J.A.; Takats, Z. Evaluation of Formalin-Fixed and FFPE Tissues for Spatially Resolved Metabolomics and Drug Distribution Studies. Pharmaceuticals 2022, 15, 1307. [Google Scholar] [CrossRef]
- Denti, V.; Piga, I.; Guarnerio, S.; Clerici, F.; Ivanova, M.; Chinello, C.; Paglia, G.; Magni, F.; Smith, A. Antigen Retrieval and Its Effect on the MALDI-MSI of Lipids in Formalin-Fixed Paraffin-Embedded Tissue. J. Am. Soc. Mass Spectrom. 2020, 31, 1619–1624. [Google Scholar] [CrossRef]
- Buck, A.; Ly, A.; Balluff, B.; Sun, N.; Gorzolka, K.; Feuchtinger, A.; Janssen, K.; Kuppen, P.J.; Van De Velde, C.J.; Weirich, G.; et al. High-resolution MALDI-FT-ICR MS Imaging for the Analysis of Metabolites from Formalin-fixed, Paraffin-embedded Clinical Tissue Samples. J. Pathol. 2015, 237, 123–132. [Google Scholar] [CrossRef] [PubMed]
- McMillen, J.C.; Fincher, J.A.; Klein, D.R.; Spraggins, J.M.; Caprioli, R.M. Effect of MALDI Matrices on Lipid Analyses of Biological Tissues Using MALDI-2 Postionization Mass Spectrometry. J. Mass. Spectrom. 2020, 55, e4663. [Google Scholar] [CrossRef] [PubMed]
- Leopold, J.; Popkova, Y.; Engel, K.M.; Schiller, J. Recent Developments of Useful MALDI Matrices for the Mass Spectrometric Characterization of Lipids. Biomolecules 2018, 8, 173. [Google Scholar] [CrossRef] [PubMed]
- Stübiger, G.; Belgacem, O.; Rehulka, P.; Bicker, W.; Binder, B.R.; Bochkov, V. Analysis of Oxidized Phospholipids by MALDI Mass Spectrometry Using 6-Aza-2-Thiothymine Together with Matrix Additives and Disposable Target Surfaces. Anal. Chem. 2010, 82, 5502–5510. [Google Scholar] [CrossRef]
- Denti, V.; Monza, N.; Bindi, G.; Porto, N.S.; L’Imperio, V.; Pagni, F.; Piga, I.; Smith, A. 6-Aza-2-Thiothymine as an Alternative Matrix for Spatial Proteomics with MALDI-MSI. Int. J. Mol. Sci. 2024, 25, 13678. [Google Scholar] [CrossRef]
- Strohalm, M.; Kavan, D.; Novák, P.; Volný, M.; Havlíček, V. mMass3: A Cross-Platform Software Environment for Precise Analysis of Mass Spectrometric Data. Anal. Chem. 2010, 82, 4648–4651. [Google Scholar] [CrossRef]
- Pang, Z.; Lu, Y.; Zhou, G.; Hui, F.; Xu, L.; Viau, C.; Spigelman, A.F.; MacDonald, P.E.; Wishart, D.S.; Li, S.; et al. MetaboAnalyst 6.0: Towards a Unified Platform for Metabolomics Data Processing, Analysis and Interpretation. Nucleic Acids Res. 2024, 52, W398–W406. [Google Scholar] [CrossRef]
- Molenaar, M.R.; Jeucken, A.; Wassenaar, T.A.; Van De Lest, C.H.A.; Brouwers, J.F.; Helms, J.B. LION/Web: A Web-Based Ontology Enrichment Tool for Lipidomic Data Analysis. GigaScience 2019, 8, giz061. [Google Scholar] [CrossRef]
- Carey, H.; Pegios, M.; Martin, L.; Saleeba, C.; Turner, A.J.; Everett, N.A.; Bjerke, I.E.; Puchades, M.A.; Bjaalie, J.G.; McMullan, S. DeepSlice: Rapid Fully Automatic Registration of Mouse Brain Imaging to a Volumetric Atlas. Nat. Commun. 2023, 14, 5884. [Google Scholar] [CrossRef]
- Puchades, M.A.; Csucs, G.; Ledergerber, D.; Leergaard, T.B.; Bjaalie, J.G. Spatial Registration of Serial Microscopic Brain Images to Three-Dimensional Reference Atlases with the QuickNII Tool. PLoS ONE 2019, 14, e0216796. [Google Scholar] [CrossRef] [PubMed]
- Engel, K.M.; Prabutzki, P.; Leopold, J.; Nimptsch, A.; Lemmnitzer, K.; Vos, D.R.N.; Hopf, C.; Schiller, J. A New Update of MALDI-TOF Mass Spectrometry in Lipid Research. Prog. Lipid Res. 2022, 86, 101145. [Google Scholar] [CrossRef] [PubMed]
- Leopold, J.; Prabutzki, P.; Engel, K.M.; Schiller, J. A Five-Year Update on Matrix Compounds for MALDI-MS Analysis of Lipids. Biomolecules 2023, 13, 546. [Google Scholar] [CrossRef]
- Zemski Berry, K.A.; Hankin, J.A.; Barkley, R.M.; Spraggins, J.M.; Caprioli, R.M.; Murphy, R.C. MALDI Imaging of Lipid Biochemistry in Tissues by Mass Spectrometry. Chem. Rev. 2011, 111, 6491–6512. [Google Scholar] [CrossRef]
- Tracey, T.J.; Kirk, S.E.; Steyn, F.J.; Ngo, S.T. The Role of Lipids in the Central Nervous System and Their Pathological Implications in Amyotrophic Lateral Sclerosis. Semin. Cell Dev. Biol. 2021, 112, 69–81. [Google Scholar] [CrossRef]
- Newton, J.; Lima, S.; Maceyka, M.; Spiegel, S. Revisiting the Sphingolipid Rheostat: Evolving Concepts in Cancer Therapy. Exp. Cell Res. 2015, 333, 195–200. [Google Scholar] [CrossRef]
- Hawkins, C.C.; Ali, T.; Ramanadham, S.; Hjelmeland, A.B. Sphingolipid Metabolism in Glioblastoma and Metastatic Brain Tumors: A Review of Sphingomyelinases and Sphingosine-1-Phosphate. Biomolecules 2020, 10, 1357. [Google Scholar] [CrossRef]
- Sousa, N.; Geiß, C.; Bindila, L.; Lieberwirth, I.; Kim, E.; Régnier-Vigouroux, A. Targeting Sphingolipid Metabolism with the Sphingosine Kinase Inhibitor SKI-II Overcomes Hypoxia-Induced Chemotherapy Resistance in Glioblastoma Cells: Effects on Cell Death, Self-Renewal, and Invasion. BMC Cancer 2023, 23, 762. [Google Scholar] [CrossRef]
- Abdel Hadi, L.; Di Vito, C.; Marfia, G.; Navone, S.E.; Campanella, R.; Riboni, L. The Role and Function of Sphingolipids in Glioblastoma Multiforme. In Bioactive Sphingolipids in Cancer Biology and Therapy; Hannun, Y.A., Luberto, C., Mao, C., Obeid, L.M., Eds.; Springer International Publishing: Cham, Switzerland, 2015; pp. 259–293. ISBN 978-3-319-20749-0. [Google Scholar]
| Matrix | Conc. (mg/mL) | Solvent | Temp. (°C) | N. Passes | Flow Rate (mL/min) | Velocity (mm/min) | Pressure (psi) |
|---|---|---|---|---|---|---|---|
| ATT | 10 | 70:30 MeOH:H2O 2 mM GUA 20 mM DAHC | 75 | 4 | 0.12 | 1200 | 10 |
| DHB | 20 | 90:10 MeOH:H2O | 60 | 16 | 0.05 | 1350 | 10 |
| NOR | 5 | 90:10 MeOH:H2O | 60 | 16 | 0.05 | 1350 | 10 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Porto, N.S.; Serrao, S.; Bindi, G.; Monza, N.; Fumagalli, C.; Denti, V.; Piga, I.; Smith, A. Exploring 6-aza-2-Thiothymine as a MALDI-MSI Matrix for Spatial Lipidomics of Formalin-Fixed Paraffin-Embedded Clinical Samples. Metabolites 2025, 15, 531. https://doi.org/10.3390/metabo15080531
Porto NS, Serrao S, Bindi G, Monza N, Fumagalli C, Denti V, Piga I, Smith A. Exploring 6-aza-2-Thiothymine as a MALDI-MSI Matrix for Spatial Lipidomics of Formalin-Fixed Paraffin-Embedded Clinical Samples. Metabolites. 2025; 15(8):531. https://doi.org/10.3390/metabo15080531
Chicago/Turabian StylePorto, Natalia Shelly, Simone Serrao, Greta Bindi, Nicole Monza, Claudia Fumagalli, Vanna Denti, Isabella Piga, and Andrew Smith. 2025. "Exploring 6-aza-2-Thiothymine as a MALDI-MSI Matrix for Spatial Lipidomics of Formalin-Fixed Paraffin-Embedded Clinical Samples" Metabolites 15, no. 8: 531. https://doi.org/10.3390/metabo15080531
APA StylePorto, N. S., Serrao, S., Bindi, G., Monza, N., Fumagalli, C., Denti, V., Piga, I., & Smith, A. (2025). Exploring 6-aza-2-Thiothymine as a MALDI-MSI Matrix for Spatial Lipidomics of Formalin-Fixed Paraffin-Embedded Clinical Samples. Metabolites, 15(8), 531. https://doi.org/10.3390/metabo15080531











