Differences in Metabolite Profiles and Bioactivities of Intra-Strain Variants of Marine Fungus Penicillium antarcticum KMM 4668
Abstract
1. Introduction
2. Materials and Methods
2.1. Fungal Intra-Strain Variant Selection
2.2. DNA Extraction, Amplification, and Sequencing
2.3. Multi-Locus Phylogenetic Analysis
2.4. Cultivation of Fungi
2.5. Extraction and HPLC MS Analysis
2.5.1. Extraction of Fungal Cultures
2.5.2. HPLC MS Analysis of Fungal Extracts
2.5.3. UHPLC-Q-TOF Data Analysis
2.6. Principal Component Analysis (PCA)
2.7. Cytotoxic Activity of Extracts
3. Results
3.1. Morphological Study
3.2. Molecular Identification of the Fungal Strain Variants
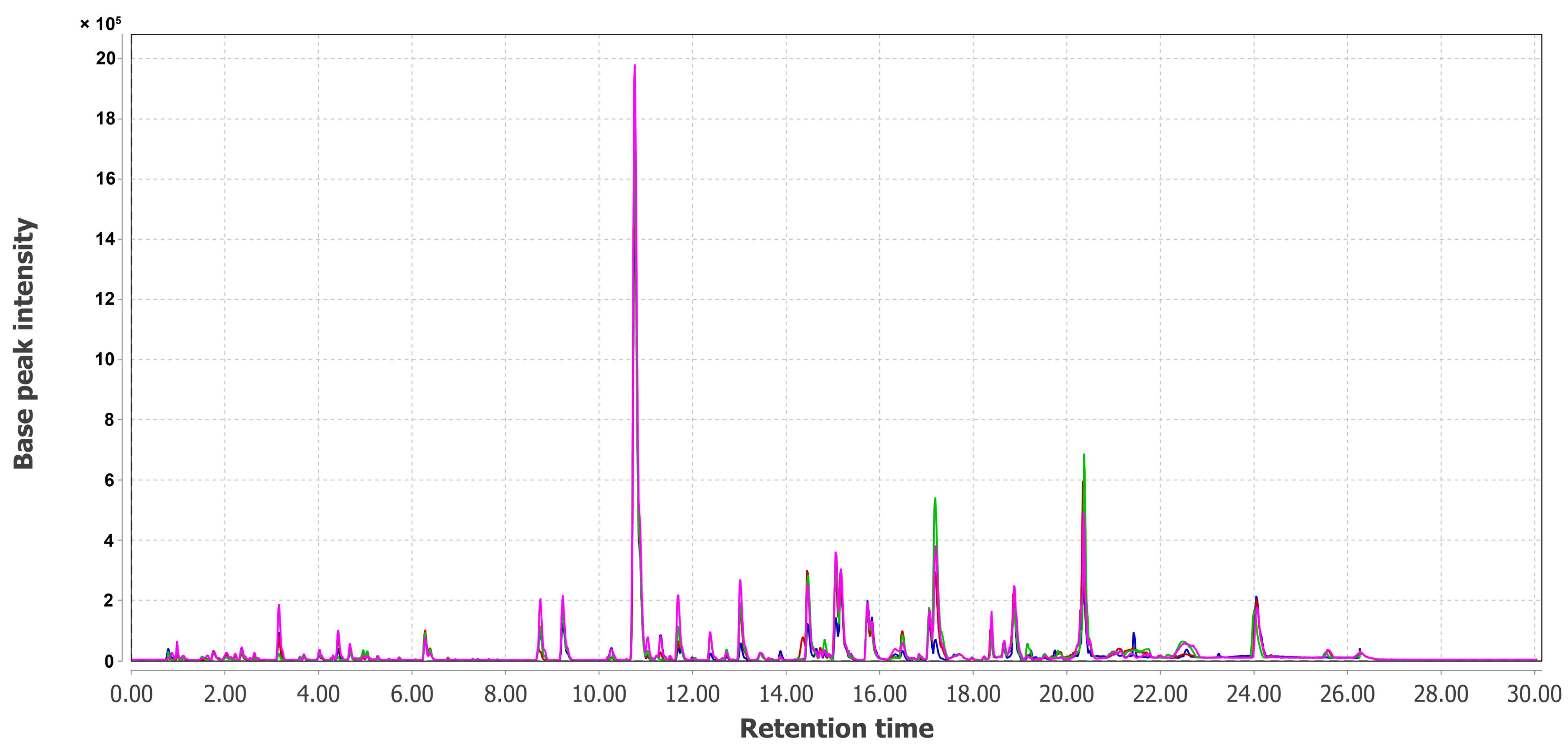
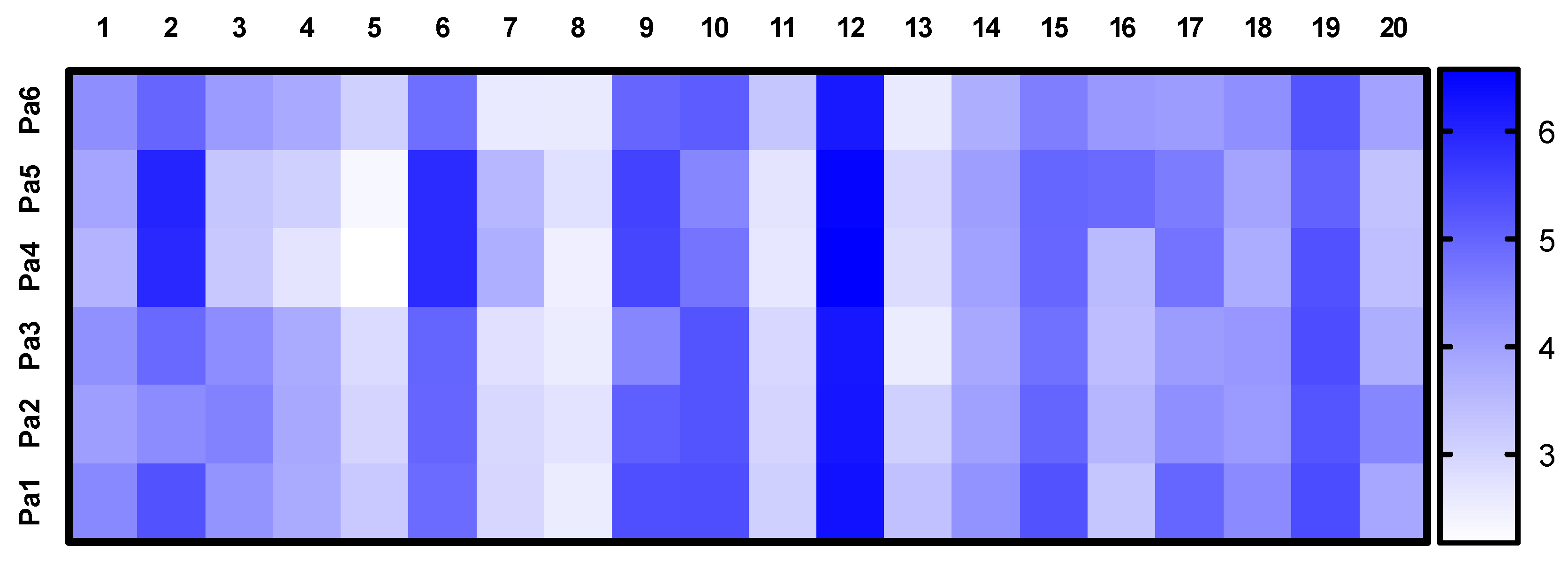
3.3. PCA of UHPLC MS Chromatogramms of Fungal Extracts
3.4. Bioassay
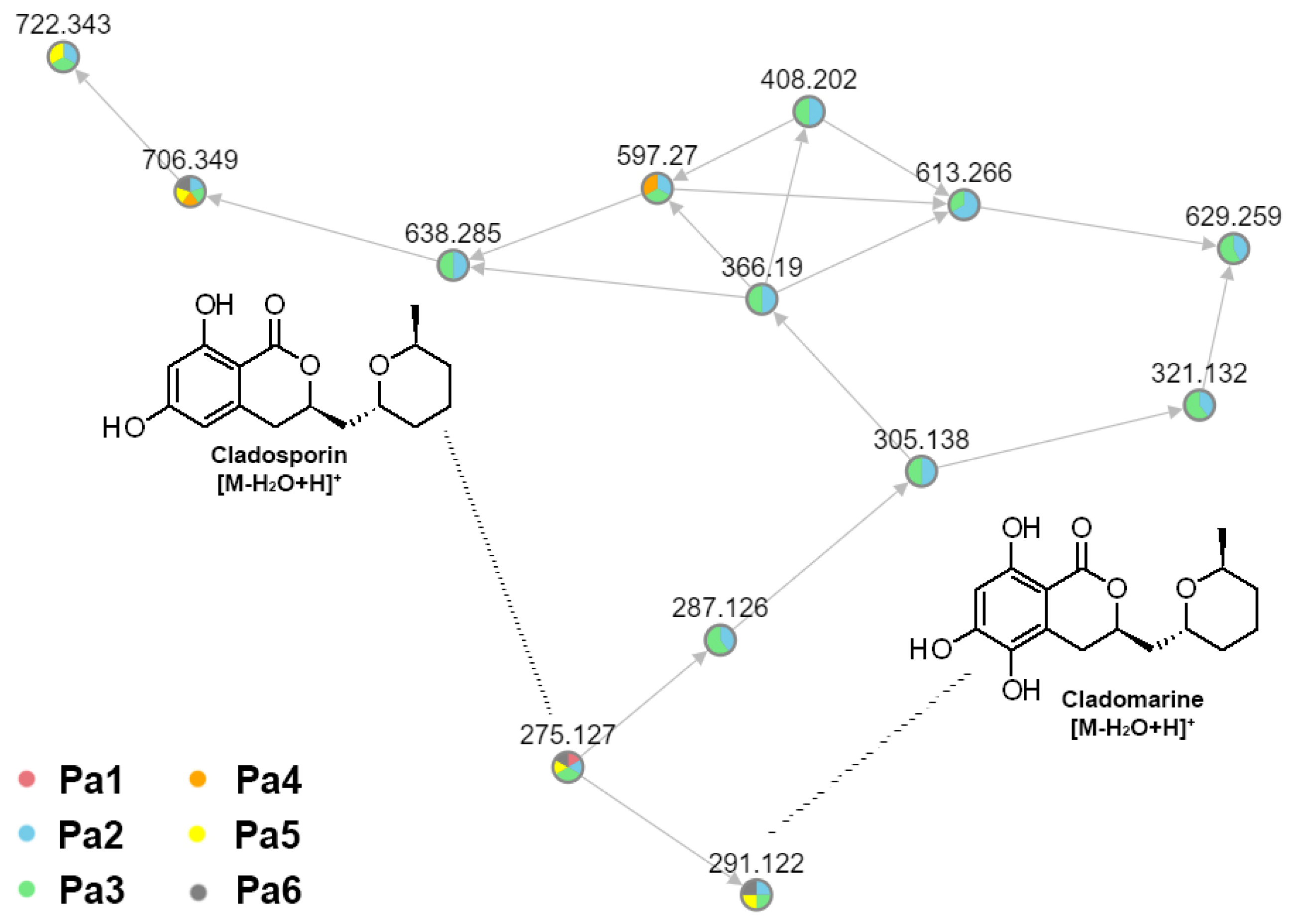
3.5. Identification of the Compounds in UHPLC MS Chromatogramms of P. antarcticum Strains
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| # | RT, min | Measured m/z | MF | Calculated m/z | Error, ppm | MQScore (GNPS) | Structure | Name and Identification Level | Ref. |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 2.03 | 195.1008 [M + H]+ | C11H14O3 | 195.1016 | −3.95 | - | - | - | - |
| 2 | 3.15 | 191.0805 [M + H]+ | C10H10N2O2 | 191.0815 | −5.25 | 0.87 |  | Chrysogine b | [15] |
| 3 | 5.05 | 325.1285 [M + H]+ | C16H20O7 | 325.1282 | 0.99 | - |  | 14-hydroxyasperentin B a | [17] |
| 4 | 5.18 | 283.1552 [M + H]+ | C15H22O5 | 283.1540 | 4.24 | - |  | Penigrisacid D c | [47] |
| - | 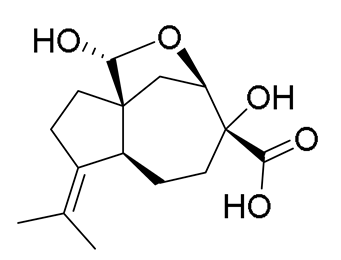 | Piltunine C c | [20] | ||||||
| 5 | 6.08 | 283.1552 [M + H]+ | C15H22O5 | 283.1540 | 4.24 | - |  | Piltunine A c | [20] |
| 6 | 6.28 | 309.1332 [M + H]+ | C16H20O6 | 309.1333 | −0.21 | - |  | Hydroxyasperentin B c (Cladomarin) | [8] |
| - | 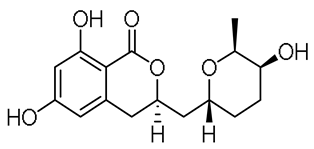 | 14-hydroxyasperentin c (14-hydroxycladosporin) | [17] | ||||||
| - | 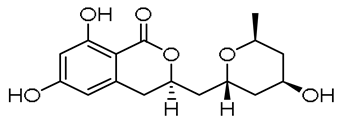 | 13-hydroxyasperentin c (13-hydroxycladosporin) | [48] | ||||||
| 7 | 7.12 | 267.1227 [M + H]+ | C14H18O5 | 267.1227 | 0.00 | - | 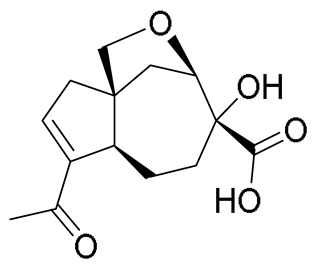 | Piltunine E c | [20] |
| 8 | 7.37 | 267.1227 [M + H]+ | C14H18O5 | 267.1227 | 0.00 | - | 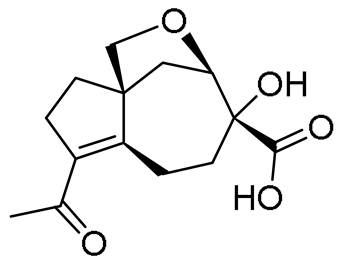 | Piltunine F c | [20] |
| 9 | 8.75 | 535.2716 [M + H]+ | C33H34N4O3 | 535.2704 | 2.24 | - | - | ||
| 10 | 9.23 | 309.1336 [M + H]+ | C16H20O6 | 309.1333 | 1.08 | - | 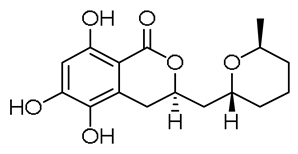 | Cladomarin c | [8] |
| - | 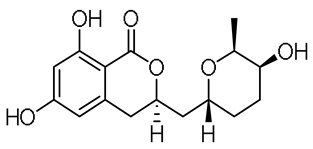 | 14-hydroxyasperentin c (14-hydroxycladosporin) | [17] | ||||||
| - | 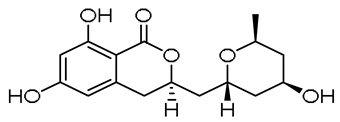 | 13-hydroxyasperentin c (13-hydroxycladosporin) | [48] | ||||||
| 11 | 9.32 | 267.1585 [M + H]+ | C15H22O4 | 267.1591 | −2.25 | - | 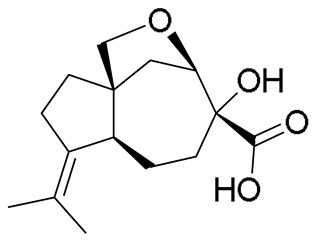 | Aspterric acid c | - |
| 12 | 10.76 | 293.1377 [M + H]+ | C16H20O5 | 293.1384 | −2.39 | - |  | Cladosporin a (Asperentin) | [19] |
| 13 | 11.69 | 501.2126 [M + H]+ | C27H32O9 | 501.2119 | 1.38 | 0.95 |  | Austine b | [49] |
| 14 | 11.69 | 523.2281 [M + Na]+ | C28H36O8 | 523.2302 | −4.09 | 0.95 | 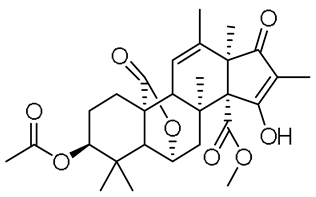 | Citreohybridonol b | [16] |
| 15 | 11.69 | 441.2272 [M + H]+ | C26H32O6 | 441.2272 | 0.08 | - | 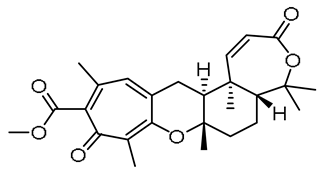 | Tropolactone C c | [50] |
| 16 | 12.72 | 318.2789 [M + H]+ | C21H35NO | 318.2791 | −0.76 | 0.81 | 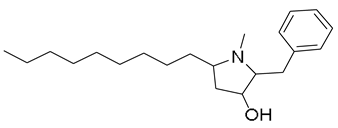 | Preussin b | [51] |
| 17 | 14.48 | 507.2282 [M + H]+ | C32H30N2O4 | 507.2278 | 0.72 | 0.91 |  | Asperphenamate b | [52] |
| 18 | 14.92 | 279.2320 [M + H]+ | C18H30O2 | 279.2319 | 0.51 | - | - | - | |
| 19 | 18.87 | 282.2793 [M + H]+ | C18H35NO | 282.2791 | 0.56 | - | - | - | |
| 20 | 19.85 | 411.3259 [M–H2O + H]+ | C28H44O3 | 411.3258 | 0.35 | 0.82 | 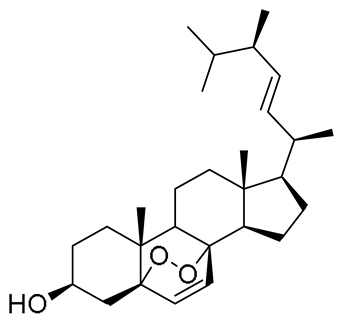 | Ergosterol peroxide a | [53] |
References
- Xiliang, Y.; Jinping, L.; Jiahui, M.; Rui, J.; Shizheng, T.; Huafeng, D.; Jing, L.; Sumei, Y.; Juan, L. Origins, Structures, and Bioactivities of Secondary Metabolites from Marine-derived Penicillium Fungi. Mini-Rev. Med. Chem. 2021, 21, 2000–2019. [Google Scholar] [CrossRef]
- Yurchenko, A.N.; Girich, E.V.; Yurchenko, E.A. Metabolites of Marine Sediment-Derived Fungi: Actual Trends of Biological Activity Studies. Mar. Drugs 2021, 19, 88. [Google Scholar] [CrossRef]
- Visagie, C.M.; Frisvad, J.C.; Houbraken, J.; Visagie, A.; Samson, R.A.; Jacobs, K. A re-evaluation of Penicillium section Canescentia, including the description of five new species. Persoonia Mol. Phylogeny Evol. Fungi 2021, 46, 163–187. [Google Scholar] [CrossRef]
- McRae, C.F.; Seppelt, R.D.; Hocking, A.D. Penicillium species from terrestrial habitats in the Windmill Islands, East Antarctica, including a new species, Penicillium antarcticum. Polar Biol. 1999, 21, 97–111. [Google Scholar] [CrossRef]
- Kirichuk, N.N.; Pivkin, M.V.; Matveeva, T.V. Three new Penicillium species from marine subaqueous soils. Mycol. Prog. 2017, 16, 15–26. [Google Scholar] [CrossRef]
- Park, M.S.; Lee, E.J.; Fong, J.J.; Sohn, J.H.; Lim, Y.W. A new record of Penicillium antarcticum from marine environments in Korea. Mycobiology 2014, 42, 109–113. [Google Scholar] [CrossRef]
- Vansteelandt, M.; Kerzaon, I.; Blanchet, E.; Fossi Tankoua, O.; Robiou Du Pont, T.; Joubert, Y.; Monteau, F.; Le Bizec, B.; Frisvad, J.C.; Pouchus, Y.F.; et al. Patulin and secondary metabolite production by marine-derived Penicillium strains. Fungal Biol. 2012, 116, 954–961. [Google Scholar] [CrossRef]
- Takahashi, K.; Sakai, K.; Nagano, Y.; Orui Sakaguchi, S.; Lima, A.O.; Pellizari, V.H.; Iwatsuki, M.; Takishita, K.; Nonaka, K.; Fujikura, K.; et al. Cladomarine, a new anti-saprolegniasis compound isolated from the deep-sea fungus, Penicillium coralligerum YK-247. J. Antibiot. 2017, 70, 911–914. [Google Scholar] [CrossRef]
- Wiese, J.; Aldemir, H.; Schmaljohann, R.; Gulder, T.A.M.; Imhoff, J.F. Asperentin B, a New Inhibitor of the Protein Tyrosine Phosphatase 1B. Mar. Drugs 2017, 15, 191. [Google Scholar] [CrossRef]
- Zang, Y.; Gong, Y.; Shi, Z.; Qi, C.; Chen, C.; Tong, Q.; Liu, J.; Wang, J.; Zhu, H.; Zhang, Y. Multioxidized aromatic polyketides produced by a soil-derived fungus Penicillium canescens. Phytochemistry 2022, 193, 113012. [Google Scholar] [CrossRef] [PubMed]
- Ishikawa, Y.; Morimoto, K.; Iseki, S. Atrovenetin as a potent antioxidant compound from Penicillium species. J. Am. Oil Chem. Soc. 1991, 68, 666–668. [Google Scholar] [CrossRef]
- Shiono, Y.; Seino, Y.; Koseki, T.; Murayama, T.; Kimura, K.-i. Antarones A and B, Two Polyketides from an Endophytic Penicillium antarcticum. Z. Für Naturforschung B 2008, 63, 909–914. [Google Scholar] [CrossRef]
- Marchese, P.; Mahajan, N.; O’Connell, E.; Fearnhead, H.; Tuohy, M.; Krawczyk, J.; Thomas, O.P.; Barry, F.; Murphy, M.J. A Novel High-Throughput Screening Platform Identifies Itaconate Derivatives from Marine Penicillium antarcticum as Inhibitors of Mesenchymal Stem Cell Differentiation. Mar. Drugs 2020, 18, 192. [Google Scholar] [CrossRef]
- Ravunni, M.; Benjamin, P.S. Penicillium verruculosum strain BS3 produces aurantioclavine and rugulosuvine B Alkaloids. Electron. J. Biol. 2016, 12, 484–489. [Google Scholar]
- Viggiano, A.; Salo, O.; Ali, H.; Szymanski, W.; Lankhorst Peter, P.; Nygård, Y.; Bovenberg Roel, A.L.; Driessen Arnold, J.M. Pathway for the Biosynthesis of the Pigment Chrysogine by Penicillium chrysogenum. Appl. Environ. Microbiol. 2018, 84, e02246-17. [Google Scholar] [CrossRef] [PubMed]
- Kosemura, S. Meroterpenoids from Penicillium citreo-viride B. IFO 4692 and 6200 hybrid. Tetrahedron 2003, 59, 5055–5072. [Google Scholar] [CrossRef]
- Leshchenko, E.V.; Antonov, A.S.; Borkunov, G.V.; Hauschild, J.; Zhuravleva, O.I.; Khudyakova, Y.V.; Menshov, A.S.; Popov, R.S.; Kim, N.Y.; Graefen, M.; et al. New Bioactive β-Resorcylic Acid Derivatives from the Alga-Derived Fungus Penicillium antarcticum KMM 4685. Mar. Drugs 2023, 21, 178. [Google Scholar] [CrossRef] [PubMed]
- Leshchenko, E.V.; Antonov, A.S.; Dyshlovoy, S.A.; Berdyshev, D.V.; Hauschild, J.; Zhuravleva, O.I.; Borkunov, G.V.; Menshov, A.S.; Kirichuk, N.N.; Popov, R.S.; et al. Meroantarctines A–C, meroterpenoids with rearranged skeletons from the alga-derived fungus Penicillium antarcticum KMM 4685 with potent p-glycoprotein inhibitory activity. J. Nat. Prod. 2022, 85, 2746–2752. [Google Scholar] [CrossRef]
- Yurchenko, A.N.; Zhuravleva, O.I.; Khmel, O.O.; Oleynikova, G.K.; Antonov, A.S.; Kirichuk, N.N.; Chausova, V.E.; Kalinovsky, A.I.; Berdyshev, D.V.; Kim, N.Y.; et al. New Cyclopiane Diterpenes and Polyketide Derivatives from Marine Sediment-Derived Fungus Penicillium antarcticum KMM 4670 and Their Biological Activities. Mar. Drugs 2023, 21, 584. [Google Scholar] [CrossRef]
- Afiyatullov, S.S.; Zhuravleva, O.I.; Antonov, A.S.; Leshchenko, E.V.; Pivkin, M.V.; Khudyakova, Y.V.; Denisenko, V.A.; Pislyagin, E.A.; Kim, N.Y.; Berdyshev, D.V.; et al. Piltunines A-F from the marine-derived fungus Penicillium piltunense KMM 4668. Mar. Drugs 2019, 17, 647. [Google Scholar] [CrossRef]
- Mayne, R.F.; Bennett, J.W.; Tallant, J. Instability of an aflatoxin-producing strain of Aspergillus parasiticus. Mycologia 1971, 63, 644–648. [Google Scholar] [CrossRef] [PubMed]
- Kucsera, J.; Yarita, K.; Takeo, K.; Yoshida, S.; Gácser, A.; Hamari, Z.; Avasi, Z.; Kevei, F. Intra-strain variability of Cryptococcus neoformans can be detected on Phloxin B medium. J. Basic Microbiol. 2002, 42, 111–119. [Google Scholar] [CrossRef]
- Pazouki, M.; Panda, T. Understanding the morphology of fungi. Bioprocess Eng. 2000, 22, 127–143. [Google Scholar] [CrossRef]
- Visagie, C.M.; Houbraken, J.; Frisvad, J.C.; Hong, S.B.; Klaassen, C.H.W.; Perrone, G.; Seifert, K.A.; Varga, J.; Yaguchi, T.; Samson, R.A. Identification and nomenclature of the genus Penicillium. Stud. Mycol. 2014, 78, 343–371. [Google Scholar] [CrossRef]
- Visagie, C.M.; Yilmaz, N.; Kocsubé, S.; Frisvad, J.C.; Hubka, V.; Samson, R.A.; Houbraken, J. A review of recently introduced Aspergillus, Penicillium, Talaromyces and other Eurotiales species. Stud Mycol. 2024, 107, 1–66. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Kimura, M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef]
- Nesterenko, L.; Popov, R.; Zhuravleva, O.; Kirichuk, N.; Chausova, V.; Krasnov, K.; Mikhail, P.; Yurchenko, E.; Isaeva, M.; Yurchenko, A. A study of the metabolic profiles of Penicillium dimorphosporum KMM 4689 which led to its re-identification as Penicillium hispanicum. Fermentation 2023, 9, 337. [Google Scholar] [CrossRef]
- Pluskal, T.; Castillo, S.; Villar-Briones, A.; Orešič, M. MZmine 2: Modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinform. 2010, 11, 395. [Google Scholar] [CrossRef]
- Jain, N.; Fries, B.C. Antigenic and phenotypic variations in fungi. Cell. Microbiol. 2009, 11, 1716–1723. [Google Scholar] [CrossRef][Green Version]
- Lan, C.-Y.; Newport, G.; Murillo, L.A.; Jones, T.; Scherer, S.; Davis, R.W.; Agabian, N. Metabolic specialization associated with phenotypic switching in Candida albicans. Proc. Natl. Acad. Sci. USA 2002, 99, 14907–14912. [Google Scholar] [CrossRef]
- Horn, B.W.; Dorner, J.W. Effect of competition and adverse culture conditions on aflatoxin production by Aspergillus flavus through successive generations. Mycologia 2002, 94, 741–751. [Google Scholar] [CrossRef]
- Yvert, G.; Ohnuki, S.; Nogami, S.; Imanaga, Y.; Fehrmann, S.; Schacherer, J.; Ohya, Y. Single-cell phenomics reveals intra-species variation of phenotypic noise in yeast. BMC Syst. Biol. 2013, 7, 54. [Google Scholar] [CrossRef] [PubMed]
- Zhgun, A.A. Fungal BGCs for Production of Secondary Metabolites: Main Types, Central Roles in Strain Improvement, and Regulation According to the Piano Principle. Int. J. Mol. Sci. 2023, 24, 11184. [Google Scholar] [CrossRef] [PubMed]
- Naranjo-Ortiz, M.A.; Gabaldón, T.A.-O. Fungal evolution: Cellular, genomic and metabolic complexity. Biol. Rev. Camb. Philos Soc. 2020, 95, 1198–1232. [Google Scholar] [CrossRef]
- Strom, N.B.; Bushley, K.E. Two genomes are better than one: History, genetics, and biotechnological applications of fungal heterokaryons. Fungal Bio. Biotechnol. 2016, 3, 4. [Google Scholar] [CrossRef]
- Jinks, J.L. Heterocaryosis in wild Penicillium. Heredity 1952, 6, 77–87. [Google Scholar] [CrossRef]
- Cochrane, R.V.K.; Sanichar, R.; Lambkin, G.R.; Reiz, B.; Xu, W.; Tang, Y.; Vederas, J.C. Production of New Cladosporin Analogues by Reconstitution of the Polyketide Synthases Responsible for the Biosynthesis of this Antimalarial Agent. Angew. Chem. Int. Ed. 2016, 55, 664–668. [Google Scholar] [CrossRef] [PubMed]
- Yan, Y.; Liu, Q.; Zang, X.; Yuan, S.; Bat-Erdene, U.; Nguyen, C.; Gan, J.; Zhou, J.; Jacobsen, S.E.; Tang, Y. Resistance-gene-directed discovery of a natural-product herbicide with a new mode of action. Nature 2018, 559, 415–418. [Google Scholar] [CrossRef]
- Wang, X.; Wedge, D.E.; Cutler, S.J. Chemical and Biological Study of Cladosporin, an Antimicrobial Inhibitor: A Review. Nat. Prod. Commun. 2016, 11, 1595–1600. [Google Scholar] [CrossRef] [PubMed]
- Hou, A.; Li, B.; Deng, Z.; Xu, M.; Dickschat, J.S. Cladosporin, A Highly Potent Antimalaria Drug? Chem. Bio. Chem. 2023, 24, e202300154. [Google Scholar] [CrossRef]
- Chambers, M.C.; MacLean, B.; Burke, R.; Amodei, D.; Ruderman, D.L.; Neumann, S.; Gatto, L.; Fischer, B.; Pratt, B.; Egertson, J.; et al. A cross-platform toolkit for mass spectrometry and proteomics. Nat. Biotechnol. 2012, 30, 918–920. [Google Scholar] [CrossRef]
- Myers, O.D.; Sumner, S.J.; Li, S.; Barnes, S.; Du, X. One Step Forward for Reducing False Positive and False Negative Compound Identifications from Mass Spectrometry Metabolomics Data: New Algorithms for Constructing Extracted Ion Chromatograms and Detecting Chromatographic Peaks. Anal. Chem. 2017, 89, 8696–8703. [Google Scholar] [CrossRef]
- Nothias, L.F.; Petras, D.; Schmid, R.; Dührkop, K.; Rainer, J.; Sarvepalli, A.; Protsyuk, I.; Ernst, M.; Tsugawa, H.; Fleischauer, M.; et al. Feature-based molecular networking in the GNPS analysis environment. Nat. Methods 2020, 17, 905–908. [Google Scholar] [CrossRef]
- Christiansen, J.V.; Larsen, T.O.; Frisvad, J.C. Production of Fungal Quinones: Problems and Prospects. Biomolecules 2022, 12, 1041. [Google Scholar] [CrossRef]
- Ruttkies, C.; Schymanski, E.L.; Wolf, S.; Hollender, J.; Neumann, S. MetFrag relaunched: Incorporating strategies beyond in silico fragmentation. J. Cheminf. 2016, 8, 3/1. [Google Scholar] [CrossRef] [PubMed]
- Xing, C.-P.; Xie, C.-L.; Xia, J.-M.; Liu, Q.-M.; Lin, W.-X.; Ye, D.-Z.; Liu, G.-M.; Yang, X.-W. Penigrisacids A–D, Four New Sesquiterpenes from the Deep-Sea-Derived Penicillium griseofulvum. Mar. Drugs 2019, 17, 507. [Google Scholar] [CrossRef]
- Grove, J.F. New metabolic products of Aspergillus flavus. Part IV. 4′-Hydroxyasperentin and 5′-hydroxyasperentin 8-methyl ether. J. Chem. Soc. Perkin Trans. 1 1973, 22, 2704–2706. [Google Scholar] [CrossRef] [PubMed]
- Chexal, K.K.; Springer, J.P.; Clardy, J.; Cole, R.J.; Kirksey, J.W.; Dorner, J.W.; Cutler, H.G.; Strawter, B.J. Austin, a novel polyisoprenoid mycotoxin from Aspergillus ustus. J. Am. Chem. Soc. 1976, 98, 6748–6750. [Google Scholar] [CrossRef]
- Cueto, M.; MacMillan, J.B.; Jensen, P.R.; Fenical, W. Tropolactones A–D, four meroterpenoids from a marine-derived fungus of the genus Aspergillus. Phytochemistry 2006, 67, 1826–1831. [Google Scholar] [CrossRef]
- Malhão, F.; Ramos, A.A.; Buttachon, S.; Dethoup, T.; Kijjoa, A.; Rocha, E. Cytotoxic and Antiproliferative Effects of Preussin, a Hydroxypyrrolidine Derivative from the Marine Sponge-Associated Fungus Aspergillus candidus KUFA 0062, in a Panel of Breast Cancer Cell Lines and Using 2D and 3D Cultures. Mar. Drugs 2019, 17, 448. [Google Scholar] [CrossRef]
- Subko, K.; Wang, X.; Nielsen, F.H.; Isbrandt, T.; Gotfredsen, C.H.; Ramos, M.C.; Mackenzie, T.; Vicente, F.; Genilloud, O.; Frisvad, J.C.; et al. Mass Spectrometry Guided Discovery and Design of Novel Asperphenamate Analogs from Penicillium astrolabium Reveals an Extraordinary NRPS Flexibility. Front Microbiol. 2021, 11, 618730. [Google Scholar] [CrossRef] [PubMed]
- Lobo, A.M.; De Abreu, M.; Prabhakar, S.; Godinho, L.S.; Jones, R.; Rzepa, H.S.; Williams, D.J. Triterpenoids of the fungus Pisolithus tinctorius. Phytochemistry 1988, 27, 3569–3574. [Google Scholar] [CrossRef]
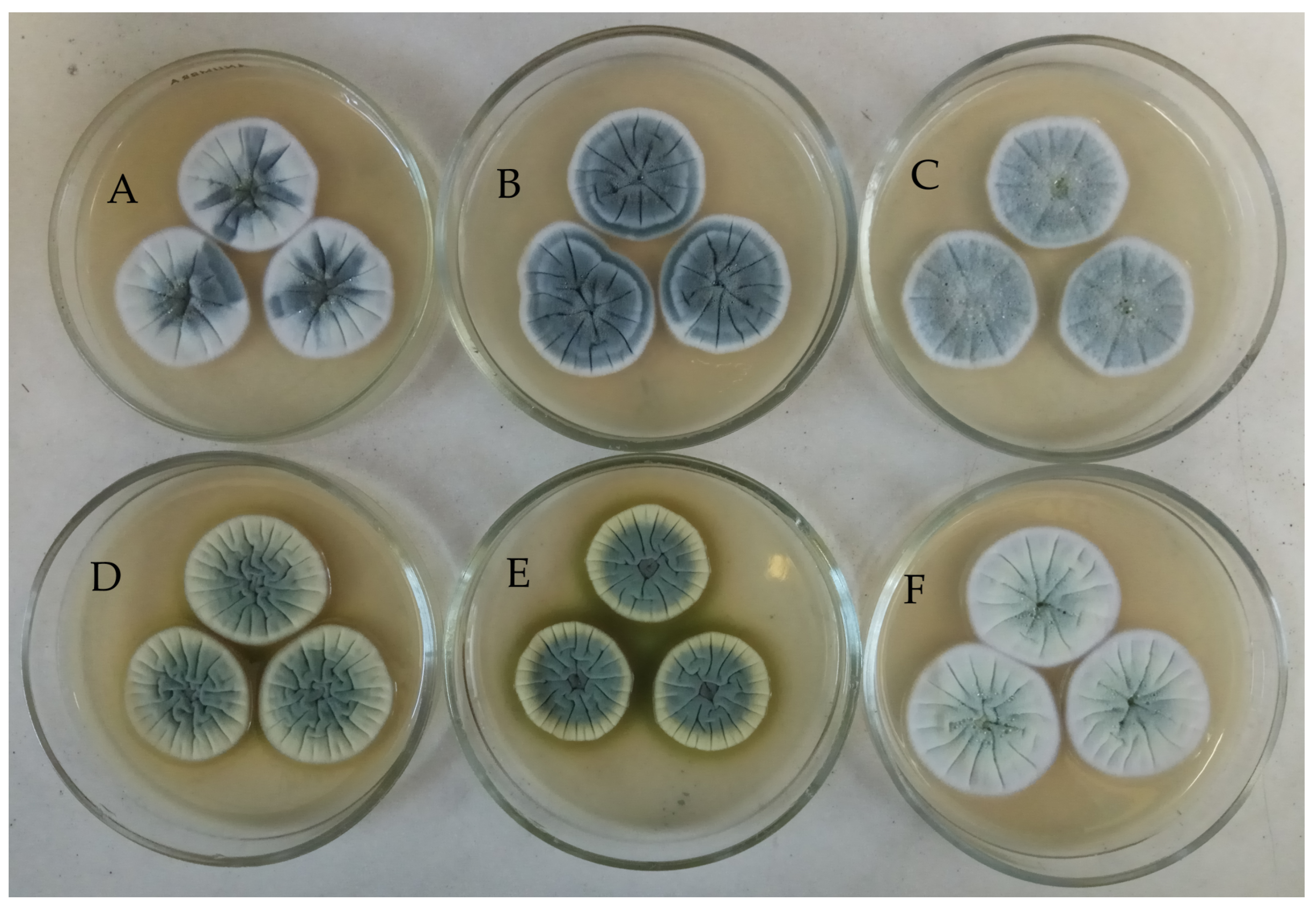




| Extracts | Cell Viability, % of Control | |||
|---|---|---|---|---|
| HeLa | PC-3 | MCF-7 | H9c2 | |
| Pa1 | 8.8 ± 0.1 | 17.1 ± 4.6 | 12.1 ± 0.1 | 19.4 ± 0.6 |
| Pa2 | 8.7 ± 0.4 | 13.9 ± 3.9 | 12.7 ± 0.6 | 20.7 ± 4.4 |
| Pa3 | 9.1 ± 0.5 | 13.3 ± 1.0 | 12.5 ± 0.2 | 19.3 ± 1.0 |
| Pa4 | 33.1 ± 3.4 | 62.3 ± 2.4 | 36.2 ± 2.9 | 31.6 ± 1.5 |
| Pa5 | 27.4 ± 3.6 | 72.8 ± 2.7 | 31.5 ± 0.6 | 41.9 ± 0.7 |
| Pa6 | 8.9 ± 0.1 | 19.9 ± 0.9 | 12.5 ± 0.6 | 22.9 ± 0.9 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Borkunov, G.V.; Kirichuk, N.N.; Chausova, V.E.; Popov, R.S.; Zhuravleva, O.I.; Chingizova, E.A.; Yurchenko, E.A.; Isaeva, M.P.; Yurchenko, A.N. Differences in Metabolite Profiles and Bioactivities of Intra-Strain Variants of Marine Fungus Penicillium antarcticum KMM 4668. Metabolites 2025, 15, 77. https://doi.org/10.3390/metabo15020077
Borkunov GV, Kirichuk NN, Chausova VE, Popov RS, Zhuravleva OI, Chingizova EA, Yurchenko EA, Isaeva MP, Yurchenko AN. Differences in Metabolite Profiles and Bioactivities of Intra-Strain Variants of Marine Fungus Penicillium antarcticum KMM 4668. Metabolites. 2025; 15(2):77. https://doi.org/10.3390/metabo15020077
Chicago/Turabian StyleBorkunov, Gleb V., Natalya N. Kirichuk, Viktoria E. Chausova, Roman S. Popov, Olesya I. Zhuravleva, Ekaterina A. Chingizova, Ekaterina A. Yurchenko, Marina P. Isaeva, and Anton N. Yurchenko. 2025. "Differences in Metabolite Profiles and Bioactivities of Intra-Strain Variants of Marine Fungus Penicillium antarcticum KMM 4668" Metabolites 15, no. 2: 77. https://doi.org/10.3390/metabo15020077
APA StyleBorkunov, G. V., Kirichuk, N. N., Chausova, V. E., Popov, R. S., Zhuravleva, O. I., Chingizova, E. A., Yurchenko, E. A., Isaeva, M. P., & Yurchenko, A. N. (2025). Differences in Metabolite Profiles and Bioactivities of Intra-Strain Variants of Marine Fungus Penicillium antarcticum KMM 4668. Metabolites, 15(2), 77. https://doi.org/10.3390/metabo15020077







