Multi-Omics Characterization of a Novel SSR4 Variant in Congenital Disorders of Glycosylation
Abstract
1. Introduction
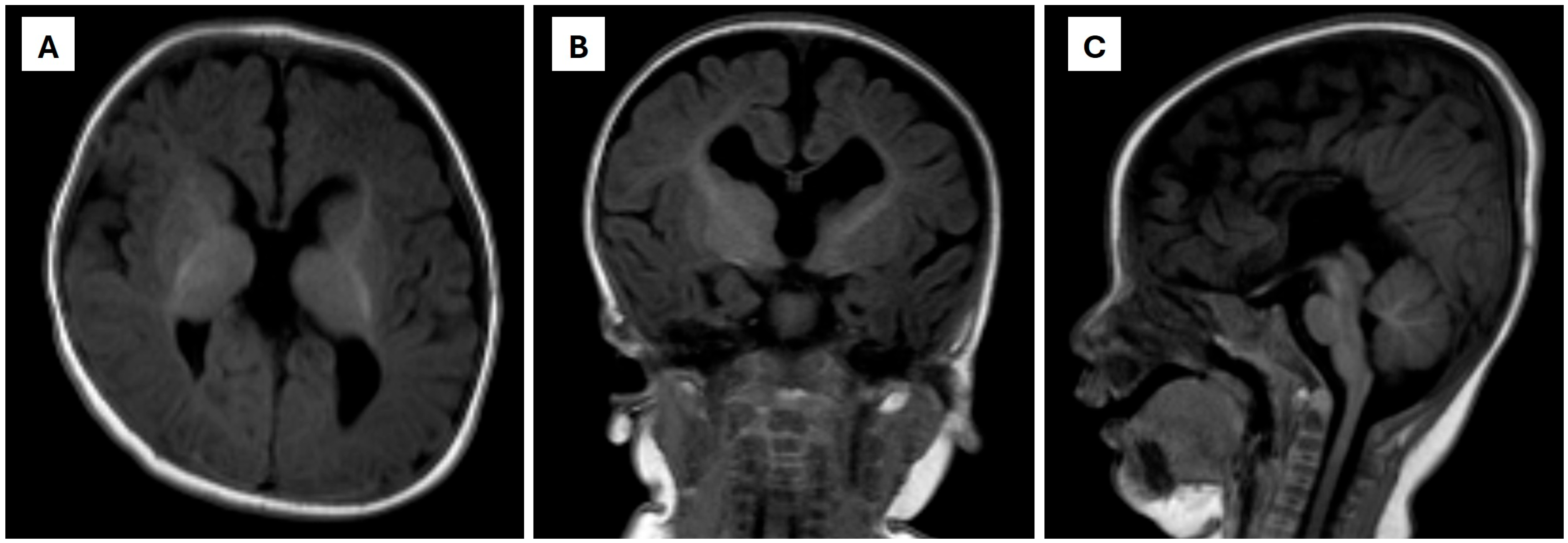
2. Case Presentation
3. Materials and Methods
3.1. Sample Collection and Ethical Considerations
3.2. Transferrin Isoform Analysis
3.3. Transferrin Glycopeptide LC–MS/MS Analysis
3.4. Targeted Metabolomics Profiling by LC–MS/MS
3.5. Whole-Exome Sequencing (WES)
4. Results
4.1. Transferrin Glycopeptide LC-MS/MS
4.2. Targeted Metabolomics Profiling
4.3. Genetic Analysis
5. Discussion
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| CDG | Congenital disorders of glycosylation (CDG) |
| ER | endoplasmic reticulum |
| δ | Delta |
| TRAP | signal sequence receptor |
| MCUG | micturating cystourethrogram |
| MRI | magnetic resonance imaging |
| WES | whole-exome sequencing |
| LC-MS/MS | Liquid Chromatography-Tandem Mass Spectrometry |
| TRFE | Human serum transferrin |
| SNFG | Symbol Nomenclature for Glycans |
| Asn | Asparagine |
| NG | non-glycosylated |
| Hex | Hexose |
| HexNAc | N-acetyl hexosamine |
| Fuc | Fucose (Fuc) |
| SA | Sialic acid |
| LPC | lysophosphatidylcholine |
Appendix A
| Parameter/Test | Patient Value/Summary | Age at Test | Reference Range/Comment |
|---|---|---|---|
| Perinatal/growth | |||
| Birth weight | 2.57 kg | At birth | 2.5–4.0 kg (term male infant) |
| Head circumference | Below 3rd centile from early infancy; decelerating head growth (microcephaly) | Infancy/follow-up | WHO growth standards for boys 0–2 years |
| Weight-for-age/length-for-age | Persistently below expected percentiles (failure to thrive) | Infancy/follow-up | WHO growth standards for boys 0–2 years |
| Basic Biochemistry | |||
| Liver function test | Total bilirubin 27.5 µmol/L; ALT 15 U/L; AST 19 U/L; ALP 248 U/L; albumin 33 g/L; globulin 17 g/L; total protein 58 g/L | 1 month | Local pediatric reference ranges |
| Venous blood gas | pH 7.38; pCO2 40 mmHg; pO2 40 mmHg; HCO3− 23.7 mmol/L; BE −1.1 | 2 months | Local pediatric reference ranges |
| Ammonia | 66.1 µmol/L | 11 months | <50 µmol/L (mildly elevated) |
| Lactate | 1.98 mmol/L | 16 months | <2.2 mmol/L |
| Thyroid function test | TSH 0.87 mIU/L; free T4 12.65 pmol/L | 16 months | Within reference ranges [10] |
| Endocrine/genital | |||
| Penile length (stretched) | 2.3 cm at 1 month; 3.6 cm at 13 months | Infancy/follow-up | Within reported Asian neonatal reference range for term males [11,12] |
| Gonadotropins/sex steroids (FSH, LH, testosterone) | Within age-appropriate reference ranges | 1 month | Based on published pediatric endocrine reference intervals [13] |
| Cortisol | Within age-appropriate reference range | 1 month | Within reference ranges [14] |
| Inborn errors of metabolism screening | |||
| Newborn IEM screening (DBS amino acids and acylcarnitines) | Isolated mild increase in arginine; other amino acids and acylcarnitines normal | 11 months | Local pediatric reference ranges |
| Plasma amino acid profile | All amino acids within normal limits | Local pediatric reference ranges | |
| Urine organic acid profile | No significant abnormal peaks | Local pediatric reference ranges |

References
- Ng, B.G.; Freeze, H.H.; Himmelreich, N.; Blau, N.; Ferreira, C.R. Clinical and biochemical footprints of congenital disorders of glycosylation: Proposed nosology. Mol. Genet. Metab. 2024, 142, 108476. [Google Scholar] [CrossRef] [PubMed]
- Losfeld, M.E.; Ng, B.G.; Kircher, M.; Buckingham, K.J.; Turner, E.H.; Eroshkin, A.; Smith, J.D.; Shendure, J.; Nickerson, D.A.; Bamshad, M.J.; et al. A new congenital disorder of glycosylation caused by a mutation in SSR4, the signal sequence receptor 4 protein of the TRAP complex. Hum. Mol. Genet. 2014, 23, 1602–1605. [Google Scholar] [CrossRef] [PubMed]
- Ng, B.G.; Raymond, K.; Kircher, M.; Buckingham, K.J.; Wood, T.; Shendure, J.; University of Washington Center for Mendelian Genomics; Wong, J.T.S.; Monteiro, F.P.; Graham, B.H.; et al. Expanding the molecular and clinical phenotype of SSR4-CDG. Hum. Mutat. 2015, 36, 1048–1051. [Google Scholar] [CrossRef] [PubMed]
- Johnsen, C.; Tabatadze, N.; Radenkovic, S.; Botzo, G.; Kuschel, B.; Melikishvili, G.; Morava, E. SSR4-CDG, an ultra-rare X-linked congenital disorder of glycosylation affecting the TRAP complex: Review of 22 affected individuals including the first adult patient. Mol. Genet. Metab. 2024, 142, 108477. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.; Jin, X.; Zhu, X. A novel SSR4 variant associated with congenital disorder of glycosylation: A case report and related analysis. Front. Genet. 2024, 15, 1402883. [Google Scholar] [CrossRef] [PubMed]
- Abu Bakar, N.; Hamzan, N.I. Advancement in clinical glycomics and glycoproteomics for congenital disorders of glycosylation: Progress and challenges ahead. Biomedicines 2025, 13, 1964. [Google Scholar] [CrossRef] [PubMed]
- Freeze, H.H.; Eklund, E.A.; Ng, B.G.; Patterson, M.C. Neurology of inherited glycosylation disorders. Lancet Neurol. 2015, 14, 923–936. [Google Scholar] [CrossRef] [PubMed]
- Lipiński, P.; Tylki-Szymańska, A. Congenital disorders of glycosylation: What clinicians need to know. Front Pediatr. 2021, 9, 715151. [Google Scholar] [CrossRef] [PubMed]
- Francisco, R.; Brasil, S.; Poejo, J.; Jaeken, J.; Pascoal, C.; Videira, P.A.; Ferreira, V.D.R. Congenital disorders of glycosylation (CDG): State of the art in 2022. Orphanet J. Rare Dis. 2023, 18, 329. [Google Scholar] [CrossRef] [PubMed]
- Fatourechi, A.; Ardakani, H.M.; Sayarifard, F.; Sheikh, M. Hypothyroidism among pediatric patients with type 1 diabetes mellitus, from patients’ characteristics to disease severity. Clin. Pediatr. Endocrinol. 2017, 26, 73–80. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Ting, T.H.; Wu, L.L. Penile length of term newborn infants in multiracial Malaysia. Singap. Med. J. 2009, 50, 817–821. [Google Scholar]
- Park, S.K.; Ergashev, K.; Chung, J.M.; Lee, S.D. Penile circumference and stretched penile length in prepubertal children: A retrospective, single-center pilot study. Investig. Clin. Urol. 2021, 62, 324–330. [Google Scholar] [CrossRef] [PubMed]
- Tomlinson, C.; Macintyre, H.; Dorrian, C.A.; Ahmed, S.F.; Wallace, A.M. Testosterone measurements in early infancy. Arch. Dis. Child. Fetal Neonatal Ed. 2004, 89, F558–F559. [Google Scholar] [CrossRef] [PubMed]
- Child and Adolescent Health Service. Cortisol Estimation and ACTH Stimulation Testing; Child and Adolescent Health Service: Perth, Australia, 2024. Available online: https://www.cahs.health.wa.gov.au/-/media/HSPs/CAHS/Documents/Health-Professionals/Neonatology-guidelines/Cortisol-Estimation-and-ACTH-Stimulation-Testing.pdf (accessed on 23 November 2025).
| Transferrin Isoform | Patient (%) | Reference Range (%) | Interpretation |
|---|---|---|---|
| Asialo-transferrin | 0.3 | 0.0–0.2 | Slightly elevated |
| Disialo-transferrin | 5.2 | 0.1–1.1 | Increased |
| Trisialo-transferrin | 14.9 | 6.3–12.4 | Mildly elevated |
| Tetrasialo-transferrin | 79.6 | 80.0–90.9 | Reduced |
| Pentasialo-transferrin | 0.0 | 0.1–1.1 | Within normal limits |
| Hexasialo-transferrin | 0.0 | 0.0–0.3 | Absent |
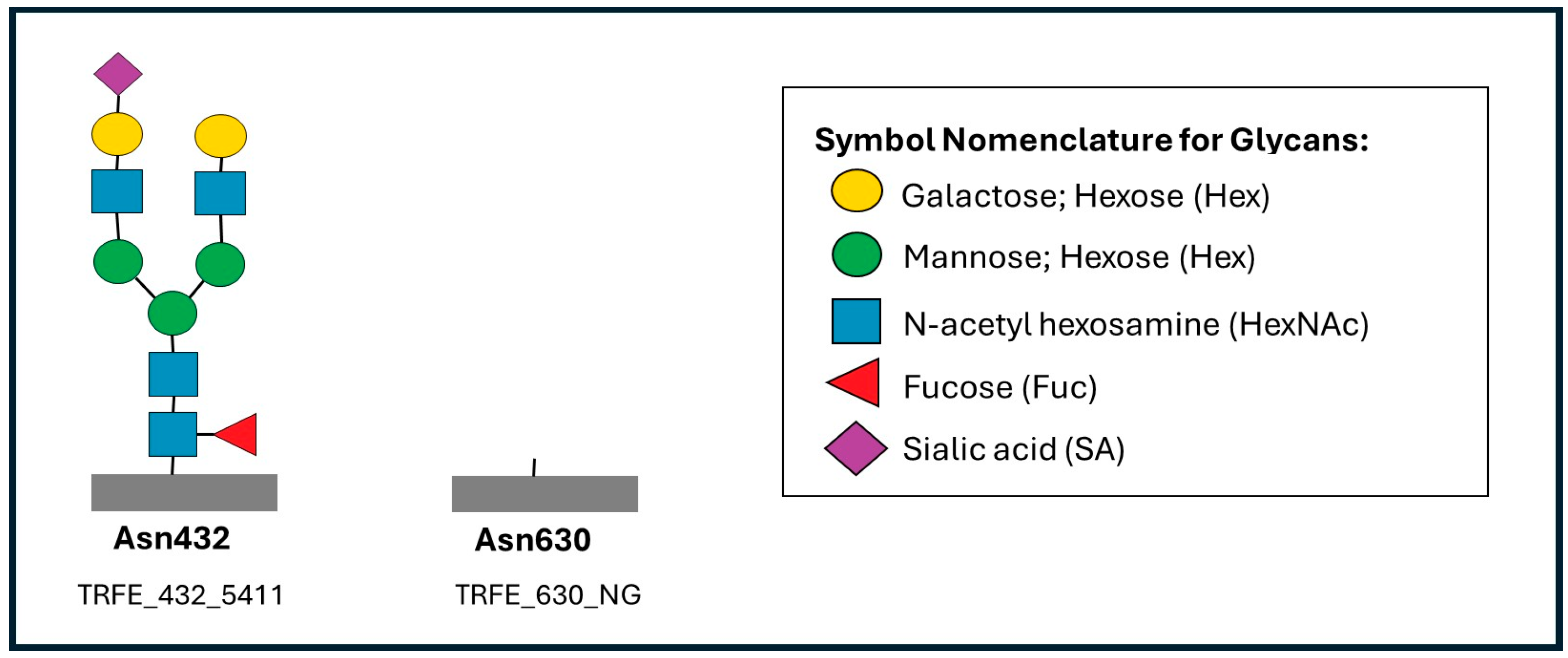
| Site | Glycopeptide | Glycoform Composition | * Finding in Patient | Interpretation |
|---|---|---|---|---|
| Asn432 | TRFE_432_5401 | Hex5HexNAc4Fuc0SA1 | ↓ Reduced | Loss of sialylated biantennary glycans (Asn432) |
| Asn432 | TRFE_432_5412 | Hex5HexNAc4Fuc1SA2 | ↑ Elevated | High fucosylated biantennary glycans (Asn432) |
| Asn432 | TRFE_432_6503 | Hex6HexNAc5Fuc0SA3 | ↑ Elevated | High sialylated triantennary glycans (Asn432) |
| Asn432 | TRFE_432_NG | Non-glycosylated peptide | ↑ Elevated | Impaired glycan occupancy (Asn 432) |
| Asn630 | TRFE_630_5401 | Hex5HexNAc4Fuc0SA1 | ↓ Reduced | Loss of sialylated biantennary glycans (Asn630) |
| Asn630 | TRFE_630_5402 | Hex5HexNAc4Fuc0SA2 | ↓ Reduced | Loss of sialylated biantennary glycans (Asn630) |
| Asn630 | TRFE_630_5411 | Hex5HexNAc4Fuc1SA1 | ↓ Reduced | Loss of fucosylated biantennary glycans (Asn630) |
| Asn630 | TRFE_630_5412 | Hex5HexNAc4Fuc1SA2 | ↓ Reduced | Loss of fucosylated biantennary glycans (Asn630) |
| Asn630 | TRFE_630_NG | Non-glycosylated peptide | ↑ Elevated | Impaired glycan occupancy (Asn630) |
| Metabolite Class | Compound | Patient Value (µM) | Control Mean (µM) | * Change |
|---|---|---|---|---|
| Amino acid | Arginine | 43.7 | 29.3 | ↑ Elevated (p < 0.05) |
| Phenylalanine | 31.9 | 23.9 | ↑ Elevated (p < 0.05) | |
| Glutamate | 32.2 | 45.3 | ↓ Reduced (p < 0.05) | |
| Lipids | C24:0-LPC | 2.15 | 0.06 | ↑ Markedly elevated (p < 0.01) |
| Carnitines | Free carnitine (C0) | 2.9 | 16.1 | ↓ Markedly reduced (p < 0.01) |
| Acylcarnitines (C2, C3, C8–C18) | Generalized reduction | Within normal range | ↓ Reduced (p < 0.05) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abu Bakar, N.; Hamzan, N.I.; Majawit, E.M.; Ahmad Ridzuan, S.N.; Hassan, N.H.; Habib, A.; Ngu, L.-H. Multi-Omics Characterization of a Novel SSR4 Variant in Congenital Disorders of Glycosylation. Metabolites 2025, 15, 786. https://doi.org/10.3390/metabo15120786
Abu Bakar N, Hamzan NI, Majawit EM, Ahmad Ridzuan SN, Hassan NH, Habib A, Ngu L-H. Multi-Omics Characterization of a Novel SSR4 Variant in Congenital Disorders of Glycosylation. Metabolites. 2025; 15(12):786. https://doi.org/10.3390/metabo15120786
Chicago/Turabian StyleAbu Bakar, Nurulamin, Nurul Izzati Hamzan, Elyssa Milus Majawit, Siti Nurwani Ahmad Ridzuan, Noor Hafizah Hassan, Anasufiza Habib, and Lock-Hock Ngu. 2025. "Multi-Omics Characterization of a Novel SSR4 Variant in Congenital Disorders of Glycosylation" Metabolites 15, no. 12: 786. https://doi.org/10.3390/metabo15120786
APA StyleAbu Bakar, N., Hamzan, N. I., Majawit, E. M., Ahmad Ridzuan, S. N., Hassan, N. H., Habib, A., & Ngu, L.-H. (2025). Multi-Omics Characterization of a Novel SSR4 Variant in Congenital Disorders of Glycosylation. Metabolites, 15(12), 786. https://doi.org/10.3390/metabo15120786






