Liquid Chromatography/Tandem Mass Spectrometry-Based Simultaneous Analysis of 32 Bile Acids in Plasma and Conventional Biomarker-Integrated Diagnostic Screening Model Development for Hepatocellular Carcinoma
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemicals and Reagents
2.2. Plasma Samples
2.3. Plasma Bile Acid Analysis by LC-MS/MS
2.4. Statistical Analyses and the Development of Diagnostic Model
3. Results and Discussion
3.1. Analysis of Serum Bile Acids in Patients with Hepatocellular Carcinoma and Other Liver Diseases
3.2. Diagnostic Screening Performance Evaluation of Bile Acids as Biomarker Candidates for HCC by Integrating with Conventional Biomarkers
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bray, F.; Laversanne, M.; Sung, H.; Ferlay, J.; Siegel, R.L.; Soerjomataram, I.; Jemal, A. Global Cancer Statistics 2022: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2024, 74, 229–263. [Google Scholar] [CrossRef] [PubMed]
- Tsuchiya, N.; Sawada, Y.; Endo, I.; Saito, K.; Uemura, Y.; Nakatsura, T. Biomarkers for the Early Diagnosis of Hepatocellular Carcinoma. World J. Gastroenterol. 2015, 21, 10573–10583. [Google Scholar] [CrossRef] [PubMed]
- Vogel, A.; Meyer, T.; Sapisochin, G.; Salem, R.; Saborowski, A. Hepatocellular Carcinoma. Lancet 2022, 400, 1345–1362. [Google Scholar] [CrossRef] [PubMed]
- Iijima, H.; Kudo, M.; Kubo, S.; Kurosaki, M.; Sakamoto, M.; Shiina, S.; Tateishi, R.; Osamu, N.; Fukumoto, T.; Matsuyama, Y.; et al. Report of the 23rd Nationwide Follow-up Survey of Primary Liver Cancer in Japan (2014–2015). Hepatol. Res. 2023, 53, 895–959. [Google Scholar] [CrossRef] [PubMed]
- Kudo, M.; Kawamura, Y.; Hasegawa, K.; Tateishi, R.; Kariyama, K.; Shiina, S.; Toyoda, H.; Imai, Y.; Hiraoka, A.; Ikeda, M.; et al. Management of Hepatocellular Carcinoma in Japan: JSH Consensus Statements and Recommendations 2021 Update. Liver Cancer 2021, 10, 181–223. [Google Scholar] [CrossRef]
- Ayuso, C.; Rimola, J.; Vilana, R.; Burrel, M.; Darnell, A.; García-Criado, Á.; Bianchi, L.; Belmonte, E.; Caparroz, C.; Barrufet, M.; et al. Diagnosis and Staging of Hepatocellular Carcinoma (HCC): Current Guidelines. Eur. J. Radiol. 2018, 101, 72–81. [Google Scholar] [CrossRef]
- Choi, J.-Y.; Lee, J.-M.; Sirlin, C.B. CT and MR Imaging Diagnosis and Staging of Hepatocellular Carcinoma: Part II. Extracellular Agents, Hepatobiliary Agents, and Ancillary Imaging Features. Radiology 2014, 273, 30–50. [Google Scholar] [CrossRef]
- Roberts, L.R.; Sirlin, C.B.; Zaiem, F.; Almasri, J.; Prokop, L.J.; Heimbach, J.K.; Murad, M.H.; Mohammed, K. Imaging for the Diagnosis of Hepatocellular Carcinoma: A Systematic Review and Meta-analysis. Hepatology 2018, 67, 401–421. [Google Scholar] [CrossRef]
- Zou, Y.-W.; Ren, Z.-G.; Sun, Y.; Liu, Z.-G.; Hu, X.-B.; Wang, H.-Y.; Yu, Z.-J. The Latest Research Progress on Minimally Invasive Treatments for Hepatocellular Carcinoma. Hepatobiliary Pancreat. Dis. Int. 2023, 22, 54–63. [Google Scholar] [CrossRef]
- Tzartzeva, K.; Obi, J.; Rich, N.E.; Parikh, N.D.; Marrero, J.A.; Yopp, A.; Waljee, A.K.; Singal, A.G. Surveillance Imaging and Alpha Fetoprotein for Early Detection of Hepatocellular Carcinoma in Patients with Cirrhosis: A Meta-Analysis. Gastroenterology 2018, 154, 1706–1718.e1. [Google Scholar] [CrossRef]
- Eisenbrey, J.R.; Gabriel, H.; Savsani, E.; Lyshchik, A. Contrast-Enhanced Ultrasound (CEUS) in HCC Diagnosis and Assessment of Tumor Response to Locoregional Therapies. Abdom. Radiol. 2021, 46, 3579–3595. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.-Y.; Chen, J.; Xia, C.-C.; Cao, L.-K.; Duan, T.; Song, B. Noninvasive Imaging of Hepatocellular Carcinoma: From Diagnosis to Prognosis. World J. Gastroenterol. 2018, 24, 2348–2362. [Google Scholar] [CrossRef] [PubMed]
- Luo, P.; Wu, S.; Yu, Y.; Ming, X.; Li, S.; Zuo, X.; Tu, J. Current Status and Perspective Biomarkers in AFP Negative HCC: Towards Screening for and Diagnosing Hepatocellular Carcinoma at an Earlier Stage. Pathol. Oncol. Res. 2020, 26, 599–603. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Chen, R.; Wei, Q.; Xu, X. The Landscape of Alpha Fetoprotein In Hepatocellular Carcinoma: Where Are We? Int. J. Biol. Sci. 2022, 18, 536–551. [Google Scholar] [CrossRef]
- Hanif, H.; Ali, M.J.; Susheela, A.T.; Khan, I.W.; Luna-Cuadros, M.A.; Khan, M.M.; Lau, D.T.-Y. Update on the Applications and Limitations of Alpha-Fetoprotein for Hepatocellular Carcinoma. World J. Gastroenterol. 2022, 28, 216–229. [Google Scholar] [CrossRef]
- Xing, X.; Cai, L.; Ouyang, J.; Wang, F.; Li, Z.; Liu, M.; Wang, Y.; Zhou, Y.; Hu, E.; Huang, C.; et al. Proteomics-Driven Noninvasive Screening of Circulating Serum Protein Panels for the Early Diagnosis of Hepatocellular Carcinoma. Nat. Commun. 2023, 14, 8392. [Google Scholar] [CrossRef]
- Park, S.J.; Jang, J.Y.; Jeong, S.W.; Cho, Y.K.; Lee, S.H.; Kim, S.G.; Cha, S.W.; Kim, Y.S.; Cho, Y.D.; Kim, H.S.; et al. Usefulness of AFP, AFP-L3, and PIVKA-II, and Their Combinations in Diagnosing Hepatocellular Carcinoma. Medicine 2017, 96, e5811. [Google Scholar] [CrossRef]
- Feng, H.; Li, B.; Li, Z.; Wei, Q.; Ren, L. PIVKA-II Serves as a Potential Biomarker That Complements AFP for the Diagnosis of Hepatocellular Carcinoma. BMC Cancer 2021, 21, 401. [Google Scholar] [CrossRef]
- Kim, D.Y.; Toan, B.N.; Tan, C.-K.; Hasan, I.; Setiawan, L.; Yu, M.-L.; Izumi, N.; Huyen, N.N.; Chow, P.K.-H.; Mohamed, R.; et al. Utility of Combining PIVKA-II and AFP in the Surveillance and Monitoring of Hepatocellular Carcinoma in the Asia-Pacific Region. Clin. Mol. Hepatol. 2023, 29, 277–292. [Google Scholar] [CrossRef]
- Zakhary, N.I.; Khodeer, S.M.; Shafik, H.E.; Abdel Malak, C.A. Impact of PIVKA-II in Diagnosis of Hepatocellular Carcinoma. J. Adv. Res. 2013, 4, 539–546. [Google Scholar] [CrossRef]
- Zhou, J.M.; Wang, T.; Zhang, K.H.; Abid, H. AFP-L3 for the Diagnosis of Early Hepatocellular Carcinoma: A Meta-Analysis. Medicine 2021, 100, E27673. [Google Scholar] [CrossRef] [PubMed]
- Norman, J.S.; Li, P.J.; Kotwani, P.; Shui, A.M.; Yao, F.; Mehta, N. AFP-L3 and DCP Strongly Predict Early Hepatocellular Carcinoma Recurrence after Liver Transplantation. J. Hepatol. 2023, 79, 1469–1477. [Google Scholar] [CrossRef] [PubMed]
- Nakagawa, S.; Beppu, T.; Okabe, H.; Sakamoto, K.; Kuroki, H.; Mima, K.; Nitta, H.; Imai, K.; Hayashi, H.; Sakamoto, Y.; et al. Triple Positive Tumor Markers Predict Recurrence and Survival in Early Stage Hepatocellular Carcinoma. Hepatol. Res. 2014, 44, 964–974. [Google Scholar] [CrossRef] [PubMed]
- Ryu, T.; Takami, Y.; Wada, Y.; Tateishi, M.; Matsushima, H.; Mikagi, K.; Saitsu, H. Double- and Triple-Positive Tumor Markers Predict Early Recurrence and Poor Survival in Patients with Hepatocellular Carcinoma within the Milan Criteria and Child-Pugh Class A. J. Gastrointest. Surg. 2017, 21, 957–966. [Google Scholar] [CrossRef] [PubMed]
- Hasegawa, K.; Takemura, N.; Yamashita, T.; Watadani, T.; Kaibori, M.; Kubo, S.; Shimada, M.; Nagano, H.; Hatano, E.; Aikata, H.; et al. Clinical Practice Guidelines for Hepatocellular Carcinoma: The Japan Society of Hepatology 2021 Version (5th JSH-HCC Guidelines). Hepatol. Res. 2023, 53, 383–390. [Google Scholar] [CrossRef]
- Hanouneh, I.A.; Alkhouri, N.; Singal, A.G. Hepatocellular Carcinoma Surveillance in the 21st Century: Saving Lives or Causing Harm? Clin. Mol. Hepatol. 2019, 25, 264–269. [Google Scholar] [CrossRef]
- Chiang, J.Y.L. Bile Acids: Regulation of Synthesis. J. Lipid Res. 2009, 50, 1955–1966. [Google Scholar] [CrossRef]
- Chiang, J.Y.L. Regulation of Bile Acid Synthesis. Front. Biosci. 1998, 3, A273. [Google Scholar] [CrossRef]
- Russell, D.W. The Enzymes, Regulation, and Genetics of Bile Acid Synthesis. Annu. Rev. Biochem. 2003, 72, 137–174. [Google Scholar] [CrossRef]
- Chiang, J.Y.L. Regulation of Bile Acid Synthesis: Pathways, Nuclear Receptors, and Mechanisms. J. Hepatol. 2004, 40, 539–551. [Google Scholar] [CrossRef]
- Matsubara, T.; Li, F.; Gonzalez, F.J. FXR Signaling in the Enterohepatic System. Mol. Cell. Endocrinol. 2013, 368, 17–29. [Google Scholar] [CrossRef] [PubMed]
- Ise, I.; Tanaka, N.; Imoto, H.; Maekawa, M.; Kohyama, A.; Watanabe, K.; Motoi, F.; Unno, M.; Naitoh, T. Changes in Enterohepatic Circulation after Duodenal–Jejunal Bypass and Reabsorption of Bile Acids in the Bilio-Pancreatic Limb. Obes. Surg. 2019, 29, 1901–1910. [Google Scholar] [CrossRef] [PubMed]
- Chiang, J.Y.L.; Ferrell, J.M. Bile Acids as Metabolic Regulators and Nutrient Sensors. Annu. Rev. Nutr. 2019, 39, 175–200. [Google Scholar] [CrossRef] [PubMed]
- Lefebvre, P.; Cariou, B.; Lien, F.; Kuipers, F.; Staels, B. Role of Bile Acids and Bile Acid Receptors in Metabolic Regulation. Physiol. Rev. 2009, 89, 147–191. [Google Scholar] [CrossRef] [PubMed]
- Shulpekova, Y.; Zharkova, M.; Tkachenko, P.; Tikhonov, I.; Stepanov, A.; Synitsyna, A.; Izotov, A.; Butkova, T.; Shulpekova, N.; Lapina, N.; et al. The Role of Bile Acids in the Human Body and in the Development of Diseases. Molecules 2022, 27, 3401. [Google Scholar] [CrossRef]
- Clayton, P.T.; Lake, B.D.; Hall, N.A.; Shortland, D.B.; Carruthers, R.A.; Lawson, A.M. Plasma Bile Acids in Patients with Peroxisomal Dysfunction Syndromes: Analysis by Capillary Gas Chromatography—Mass Spectrometry. Eur. J. Pediatr. 1987, 146, 166–173. [CrossRef]
- Shao, Y.; Chen, S.; Li, H.; Tang, Q.; Xu, D. Maternal Bile Acid Profile and Subtype Analysis of Intrahepatic Cholestasis of Pregnancy. Orphanet J. Rare Dis. 2021, 16, 259. [Google Scholar] [CrossRef]
- Murai, T.; Oda, K.; Toyo, T.; Nittono, H.; Takei, H.; Muto, A.; Kimura, A.; Kurosawa, T. Determination of 3β-Hydroxy-Δ5-Bile Acids and Related Compounds in Biological Fluids of Patients with Cholestasis by Liquid Chromatography-Tandem Mass Spectrometry. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2013, 923–924, 120–127. [Google Scholar] [CrossRef]
- Clayton, P.T. Inborn Errors of Bile Acid Metabolism. J. Inherit. Metab. Dis. 1991, 14, 478–496. [Google Scholar] [CrossRef]
- Marra, F.; Svegliati-Baroni, G. Lipotoxicity and the Gut-Liver Axis in NASH Pathogenesis. J. Hepatol. 2018, 68, 280–295. [Google Scholar] [CrossRef]
- Xie, G.; Wang, X.; Huang, F.; Zhao, A.; Chen, W.; Yan, J.; Zhang, Y.; Lei, S.; Ge, K.; Zheng, X.; et al. Dysregulated Hepatic Bile Acids Collaboratively Promote Liver Carcinogenesis. Int. J. Cancer 2016, 139, 1764–1775. [Google Scholar] [CrossRef] [PubMed]
- Nimer, N.; Choucair, I.; Wang, Z.; Nemet, I.; Li, L.; Gukasyan, J.; Weeks, T.L.; Alkhouri, N.; Zein, N.; Tang, W.H.W.; et al. Bile Acids Profile, Histopathological Indices and Genetic Variants for Non-Alcoholic Fatty Liver Disease Progression. Metabolism 2021, 116, 154457. [Google Scholar] [CrossRef] [PubMed]
- Puri, P.; Daita, K.; Joyce, A.; Mirshahi, F.; Santhekadur, P.K.; Cazanave, S.; Luketic, V.A.; Siddiqui, M.S.; Boyett, S.; Min, H.K.; et al. The Presence and Severity of Nonalcoholic Steatohepatitis Is Associated with Specific Changes in Circulating Bile Acids. Hepatology 2018, 67, 534–548. [Google Scholar] [CrossRef] [PubMed]
- Suga, T.; Yamaguchi, H.; Ogura, J.; Shoji, S.; Maekawa, M.; Mano, N. Altered Bile Acid Composition and Disposition in a Mouse Model of Non-Alcoholic Steatohepatitis. Toxicol. Appl. Pharmacol. 2019, 379, 114664. [Google Scholar] [CrossRef] [PubMed]
- Shoji, S.; Maekawa, M.; Ogura, J.; Sato, T.; Mano, N. Identification Cholesterol Metabolites Altered before the Onset of Nonalcoholic Steatohepatitis by Targeted Metabolomics. Biochim. Biophys. Acta (BBA)-Mol. Cell Biol. Lipids 2022, 1867, 159135. [Google Scholar] [CrossRef] [PubMed]
- Lake, A.D.; Novak, P.; Shipkova, P.; Aranibar, N.; Robertson, D.; Reily, M.D.; Lu, Z.; Lehman-McKeeman, L.D.; Cherrington, N.J. Decreased Hepatotoxic Bile Acid Composition and Altered Synthesis in Progressive Human Nonalcoholic Fatty Liver Disease. Toxicol. Appl. Pharmacol. 2013, 268, 132–140. [Google Scholar] [CrossRef]
- Colosimo, S.; Tomlinson, J.W. Bile Acids as Drivers and Biomarkers of Hepatocellular Carcinoma. World J. Hepatol. 2022, 14, 1730–1738. [Google Scholar] [CrossRef]
- Shen, E.Y.L.; Mei Ran Abellona, U.; Cox, I.J.; Taylor-Robinson, S.D. The Role of Mass Spectrometry in Hepatocellular Carcinoma Biomarker Discovery. Metabolites 2023, 13, 1059. [Google Scholar] [CrossRef]
- Goicoechea, L.; Conde de la Rosa, L.; Torres, S.; García-Ruiz, C.; Fernández-Checa, J.C. Mitochondrial Cholesterol: Metabolism and Impact on Redox Biology and Disease. Redox Biol. 2023, 61, 102643. [Google Scholar] [CrossRef]
- Stepien, M.; Keski-Rahkonen, P.; Kiss, A.; Robinot, N.; Duarte-Salles, T.; Murphy, N.; Perlemuter, G.; Viallon, V.; Tjønneland, A.; Rostgaard-Hansen, A.L.; et al. Metabolic Perturbations Prior to Hepatocellular Carcinoma Diagnosis: Findings from a Prospective Observational Cohort Study. Int. J. Cancer 2021, 148, 609–625. [Google Scholar] [CrossRef]
- Stepien, M.; Lopez-Nogueroles, M.; Lahoz, A.; Kühn, T.; Perlemuter, G.; Voican, C.; Ciocan, D.; Boutron-Ruault, M.; Jansen, E.; Viallon, V.; et al. Prediagnostic Alterations in Circulating Bile Acid Profiles in the Development of Hepatocellular Carcinoma. Int. J. Cancer 2022, 150, 1255–1268. [Google Scholar] [CrossRef] [PubMed]
- Khalil, A.; ElSheashaey, A.; Abdelsameea, E.; Obada, M.; Bayomy, F.F.M.; El-Said, H. Value of Bile Acids in Diagnosing Hepatitis C Virus-Induced Liver Cirrhosis and Hepatocellular Carcinoma. Br. J. Biomed. Sci. 2022, 79, 10191. [Google Scholar] [CrossRef] [PubMed]
- Ressom, H.W.; Xiao, J.F.; Tuli, L.; Varghese, R.S.; Zhou, B.; Tsai, T.H.; Nezami Ranjbar, M.R.; Zhao, Y.; Wang, J.; Di Poto, C.; et al. Utilization of Metabolomics to Identify Serum Biomarkers for Hepatocellular Carcinoma in Patients with Liver Cirrhosis. Anal. Chim. Acta 2012, 743, 90–100. [Google Scholar] [CrossRef] [PubMed]
- Han, J.; Qin, W.X.; Li, Z.L.; Xu, A.J.; Xing, H.; Wu, H.; Zhang, H.; Wang, M.D.; Li, C.; Liang, L.; et al. Tissue and Serum Metabolite Profiling Reveals Potential Biomarkers of Human Hepatocellular Carcinoma. Clin. Chim. Acta 2019, 488, 68–75. [Google Scholar] [CrossRef]
- Maekawa, M.; Shimada, M.; Iida, T.; Goto, J.; Mano, N. Tandem Mass Spectrometric Characterization of Bile Acids and Steroid Conjugates Based on Low-Energy Collision-Induced Dissociation. Steroids 2014, 80, 80–91. [Google Scholar] [CrossRef]
- Kakiyama, G.; Muto, A.; Shimada, M.; Mano, N.; Goto, J.; Hofmann, A.F.; Iida, T. Chemical Synthesis of 3beta-Sulfooxy-7beta-Hydroxy-24-nor-5-Cholenoic Acid: An Internal Standard for Mass Spectrometric Analysis of the Abnormal Delta5-Bile Acids Occurring in Niemann-Pick Disease. Steroids 2009, 74, 766–772. [Google Scholar] [CrossRef]
- Steiner, C.; Othman, A.; Saely, C.H.; Rein, P.; Drexel, H.; von Eckardstein, A.; Rentsch, K.M. Bile Acid Metabolites in Serum: Intraindividual Variation and Associations with Coronary Heart Disease, Metabolic Syndrome and Diabetes Mellitus. PLoS ONE 2011, 6, e25006. [Google Scholar] [CrossRef]
- Danese, E.; Negrini, D.; Pucci, M.; De Nitto, S.; Ambrogi, D.; Donzelli, S.; Lievens, P.M.J.; Salvagno, G.L.; Lippi, G. Bile Acids Quantification by Liquid Chromatography-Tandem Mass Spectrometry: Method Validation, Reference Range, and Interference Study. Diagnostics 2020, 10, 462. [Google Scholar] [CrossRef]
- Yin, P.; Wan, D.; Zhao, C.; Chen, J.; Zhao, X.; Wang, W.; Lu, X.; Yang, S.; Gu, J.; Xu, G. A Metabonomic Study of Hepatitis B-Induced Liver Cirrhosis and Hepatocellular Carcinoma by Using RP-LC and HILIC Coupled with Mass Spectrometry. Mol. Biosyst. 2009, 5, 868. [Google Scholar] [CrossRef]
- Park, J.Y.; Park, B.K.; Ko, J.S.; Bang, S.; Song, S.Y.; Chung, J.B. Bile Acid Analysis in Biliary Tract Cancer. Yonsei Med. J. 2006, 47, 817–825. [Google Scholar] [CrossRef]
- Alonso-Peña, M.; Espinosa-Escudero, R.; Hermanns, H.M.; Briz, O.; Herranz, J.M.; Garcia-Ruiz, C.; Fernandez-Checa, J.C.; Juamperez, J.; Avila, M.; Argemi, J.; et al. Impact of Liver Inflammation on Bile Acid Side Chain Shortening and Amidation. Cells 2022, 11, 3983. [Google Scholar] [CrossRef] [PubMed]
- Ge, X.; Huang, S.; Ren, C.; Zhao, L. Taurocholic Acid and Glycocholic Acid Inhibit Inflammation and Activate Farnesoid X Receptor Expression in LPS-Stimulated Zebrafish and Macrophages. Molecules 2023, 28, 2005. [Google Scholar] [CrossRef] [PubMed]
- Chieco, P.; Romagnoli, E.; Aicardi, G.; Suozzi, A.; Cantelli Forti, G.; Roda, A. Apoptosis Induced in Rat Hepatocytes by in Vivo Exposure to Taurochenodeoxycholate. Histochem. J. 1997, 29, 875–883. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Niu, K.; Wang, R.; Liang, X.; Lin, C.; Wu, X.; Zhai, Z. Taurochenodeoxycholic Acid Inhibits Intestinal Epithelial Cell Proliferation and Induces Apoptosis Independent of the Farnesoid X Receptor. Food Funct. 2023, 14, 5277–5289. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Yang, M.; Zhao, J.; Li, X.; Xiao, X.; Zhang, Y.; Jin, X.; Liao, M. Bile Salt (Glycochenodeoxycholate Acid) Induces Cell Survival and Chemoresistance in Hepatocellular Carcinoma. J. Cell. Physiol. 2019, 234, 10899–10906. [Google Scholar] [CrossRef]
- Parikh, N.D.; Tayob, N.; Singal, A.G. Blood-Based Biomarkers for Hepatocellular Carcinoma Screening: Approaching the End of the Ultrasound Era? J. Hepatol. 2023, 78, 207–216. [Google Scholar] [CrossRef]
- Zhu, W.W.; Guo, J.J.; Guo, L.; Jia, H.L.; Zhu, M.; Zhang, J.B.; Loffredo, C.A.; Forgues, M.; Huang, H.; Xing, X.J.; et al. Evaluation of Midkine as a Diagnostic Serum Biomarker in Hepatocellular Carcinoma. Clin. Cancer Res. 2013, 19, 3944–3954. [Google Scholar] [CrossRef]
- Shang, S.; Plymoth, A.; Ge, S.; Feng, Z.; Rosen, H.R.; Sangrajrang, S.; Hainaut, P.; Marrero, J.A.; Beretta, L. Identification of Osteopontin as a Novel Marker for Early Hepatocellular Carcinoma. Hepatology 2012, 55, 483–490. [Google Scholar] [CrossRef]
- Wan, H.G.; Xu, H.; Gu, Y.M.; Wang, H.; Xu, W.; Zu, M.H. Comparison Osteopontin vs. AFP for the Diagnosis of HCC: A Meta-Analysis. Clin. Res. Hepatol. Gastroenterol. 2014, 38, 706–714. [Google Scholar] [CrossRef]
- Sun, T.; Tang, Y.; Sun, D.; Bu, Q.; Li, P. Osteopontin versus Alpha-Fetoprotein as a Diagnostic Marker for Hepatocellular Carcinoma: A Meta-Analysis. Onco Targets Ther. 2018, 11, 8925–8935. [Google Scholar] [CrossRef]
- Kim, J.Y.; Lee, G.B.; Lee, J.C.; Moon, M.H. High-Speed Screening of Lipoprotein Components Using Online Miniaturized Asymmetrical Flow Field-Flow Fractionation and Electrospray Ionization Tandem Mass Spectrometry: Application to Hepatocellular Carcinoma Plasma Samples. Anal. Chem. 2021, 93, 4867–4875. [Google Scholar] [CrossRef] [PubMed]
- Buechler, C.; Aslanidis, C. Role of Lipids in Pathophysiology, Diagnosis and Therapy of Hepatocellular Carcinoma. Biochim. Biophys. Acta Mol. Cell. Biol. Lipids 2020, 1865, 158658. [Google Scholar] [CrossRef] [PubMed]
- Takaki, S.; Fukuhara, T.; Mori, N.; Tsuji, K. High Cholinesterase Predicts Tolerance to Sorafenib Treatment and Improved Prognosis in Patients with Transarterial Chemoembolization Refractory Intermediate Stage Hepatocellular Carcinoma. Mol. Clin. Oncol. 2020, 12, 60–68. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.X.; Yang, F.; Ge, N.; Guo, J.T.; Sun, S.Y. Role of Albumin-Bilirubin Score in Non-Malignant Liver Disease. World J. Gastroenterol. 2024, 30, 999–1004. [Google Scholar] [CrossRef] [PubMed]
- Tian, Y.B.; Niu, H.; Xu, F.; Shang-Guan, P.W.; Song, W.W. ALBI Score Combined with FIB-4 Index to Predict Post-Hepatectomy Liver Failure in Patients with Hepatocellular Carcinoma. Sci. Rep. 2024, 14, 8034. [Google Scholar] [CrossRef] [PubMed]
- Ruoslathi, E.; Terry, W.D. α Foetoprotein and Serum Albumin Show Sequence Homology. Nature 1976, 260, 804–805. [Google Scholar] [CrossRef]
- Hagström, H.; Yan, J.; Talbäck, M.; Andreasson, A.; Walldius, G.; Bottai, M.; Hammar, N. Improved Prediction of 10-Year Risk of Severe Liver Disease in the General Population Using Commonly Available Biomarkers. Aliment. Pharmacol. Ther. 2023, 57, 418–425. [Google Scholar] [CrossRef]
- Elawdi, H.A.; Franzini, M.; Paolicchi, A.; Emdin, M.; Fornaciari, I.; Fierabracci, V.; De Simone, P.; Carrai, P.; Filipponi, F. Circulating Gamma-Glutamyltransferase Fractions in Cirrhosis. Liver Int. 2014, 34, 191–199. [Google Scholar] [CrossRef]
- Lemoine, M.; Shimakawa, Y.; Nayagam, S.; Khalil, M.; Suso, P.; Lloyd, J.; Goldin, R.; Njai, H.F.; Ndow, G.; Taal, M.; et al. The Gamma-Glutamyl Transpeptidase to Platelet Ratio (GPR) Predicts Significant Liver Fibrosis and Cirrhosis in Patients with Chronic HBV Infection in West Africa. Gut 2016, 65, 1369–1376. [Google Scholar] [CrossRef]
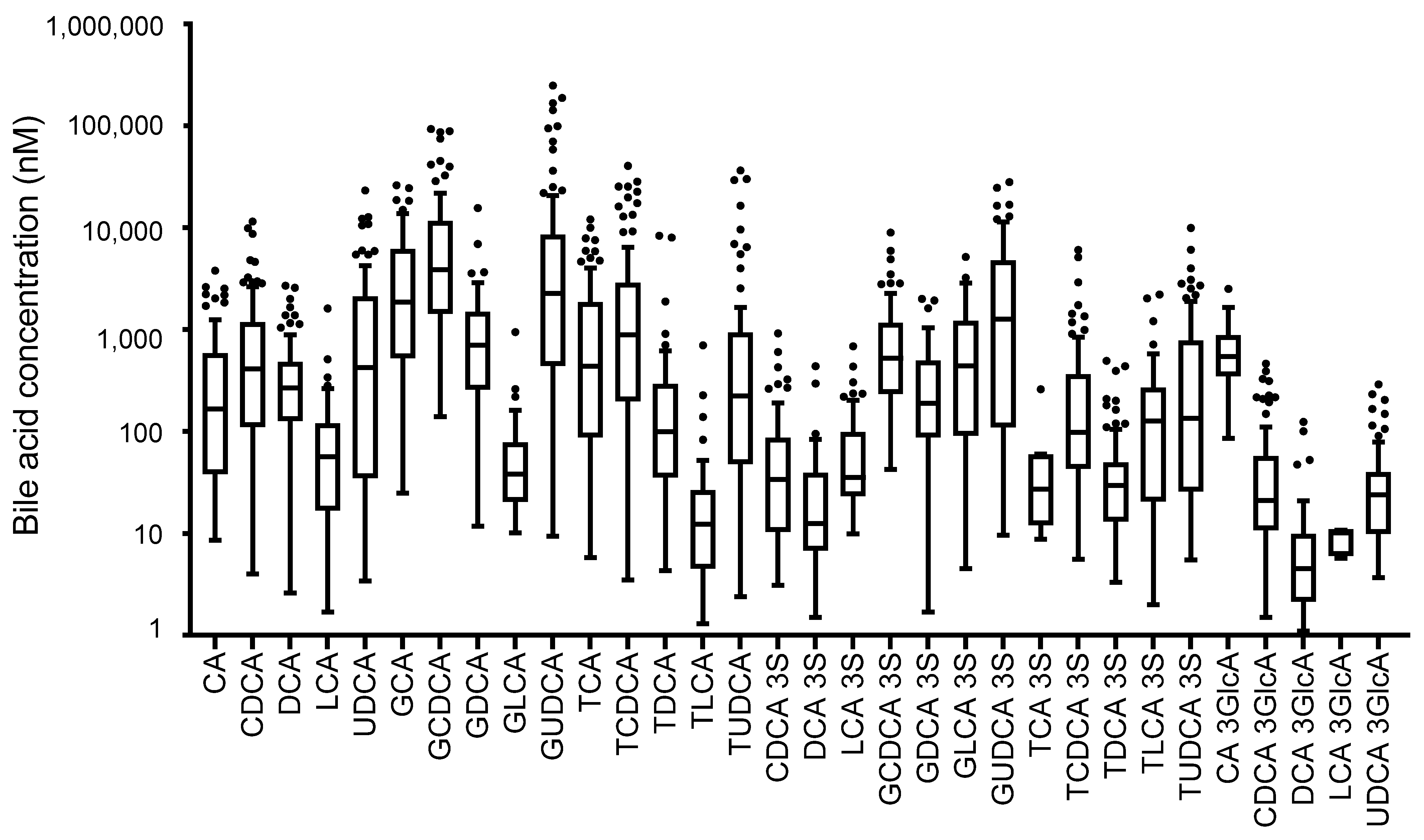
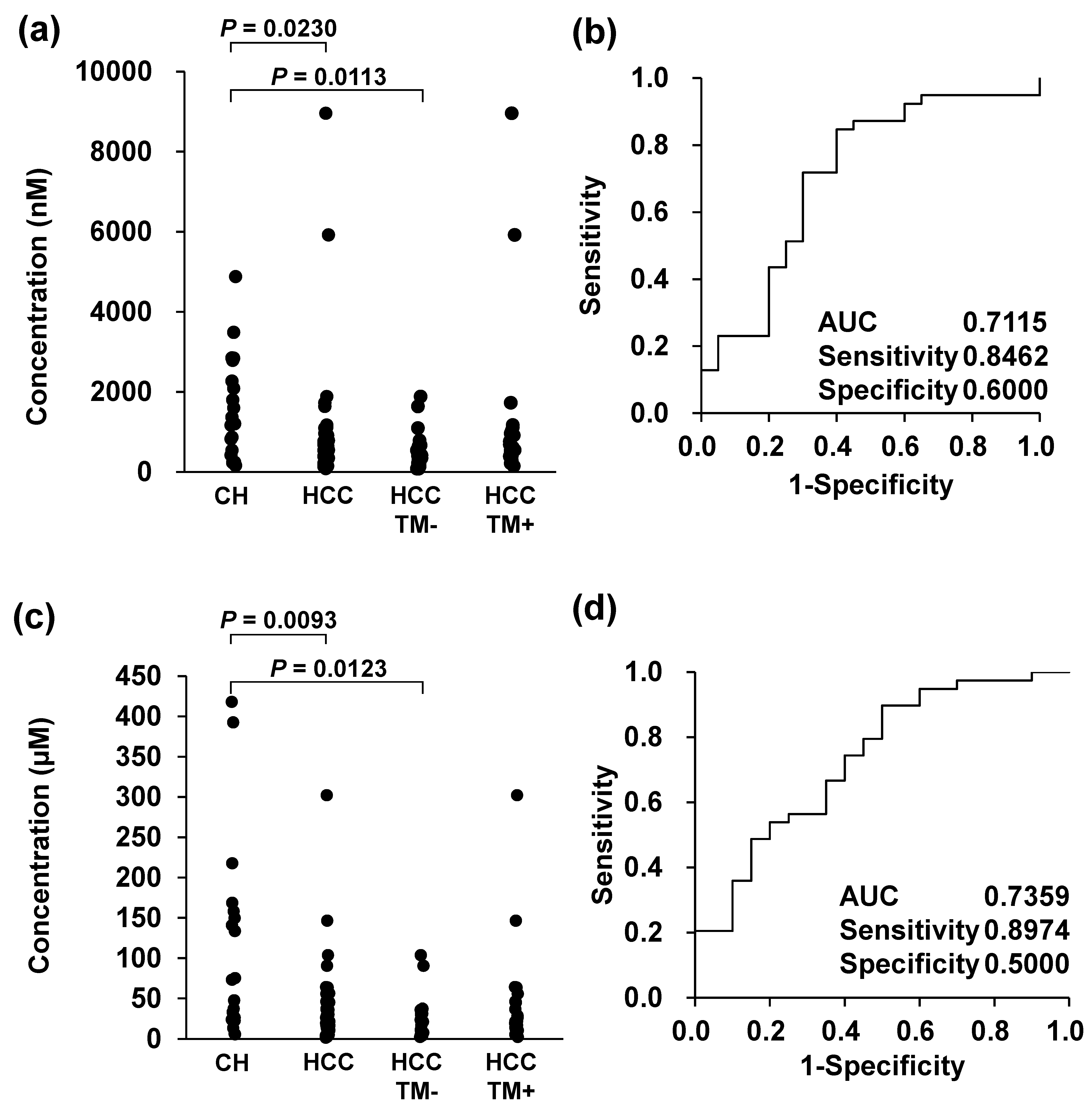
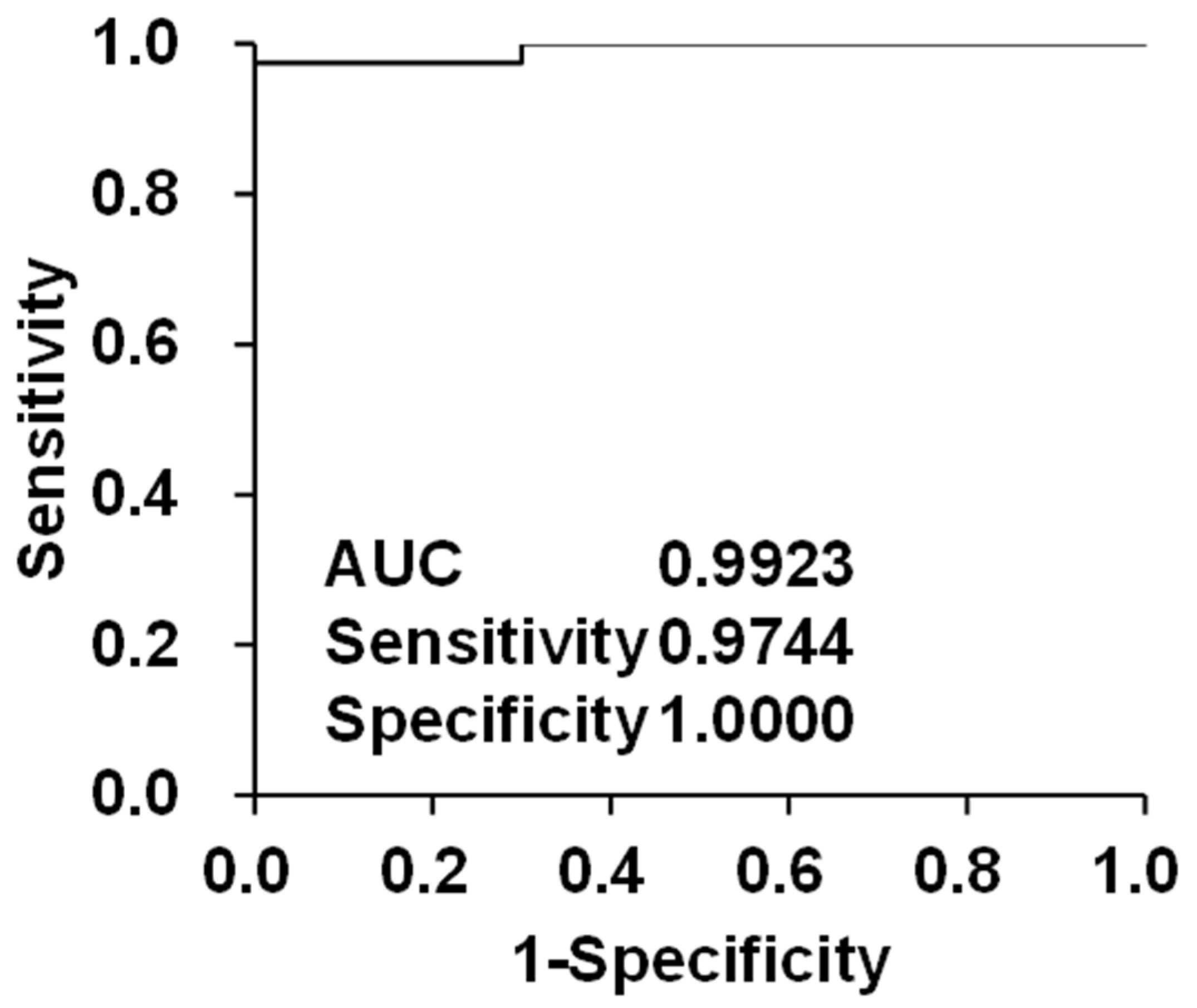
| Chronic Hepatitis (n = 20) | Hepatic Cirrhosis (n = 20) | HCC | p Value | |||
|---|---|---|---|---|---|---|
| Total (n = 39) | TM− (n = 19) | TM+ (n = 20) | ||||
| Sex, n (male/female) | 10/10 | 12/8 | 24/15 | 12/7 | 12/8 | 0.6976 a |
| Age, median (IQR) | 56 (48.5–65) | 53.5 (49.75–62.75) | 70 (62–76) | 68 (60–76) | 70.5 (64.5–78.25) | <0.0001 b |
| Primary disease, n (%) | 0.0122 b | |||||
| HBV | 7 (35) | 4 (20) | 6 (15) | 3 (16) | 3 (15) | |
| HCV | 5 (25) | 4 (20) | 20 (51) | 12 (63) | 8 (40) | |
| NASH | 3 (15) | 3 (15) | 3 (8) | 1 (5) | 2 (10) | |
| PBC | 3 (15) | 5 (20) | 0 (0) | 0 (0) | 0 (0) | |
| Alcohol | 1 (5) | 4 (20) | 8 (21) | 3 (16) | 5 (25) | |
| Others | AIH, 2 (10) Unknown, 1 (5) | Wilson disease, 1 (5) Unknown, 1 (5) | Unknown, 2 (5) | 0 (0) | Unknown, 2 (10) | |
| GGT, median (IQR) (U/L) | 62 (41.25–116.5) | 35 (18.25–158.75) | 37 (20–85) | 25 (18–42) | 83.5 (33.25–138.5) | 0.1756 a |
| AST, median (IQR) (U/L) | 49.5 (27.5–79) | 49 (34.25–62.25) | 35 (26–50) | 28 (18–42) | 45.5 (30.25–56.25) | 0.0401 a |
| ALT, median (IQR) (U/L) | 62 (29–99.75) | 33 (15.5–50) | 28 (17–35) | 20 (16–31) | 30.5 (17.5–47.25) | 0.0005 a |
| Albumin, median (IQR) (g/dL) | 4 (3.7–4.3) | 2.95 (1.95–3.425) | 3.5 (3.1–3.7) | 3.6 (3.4–4.2) | 3.3 (3–3.6) | <0.0001 a |
| ChE, median (IQR) (U/L) | 288 (238.75–335.5) | 144 (54.5–192.5) | 199 (142–253) | 221 (176–323) | 175 (136–220.5) | <0.0001 a |
| Scr, median (IQR) (mg/dL) | 0.675 (0.58–0.8375) | 0.765 (0.5825–1.07) | 0.73 (0.64–0.93) | 0.77 (0.64–1.06) | 0.73 (0.635–0.815) | 0.6146 a |
| UDCA take, n (Yes/No) | 8/12 | 11/9 | 16/23 | 7/12 | 9/11 | 0.5601 b |
| AFP, median (IQR) (ng/mL) c | - | 4.1 (2.425–14.4) | 5.4 (3.2–45.8) | 4.4 (3.1–5.4) | 37.7 (5.175–1570.975) | 0.1811 a |
| PIVKA-II, median (IQR) (mAU/mL) d | - | 59.5 (37.5–457.5) | 23 (18–134) | 19 (16–21) | 122.5 (45.25–1749.25) | 0.1560 a |
| AFP-L3, median (IQR) (%) e | - | 0.5 (0.5–12.45) | 0.5 (0.5–28.2) | 0.5 (0.5–0.5) | 21.3 (1.575–43.75) | 0.8027 a |
| Chronic Hepatitis | HC | HCC | p Value | ||||
|---|---|---|---|---|---|---|---|
| Total | TM− | TM+ | All Group b | CH vs. HCC a | |||
| CA (nM) [median (IQR)] | 39.0 (27.2–173.1) | 189.6 (64.4–619.8) | 314.3 (41.5–909.1) | 136.8 (37.8–590.1) | 426.8 (87.6–1116.9) | 0.0721 | 0.9666 |
| CDCA (nM) [median (IQR)] | 127.1 (47.4–452.7) | 572.6 (284.8–2044.1) | 634.7 (197.1–1723.6) | 405.2 (109.9–1532.8) | 741.5 (362.6–2504.1) | 0.0884 | 0.9249 |
| DCA (nM) [median (IQR)] | 226.4 (126.3–459.7) | 358.9 (208.5–1024.1) | 259.7 (106.8–407.4) | 293.9 (192.3–613.3) | 259.7 (37.6–384.4) | 0.3202 | 0.3450 |
| LCA (nM) [median (IQR)] | 23.2 (14.8–60.6) | 72.7 (22.6–120.0) | 103.6 (15.3–154.3) | 80.1 (15.3–125.9) | 135.8 (16.2–258.3) | 0.2012 | 0.7907 |
| UDCA (nM) [median (IQR)] | 135.5 (9.2–998.5) | 492.2 (35.8–5296.0) | 572.2 (39.3–2440.0) | 272.7 (21.3–1971.0) | 1327.5 (121.5–3178.2) | 0.0242 | 0.9528 |
| GCA (nM) [median (IQR)] | 842.6 (309.4–1845.4) | 6060.0 (1952.4–12,455.0) | 1725.5 (524.5–5480.0) | 1104.6 (283.5–2796.0) | 2515.7 (1005.0–7615.3) | 0.0008 | 0.0327 |
| GCDCA (nM) [median (IQR)] | 1837.4 (601.6–3608.0) | 15,534.5 (2147.6–41,090.0) | 5135.0 (1706.2–9624.0) | 2896.6 (1201.3–5881.0) | 6549.0 (2111.5–12,073.5) | < 0.0001 | 0.0561 |
| GDCA (nM) [median (IQR)] | 622.7 (232.5–1199.6) | 750.3 (261.2–1913.4) | 755.3 (255.8–1574.6) | 836.2 (257.9–1531.3) | 379.3 (110.0–2738.4) | 0.3466 | 0.9818 |
| GLCA (nM) [median (IQR)] | 21.5 (18.7–30.9) | 37.9 (21.4–59.0) | 66.6 (21.9–119.6) | 68.6 (11.7–127.1) | 58.0 (22.7–106.9) | 0.1346 | 0.5084 |
| GUDCA (nM) [median (IQR)] | 1201.2 (248.4–4011.0) | 2681.5 (824.3–67,072.5) | 2281.0 (432.7–11,582.0) | 1306.5 (66.9–13985.0) | 2666.2 (599.4–8557.0) | 0.0158 | 0.5561 |
| TCA (nM) [median (IQR)] | 136.7 (54.8–698.6) | 1626.2 (375.0–2698.5) | 338.6 (57.2–1756.0) | 121.9 (56.6–1145.5) | 475.4 (137.1–2404.8) | 0.1248 | 0.0607 |
| TCDCA (nM) [median (IQR)] | 391.9 (106.0–1222.5) | 3880.0 (578.4–15,372.5) | 1126.0 (196.0–2716.9) | 437.6 (96.2–2585.5) | 1288.5 (263.9–2844.9) | 0.0040 | 0.0313 |
| TDCA (nM) [median (IQR)] | 166.0 (49.6–254.8) | 77.2 (27.3–380.8) | 77.5 (21.3–293.9) | 164.4 (48.2–333.5) | 56.0 (6.2–160.6) | 0.5894 | 0.9557 |
| TLCA (nM) [median (IQR)] | 5.5 (3.8–24.7) | 8.3 (5.2–20.8) | 18.7 (7.5–35.1) | 23.6 (10.4–35.0) | 8.5 (3.3–52.1) | 0.3826 | 0.3604 |
| TUDCA (nM) [median (IQR)] | 74.6 (19.1–222.2) | 473.8 (139.4–6565.0) | 241.6 (40.3–931.3) | 363.2 (15.0–1171.5) | 241.6 (48.4–927.2) | 0.0681 | 0.2966 |
| CDCA 3S (nM) [median (IQR)] | 9.0 (6.9–18.8) | 72.4 (16.7–88.4) | 37.5 (10.7–104.5) | 55.4 (9.7–87.7) | 33.2 (11.0–135.7) | 0.2542 | 0.6424 |
| DCA 3S (nM) [median (IQR)] | 10.9 (6.2–14.8) | 16.2 (5.8–82.2) | 15.1 (7.0–41.5) | 15.1 (6.6–80.1) | 14.9 (7.8–33.7) | 0.2262 | 0.9244 |
| LCA 3S (nM) [median (IQR)] | 23.4 (13.6–36.5) | 37.0 (20.9–175.3) | 40.3 (24.3–125.2) | 56.0 (27.9–196.5) | 32.6 (24.3–96.8) | 0.2081 | 0.9420 |
| GCDCA 3S (nM) [median (IQR)] | 284.2 (157.1–504.6) | 1285.3 (448.6–2660.0) | 538.7 (285.2–960.9) | 395.0 (193.1–670.1) | 649.6 (422.5–1091.1) | 0.0266 | 0.0230 |
| GDCA 3S (nM) [median (IQR)] | 224.7 (85.9–524.2) | 413.1 (124.5–820.3) | 170.2 (81.1–286.7) | 165.2 (107.8–409.2) | 189.9 (44.5–266.6) | 0.0324 | 0.2418 |
| GLCA 3S (nM) [median (IQR)] | 517.0 (205.5–1243.2) | 395.9 (59.8–1527.8) | 419.0 (91.0–1216.2) | 655.0 (326.8–1366.0) | 280.1 (15.4–599.7) | 0.9949 | 0.8369 |
| GUDCA 3S (nM) [median (IQR)] | 338.0 (54.3–2787.6) | 2803.8 (205.9–10,962.5) | 1842.0 (110.2–3140.0) | 544.0 (60.8–3140.0) | 2016.4 (821.9–3192.5) | 0.0040 | 0.3658 |
| TCA 3S (nM) [median (IQR)] | 30.0 (24.8–59.7) | 14.5 (11.2–60.1) | 133.9 (8.8–259.0) | Not detected | 133.9 (8.8–259.0) | 0.3906 | 1.0000 |
| TCDCA 3S (nM) [median (IQR)] | 52.0 (18.9–109.2) | 153.9 (79.2–1074.5) | 103.8 (43.9–365.3) | 61.1 (27.8–129.7) | 189.7 (99.5–385.1) | 0.1153 | 0.3320 |
| TDCA 3S (nM) [median (IQR)] | 31.7 (17.6–97.7) | 40.1 (13.4–150.2) | 23.4 (11.4–35.4) | 23.4 (13.4–35.2) | 19.2 (6.8–49.4) | 0.2611 | 0.1485 |
| TLCA 3S (nM) [median (IQR)] | 155.1 (36.3–310.8) | 137.6 (33.2–281.7) | 95.9 (17.5–260.7) | 151.8 (70.7–297.2) | 70.2 (3.1–160.8) | 0.7421 | 0.8452 |
| TUDCA 3S (nM) [median (IQR)] | 83.5 (12.1–490.1) | 869.0 (25.9–2180.0) | 134.8 (39.7–555.4) | 91.2 (19.1–545.4) | 162.9 (78.1–726.4) | 0.1929 | 0.4887 |
| CA 3GlcA (nM) [median (IQR)] | 548.4 (345.4–637.7) | 657.7 (252.1–1100.9) | 532.5 (377.4–1018.6) | 528.7 (260.8–1018.6) | 583.3 (382.9–1042.4) | 0.1740 | 0.9423 |
| CDCA 3GlcA (nM) [median (IQR)] | 9.8 (3.7–20.7) | 37.2 (12.7–139.0) | 23.6 (16.4–57.5) | 23.6 (16.4–50.8) | 23.1 (16.2–73.7) | 0.0079 | 0.7513 |
| DCA 3GlcA (nM) [median (IQR)] | 4.0 (2.6–6.2) | 10.3 (2.2–52.5) | 4.4 (1.9–6.3) | 4.6 (1.7–11.5) | 4.2 (2.1–5.0) | 0.0209 | 0.3210 |
| LCA 3GlcA (nM) [median (IQR)] | Not detected | 10.4 (10.4–10.4) | 8.5 (5.9–10.8) | 8.6 (6.4–10.8) | 8.2 (5.7–10.6) | 0.5505 | 1.0000 |
| UDCA 3GlcA (nM) [median (IQR)] | 28.6 (9.3–55.5) | 21.7 (7.5–35.7) | 25.4 (10.9–42.0) | 28.7 (12.9–45.9) | 16.5 (9.4–50.9) | 0.8427 | 0.7473 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yamauchi, M.; Maekawa, M.; Sato, T.; Sato, Y.; Kumondai, M.; Tsuruoka, M.; Inoue, J.; Masamune, A.; Mano, N. Liquid Chromatography/Tandem Mass Spectrometry-Based Simultaneous Analysis of 32 Bile Acids in Plasma and Conventional Biomarker-Integrated Diagnostic Screening Model Development for Hepatocellular Carcinoma. Metabolites 2024, 14, 513. https://doi.org/10.3390/metabo14090513
Yamauchi M, Maekawa M, Sato T, Sato Y, Kumondai M, Tsuruoka M, Inoue J, Masamune A, Mano N. Liquid Chromatography/Tandem Mass Spectrometry-Based Simultaneous Analysis of 32 Bile Acids in Plasma and Conventional Biomarker-Integrated Diagnostic Screening Model Development for Hepatocellular Carcinoma. Metabolites. 2024; 14(9):513. https://doi.org/10.3390/metabo14090513
Chicago/Turabian StyleYamauchi, Minami, Masamitsu Maekawa, Toshihiro Sato, Yu Sato, Masaki Kumondai, Mio Tsuruoka, Jun Inoue, Atsushi Masamune, and Nariyasu Mano. 2024. "Liquid Chromatography/Tandem Mass Spectrometry-Based Simultaneous Analysis of 32 Bile Acids in Plasma and Conventional Biomarker-Integrated Diagnostic Screening Model Development for Hepatocellular Carcinoma" Metabolites 14, no. 9: 513. https://doi.org/10.3390/metabo14090513
APA StyleYamauchi, M., Maekawa, M., Sato, T., Sato, Y., Kumondai, M., Tsuruoka, M., Inoue, J., Masamune, A., & Mano, N. (2024). Liquid Chromatography/Tandem Mass Spectrometry-Based Simultaneous Analysis of 32 Bile Acids in Plasma and Conventional Biomarker-Integrated Diagnostic Screening Model Development for Hepatocellular Carcinoma. Metabolites, 14(9), 513. https://doi.org/10.3390/metabo14090513









