Tolerance Mechanisms and Removal Efficiency of Chlorella pyrenoidosa in Treating 3-Fluorophenol Pollution
Abstract
1. Introduction
2. Materials and Methods
2.1. Exposure of C. pyrenoidosa to 3-Fluorophenol
2.2. Determination of Algal Growth and Biochemical Indicators
2.3. Determination of Algal Metabolic Profile
2.4. Determination of 3-Fluorophenol
3. Results and Discussion
3.1. Effects of 3-Fluorophenol on the Growth and Biochemical Indicators of C. pyrenoidosa
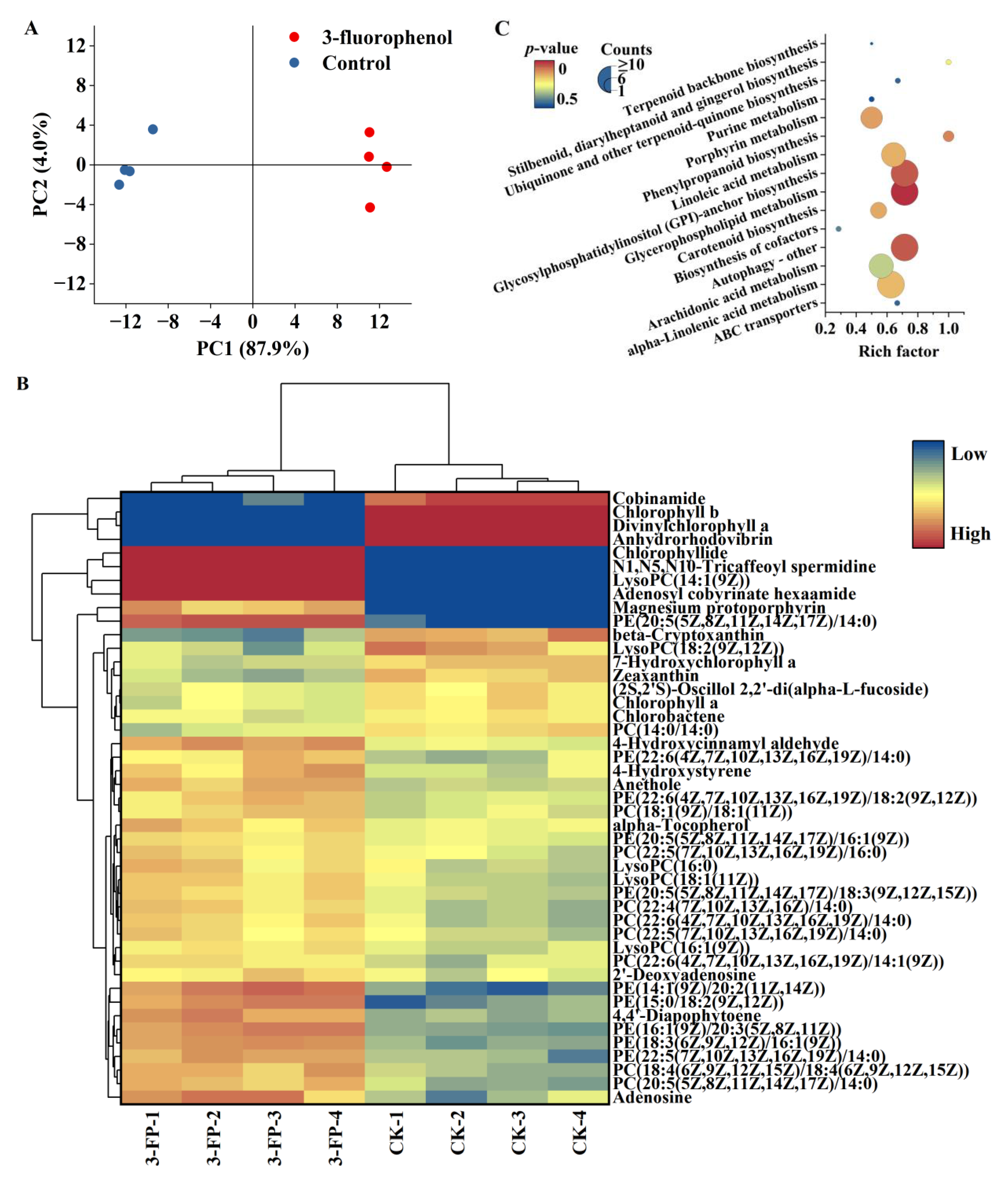
3.2. Effects of 3-Fluorophenol on the Metabolic Profiles of C. pyrenoidosa
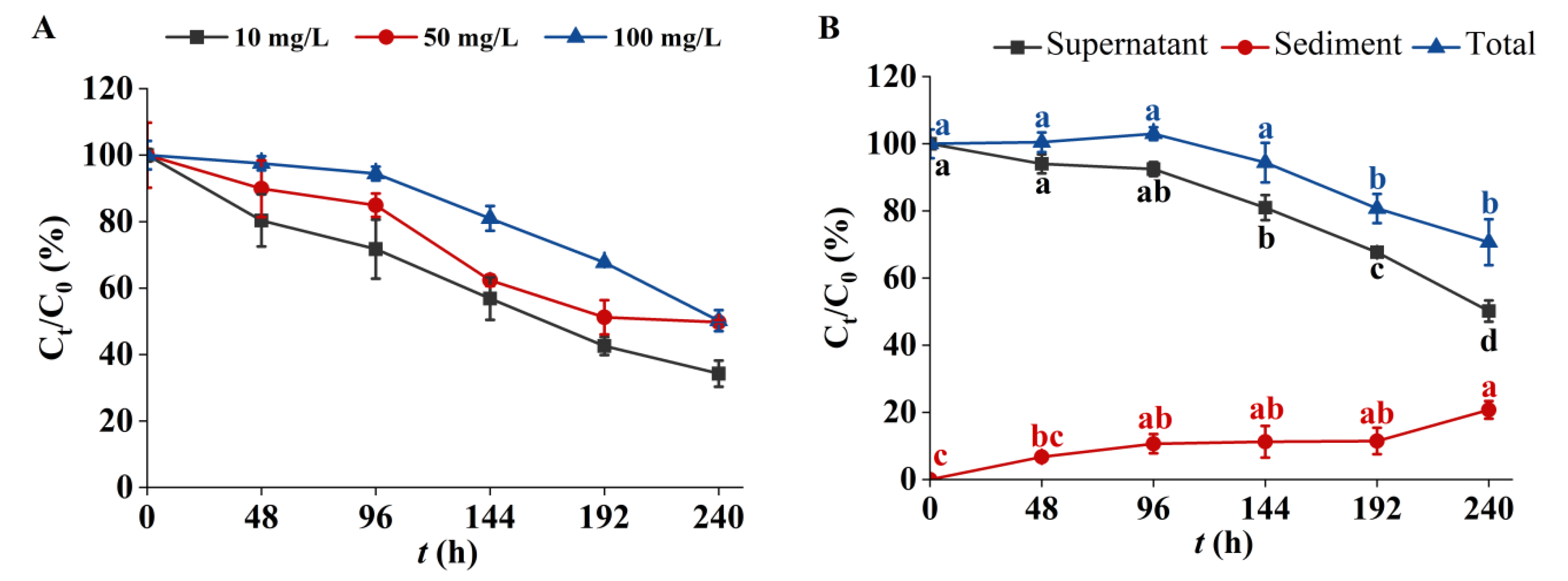
3.3. Removal of 3-Fluorophenol in the C. pyrenoidosa Solution
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Shah, P.; Westwell, A.D. The role of fluorine in medicinal chemistry. J. Enzyme Inhib. Med. Chem. 2007, 22, 527–540. [Google Scholar] [CrossRef]
- Liu, N.-R.; Yang, K.; Li, W.-T.; Pang, Z.-H.; Zhang, Q.; Wang, J.-J.; Dang, W.-X.; Jia, R.-Y.; Fu, Z.-W.; Li, Y.-X.; et al. Evaluation of the inhibition of chlorophenols towards human cytochrome P450 3A4 and differences among various species. Sci. Total Environ. 2020, 724, 138187. [Google Scholar] [CrossRef] [PubMed]
- Inoue, M.; Sumii, Y.; Shibata, N. Contribution of Organofluorine Compounds to Pharmaceuticals. ACS Omega 2020, 5, 10633–10640. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.-Q.; Zheng, T.; Zhang, W.J.; Shen, X.; Lv, L.; Li, Y.-M. Degradation of 3-fluoroanilne by Rhizobium sp. JF-3. Biodegradation 2019, 30, 433–445. [Google Scholar] [CrossRef] [PubMed]
- Franco, A.R.; Ferreira, A.C.; Castro, P.M.L. Co-metabolic degradation of mono-fluorophenols by the ectomycorrhizal fungi Pisolithus tinctorius. Chemosphere 2014, 111, 260–265. [Google Scholar] [CrossRef] [PubMed]
- Mu, Y.; Xing, J.; Shen, J.; Ying, L.; Mao, L.; Xu, X.; Lou, Y.; Wu, X. Determination of 19 chlorophenols in fish by QuEChERS-gas chromatography-mass spectrometry. Chin. J. Chromatogr. 2022, 40, 477–487. [Google Scholar] [CrossRef] [PubMed]
- Svihlikova, V.; Lankova, D.; Poustka, J.; Tomaniova, M.; Hajslova, J.; Pulkrabova, J. Perfluoroalkyl substances (PFASs) and other halogenated compounds in fish from the upper Labe River basin. Chemosphere 2015, 129, 170–178. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Peng, R.; Tian, Y.; Xu, J.; Wang, B.; Han, H.; Fu, X.; Gao, J.; Yao, Q. Metabolic engineering of Escherichia coli for efficient degradation of 4-fuorophenol. AMB Express 2022, 12, 55. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Chang, M.; Li, M.; An, Z.; Zhang, C.; Liu, J.; He, M. Ozone mechanism; kinetics, and toxicity studies of halophenols: Theoretical calculation combined with toxicity experiment. Sci. Total Environ. 2023, 858, 160101. [Google Scholar] [CrossRef] [PubMed]
- Folle, N.M.T.; Azevedo-Linhares, M.; Garcia, J.R.E.; da Costa Souza, A.T.; Grotzner, S.R.; de Oliveira, E.C.; Paulin, A.F.; Leite, N.F.; Neto, F.F.; de Oliveira Ribeiro, C.A. Low concentration of 2,4,6-tribromophenol (TBP) represents a risk to South American silver catfish Ramdia quelen (Quoy and Gaimard, 1824) population. Ecotox. Environ. Safe 2020, 187, 109815. [Google Scholar] [CrossRef]
- Han, C.; Zhu, W.; Ma, G.; Chen, Y.; Li, X.; Wei, X.; Yu, H. Computational insight into biotransformation of halophenols by cytochrome P450: Mechanism and reactivity for epoxidation. Chemosphere 2022, 286, 131708. [Google Scholar] [CrossRef] [PubMed]
- Kinani, S.; Souissi, Y.; Kinani, A.; Vujović, S.; Aït-Aïssa, S.; Bouchonnet, S. Photodegradation of fluorene in aqueous solution: Identification and biological activity testing of degradation products. J. Chromatogr. A 2016, 1442, 118–128. [Google Scholar] [CrossRef] [PubMed]
- Wei, J.; Ma, D.; Ma, X.; Sheng, Q.; Sun, X.; Li, J.; Liu, X.; Shen, J.; Zheng, M.; Wang, L. New insight into increased toxicity during ozonation of chlorophenol: The significant contribution of oxidizing intermediates. Sci. Total Environ. 2021, 769, 144569. [Google Scholar] [CrossRef] [PubMed]
- Ma, D.; Wei, J.; Zhang, H.; Zhou, Y.; Shen, J.; Wang, L.; Zhang, P. Acute toxicity evolution during ozonation of mono-chlorophenols and initial identification of highly toxic intermediates. Environ. Sci. Process. Impacts 2019, 21, 1509–1518. [Google Scholar] [CrossRef] [PubMed]
- Miller, E.; Menashe, O.; Dosoretz, C.G. A tailored permeable reactive bio-barrier forin situgroundwater remediation: Removal of 3-chlorophenol as a case study. Environ. Technol. 2022, 43, 1200–1210. [Google Scholar] [CrossRef]
- Gallego, A.; Soule, J.L.; Napolitano, H.; Rossi, S.L.; Vescina, C.; Korol, S.E. Biodegradability of chlorophenols in surface waters from the urban area of buenos aires. Bull. Environ. Contam. Toxicol. 2018, 100, 541–547. [Google Scholar] [CrossRef]
- Li, B.; Wu, D.; Li, Y.; Shi, Y.; Wang, C.; Sun, J.; Song, C. Metabolic Mechanism of Sulfadimethoxine Biodegradation by Chlorella sp. L38 and Phaeodactylum tricornutum MASCC-0025. Front. Microbiol. 2022, 13, 840562. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Zhang, X.; Bao, Y.; Zhang, K.; Qiu, J.; He, Q.; Zhu, J.; He, J. Enhanced degradation of dicamba by an anaerobic sludge acclimated from river sediment. Sci. Total Environ. 2021, 777, 145931. [Google Scholar] [CrossRef]
- Olaniran, A.O.; Igbinosa, E.O. Chlorophenols and other related derivatives of environmental concern: Properties, distribution and microbial degradation processes. Chemosphere 2011, 83, 1297–1306. [Google Scholar] [CrossRef]
- Escudero, A.; Hunter, C.; Roberts, J.; Helwig, K.; Pahl, O. Pharmaceuticals removal and nutrient recovery from wastewaters by Chlamydomonas acidophila. Biochem. Eng. J. 2020, 156, 107517. [Google Scholar] [CrossRef]
- Sawaya, C.; El Khoury, C.; Ramadan, L.; Deeb, R.; Harb, M. Effects of influent municipal wastewater microbial community and antibiotic resistance gene profiles on anaerobic membrane bioreactor effluent. Water Reuse 2022, 12, 304–318. [Google Scholar] [CrossRef]
- Andreotti, V.; Solimeno, A.; Rossi, S.; Ficara, E.; Marazzi, F.; Mezzanotte, V.; Garcia, J. Bioremediation of aquaculture wastewater with the microalgae Tetraselmis suecica: Semi-continuous experiments, simulation and photo-respirometric tests. Sci. Total Environ. 2020, 738, 139859. [Google Scholar] [CrossRef] [PubMed]
- Jalilian, N.; Najafpour, G.D.; Khajouei, M. Macro and Micro Algae in Pollution Control and Biofuel Production—A Review. ChemBioEng Rev. 2020, 7, 18–33. [Google Scholar] [CrossRef]
- Cheng, P.; Chu, R.; Zhang, X.; Song, L.; Chen, D.; Zhou, C.; Yan, X.; Cheng, J.J.; Ruan, R. Screening of the dominant Chlorella pyrenoidosa for biofilm attached culture and feed production while treating swine wastewater. Bioresour. Technol. 2020, 318, 124054. [Google Scholar] [CrossRef] [PubMed]
- Ran, Y.; Sun, D.; Liu, X.; Zhang, L.; Niu, Z.; Chai, T.; Hu, Z.; Qiao, K. Chlorella pyrenoidosa as a potential bioremediator: Its tolerance and molecular responses to cadmium and lead. Sci. Total Environ. 2024, 912, 168712. [Google Scholar] [CrossRef] [PubMed]
- Gojkovic, Z.; Lindberg, R.H.; Tysklind, M.; Funk, C. Northern green algae have the capacity to remove active pharmaceutical ingredients. Ecotox. Environ. Safe 2019, 170, 644–656. [Google Scholar] [CrossRef] [PubMed]
- Escapa, C.; Coimbra, R.N.; Paniagua, S.; García, A.I.; Otero, M. Nutrients and pharmaceuticals removal from wastewater by culture and harvesting of Chlorella sorokiniana. Bioresour. Technol. 2015, 185, 276–284. [Google Scholar] [CrossRef] [PubMed]
- Shivagangaiah, C.P.; Sanyal, D.; Dasgupta, S.; Banik, A. Phycoremediation and photosynthetic toxicity assessment of lead by two freshwater microalgae Scenedesmus acutus and Chlorella pyrenoidosa. Physiol. Plant. 2021, 173, 246–258. [Google Scholar] [CrossRef]
- Tao, M.-T.; Bian, Z.-Q.; Zhang, J.; Wang, T.; Shen, H. Quantitative evaluation and the toxicity mechanism of synergism within three organophosphorus pesticide mixtures to Chlorella pyrenoidosa. Environ. Sci. Process. Impacts 2020, 22, 2095–2103. [Google Scholar] [CrossRef]
- Li, M.; Ma, L.; An, Y.; Wei, D.; Ma, H.; Zhu, J.; Wang, C. Bioremoval efficiency and metabolomic profiles of cellular responses of Chlorella pyrenoidosa to phenol and 4-fluorophenol. Water Reuse 2023, 13, 97–106. [Google Scholar] [CrossRef]
- Perez-Caselles, C.; Burgos, L.; Sanchez-Balibrea, I.; Egea, J.A.; Faize, L.; Martin-Valmaseda, M.; Bogdanchikova, N.; Pestryakov, A.; Alburquerque, N. The Effect of Silver Nanoparticle Addition on Micropropagation of Apricot Cultivars (Prunus armeniaca L.) in Semisolid and Liquid Media. Plants 2023, 12, 1547. [Google Scholar] [CrossRef] [PubMed]
- Calabrese, E.J. Hormesis: Why it is important to toxicology and toxicologists. Environ. Toxicol. Chem. 2008, 27, 1451–1474. [Google Scholar] [CrossRef] [PubMed]
- Hashmi, M.Z.; Naveedullah; Shen, H.; Zhu, S.; Yu, C.; Shen, C. Growth, bioluminescence and shoal behavior hormetic responses to inorganic and/or organic chemicals: A review. Environ. Int. 2014, 64, 28–39. [Google Scholar] [CrossRef] [PubMed]
- Munro, V.; Kelly, V.; Messner, C.B.; Kustatscher, G. Cellular control of protein levels: A systems biology perspective. Proteomics 2023, 24, e2200220. [Google Scholar] [CrossRef] [PubMed]
- Shikov, A.; Chirkova, T.V.; Yemelyanov, V.V. Functions of reactive oxygen species in plant cells under normal conditions and during adaptation. Ecol. Genet. 2021, 19, 343–363. [Google Scholar] [CrossRef]
- Mansoor, S.; Wani, O.A.; Lone, J.K.; Manhas, S.; Kour, N.; Alam, P.; Ahmad, A.; Ahmad, P. Reactive oxygen species in plants: From source to sink. Antioxidants 2022, 11, 225. [Google Scholar] [CrossRef]
- Rajput, V.D.; Harish; Singh, R.K.; Verma, K.K.; Sharma, L.; Quiroz-Figueroa, F.R.; Meena, M.; Gour, V.S.; Minkina, T.; Sushkova, S.; et al. Recent developments in enzymatic antioxidant defence mechanism in plants with special reference to abiotic stress. Biology 2021, 10, 267. [Google Scholar] [CrossRef]
- Wang, X.D.; Lu, Y.C.; Xiong, X.H.; Yuan, Y.; Lu, L.X.; Liu, Y.J.; Mao, J.H.; Xiao, W.W. Toxicological responses; bioaccumulation, and metabolic fate of triclosan in Chlamydomonas reinhardtii. Environ. Sci. Pollut. Res. 2020, 27, 11246–11259. [Google Scholar] [CrossRef] [PubMed]
- Hishikawa, D.; Hashidate, T.; Shimizu, T.; Shindou, H. Diversity and function of membrane glycerophospholipids generated by the remodeling pathway in mammalian cells. J. Lipid Res. 2014, 55, 799–807. [Google Scholar] [CrossRef]
- White, D.A.; Rooks, P.A.; Kimmance, S.; Tait, K.; Jones, M.; Tarran, G.A.; Cook, C.; Llewellyn, C.A. Modulation of Polar Lipid Profiles in Chlorella sp. in Response to Nutrient Limitation. Metabolites 2019, 9, 39. [Google Scholar] [CrossRef]
- Qian, L.; Qi, S.; Cao, F.; Zhang, J.; Zhao, F.; Li, C.; Wang, C. Toxic effects of boscalid on the growth, photosynthesis, antioxidant system and metabolism of Chlorella vulgaris. Environ. Pollut. 2018, 242, 171–181. [Google Scholar] [CrossRef]
- Mei, J.; Ning, N.; Wu, H.; Chen, X.; Li, Z.; Liu, W. Glycosylphosphatidylinositol Anchor Biosynthesis Pathway-Related Protein GPI7 Is Required for the Vegetative Growth and Pathogenicity of Colletotrichum graminicola. Int. J. Mol. Sci. 2022, 23, 2985. [Google Scholar] [CrossRef]
- Liu, S.; Yao, S.; Yang, H.; Liu, S.; Wang, Y. Autophagy: Regulator of cell death. Cell Death Dis. 2023, 14, 648. [Google Scholar] [CrossRef]
- Zhuang, X.; Chung, K.P.; Cui, Y.; Lin, W.; Gao, C.; Kang, B.-H.; Jiang, L. ATG9 regulates autophagosome progression from the endoplasmic reticulum in Arabidopsis. Proc. Natl. Acad. Sci. USA 2017, 114, E426–E435. [Google Scholar] [CrossRef]
- Huang, H.-L.; Wang, B.-G. Antioxidant capacity and lipophilic content of seaweeds collected from the Qingdao coastline. J. Agric. Food. Chem. 2004, 52, 4993–4997. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.; Shahzad, B.; Rehman, A.; Bhardwaj, R.; Landi, M.; Zheng, B. Response of phenylpropanoid pathway and the role of polyphenols in plants under abiotic stress. Molecules 2019, 24, 2452. [Google Scholar] [CrossRef] [PubMed]
- Barrena, L.E.P.; Mats, L.; Earl, H.J.; Bozzo, G.G. Phenylpropanoid Metabolism in Phaseolus vulgaris during Growth under Severe Drought. Metabolites 2024, 14, 319. [Google Scholar] [CrossRef] [PubMed]
- Rui, Z.; Li, L.; Xue, H.; Bi, Y.; Raza, H.; Si, M.; Peng, H.; Nan, M.; Zong, Y.; Prusky, D. Ca2+ applications affect the phenylpropanoid metabolism in potato tubers induced by T-2 toxin. Postharvest Biol. Technol. 2021, 180, 111616. [Google Scholar] [CrossRef]
- Wang, B.; Wu, C.; Wang, G.; He, J.; Zhu, S. Transcriptomic analysis reveals a role of phenylpropanoid pathway in the enhancement of chilling tolerance by pre-storage cold acclimation in cucumber fruit. Sci. Hortic. 2021, 288, 110282. [Google Scholar] [CrossRef]
- Dong, N.-Q.; Lin, H.-X. Contribution of phenylpropanoid metabolism to plant development and plant-environment interactions. J. Integr. Plant Biol. 2020, 63, 180–209. [Google Scholar] [CrossRef]
- Meguro, M.; Ito, H.; Takabayashi, A.; Tanaka, R.; Tanaka, A. Identification of the 7-Hydroxymethyl Chlorophyll a Reductase of the Chlorophyll Cycle in Arabidopsis. Plant Cell 2011, 23, 3442–3453. [Google Scholar] [CrossRef] [PubMed]
- Lim, H.; Tanaka, A.; Tanaka, R.; Ito, H. In Vitro Enzymatic Activity Assays Implicate the Existence of the Chlorophyll Cycle in Chlorophyll b-Containing Cyanobacteria. Plant Cell Physiol. 2020, 60, 2672–2683. [Google Scholar] [CrossRef] [PubMed]
- Takaichi, S. Carotenoids in Algae: Distributions, Biosyntheses and Functions. Mar. Drugs 2011, 9, 1101–1118. [Google Scholar] [CrossRef] [PubMed]
- Sun, T.; Tadmor, Y.; Li, L. Pathways for Carotenoid Biosynthesis, Degradation, and Storage. Methods Mol. Biol. 2020, 2083, 3–23. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, H.; Prajapati, S.K. Allelopathic effect of benzoic acid (hydroponics root exudate) on microalgae growth. Environ. Res. 2023, 219, 115020. [Google Scholar] [CrossRef] [PubMed]
- Liang, C.; Lv, H.; Liu, W.; Wang, Q.; Yao, X.; Li, X.; Hu, Z.; Wang, J.; Zhu, L.; Wang, J. Mechanism of the adverse outcome of Chlorella vulgaris exposure to diethyl phthalate: Water environmental health reflected by primary producer toxicity. Sci. Total Environ. 2024, 912, 168876. [Google Scholar] [CrossRef] [PubMed]
- León-Vaz, A.; León, R.; Giráldez, I.; Vega, J.M.; Vigara, J. Impact of heavy metals in the microalga Chlorella sorokiniana and assessment of its potential use in cadmium bioremediation. Aquat. Toxicol. 2021, 239, 105941. [Google Scholar] [CrossRef] [PubMed]
- Hamed, S.M.; Zinta, G.; Klöck, G.; Asard, H.; Selim, S.; AbdElgawad, H. Zinc-induced differential oxidative stress and antioxidant responses in Chlorella sorokiniana and Scenedesmus acuminatus. Ecotoxicol. Environ. Saf. 2017, 140, 256–263. [Google Scholar] [CrossRef]
- Zhang, W.; Tan, N.G.J.; Fu, B.; Li, S.F.Y. Metallomics and NMR-based metabolomics of Chlorella sp. reveal the synergistic role of copper and cadmium in multi-metal toxicity and oxidative stress. Metallomics 2015, 7, 426–438. [Google Scholar] [CrossRef] [PubMed]
- Deng, D.; Chen, H.X.; Wong, Y.S.; Tam, N.F.Y. Physiological response and oxidative transformation of 2,2′,4,4′-tetrabromodiphenyl ether (BDE-47) by a Chlorella isolate. Sci. Total Environ. 2020, 744, 140869. [Google Scholar] [CrossRef]
- Ding, N.; Wang, L.; Kang, Y.; Luo, K.; Zeng, D.; Man, Y.; Zhang, Q.; Zeng, L.; Luo, J.; Jiang, F. The comparison of transcriptomic response of green microalga Chlorella sorokiniana exposure to environmentally relevant concentration of cadmium(II) and 4-n-nonylphenol. Environ. Geochem. Health 2020, 42, 2881–2894. [Google Scholar] [CrossRef] [PubMed]
- Chernysheva, M.; Bulatova, M.; Ding, X.; Catalonia, B.O.; Ding, X.; Haukka, M. Influence of substituents in aromatic ring on the strength of halogen bonding in iodobenzene derivatives. Cryst. Growth Des. 2020, 20, 7197–7210. [Google Scholar] [CrossRef]
- Liu, J.; Liu, Y.; Tian, Y.; Feng, L.; Zhang, L. Comparison of the oxidation of halogenated phenols in UV/PDS and UV/H2O2 advanced oxidation processes. RSC Adv. 2020, 10, 6464–6472. [Google Scholar] [CrossRef] [PubMed]
- Fawzy, M.A.; Alharthi, S. Cellular responses and phenol bioremoval by green alga Scenedesmus abundans: Equilibrium, kinetic and thermodynamic studies. Environ. Technol. Innov. 2021, 22, 101463. [Google Scholar] [CrossRef]
- Dubey, S.; Chen, C.-W.; Haldar, D.; Tambat, V.S.; Kumar, P.; Tiwari, A.; Singhania, R.R.; Dong, C.-D.; Patel, A.K. Advancement in algal bioremediation for organic, inorganic, and emerging pollutants. Environ. Pollut. 2023, 317, 120840. [Google Scholar] [CrossRef] [PubMed]
- El-Naggar, N.E.-A.; Hamouda, R.A.; Mousa, I.E.; Abdel-Hamid, M.S.; Rabei, N.H. Statistical optimization for cadmium removal using Ulva fasciata biomass: Characterization, immobilization and application for almost-complete cadmium removal from aqueous solutions. Sci. Rep. 2018, 8, 12456. [Google Scholar] [CrossRef] [PubMed]
- Gondi, R.; Kavitha, S.; Kannah, R.Y.; Karthikeyan, O.P.; Kumar, G.; Tyagi, V.K.; Banu, J.R. Algal-based system for removal of emerging pollutants from wastewater: A review. Bioresour. Technol. 2021, 344, 126245. [Google Scholar] [CrossRef]
- Ding, T.; Wang, S.; Yang, B.; Li, J. Biological removal of pharmaceuticals by Navicula sp. and biotransformation of bezafibrate. Chemosphere 2020, 240, 124949. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, M.; Shang, Z.; Ma, Y.; Zhao, H.; Ni, Z.; Wei, Z.; Zhang, X. Tolerance Mechanisms and Removal Efficiency of Chlorella pyrenoidosa in Treating 3-Fluorophenol Pollution. Metabolites 2024, 14, 449. https://doi.org/10.3390/metabo14080449
Li M, Shang Z, Ma Y, Zhao H, Ni Z, Wei Z, Zhang X. Tolerance Mechanisms and Removal Efficiency of Chlorella pyrenoidosa in Treating 3-Fluorophenol Pollution. Metabolites. 2024; 14(8):449. https://doi.org/10.3390/metabo14080449
Chicago/Turabian StyleLi, Min, Zhenfang Shang, Yonglan Ma, Huijun Zhao, Zhijing Ni, Zhaojun Wei, and Xiu Zhang. 2024. "Tolerance Mechanisms and Removal Efficiency of Chlorella pyrenoidosa in Treating 3-Fluorophenol Pollution" Metabolites 14, no. 8: 449. https://doi.org/10.3390/metabo14080449
APA StyleLi, M., Shang, Z., Ma, Y., Zhao, H., Ni, Z., Wei, Z., & Zhang, X. (2024). Tolerance Mechanisms and Removal Efficiency of Chlorella pyrenoidosa in Treating 3-Fluorophenol Pollution. Metabolites, 14(8), 449. https://doi.org/10.3390/metabo14080449






