Metabolic Changes in Pseudomonas oleovorans Isolated from Contaminated Construction Material Exposed to Varied Biocide Treatments
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemicals
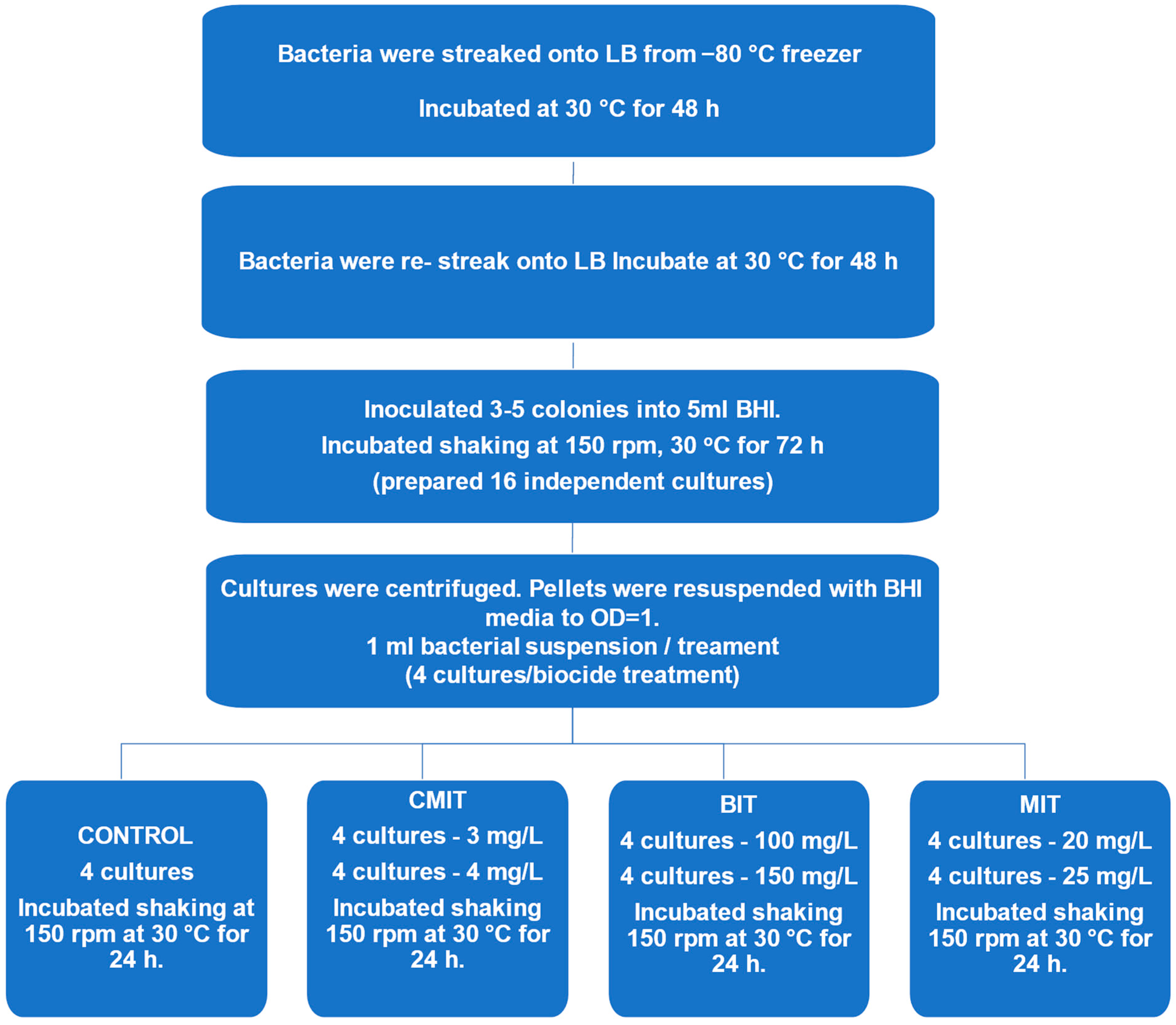
2.2. Biocide Exposure
2.3. LC–MS Analysis
2.4. Data Preprocessing
- Mass Detection: Mass detection utilized a noise level threshold of 750.
- ADAP Chromatogram Building: Chromatograms were built with a minimum group size of 5, a group intensity threshold of 200, a minimum height of 1000, and an m/z tolerance of 0.007 m/z or 7 ppm.
- Chromatogram Deconvolution: Deconvolution applied a 70% chromatographic threshold, 0.05 min minimum RT range, 5% minimum relative height, 1200 minimum absolute height, a minimum peak top/edge ratio of 1.2, and a peak duration range of 0.08–5.0.
- Isotopic Peak Grouper: Isotopic peaks were grouped with an m/z tolerance of 5.0 ppm and an RT tolerance of 0.05 min, and the most intense isotope was selected as the representative.
- Join Aligner: Data alignment used a m/z tolerance of 0.008 or 8 ppm and a weight of 2, RT tolerance of 0.15 min and a weight of 1, without requiring charge state or identification, and without comparing isotope patterns.
- Peak List Row Filter: Rows in the peak list were filtered, requiring a minimum presence in 10% of the samples.
- Gap Filling: Gap filling was conducted using the same RT and m/z range with an m/z tolerance of 0.009 m/z or 11.0 ppm.
- Identification: Compounds were identified using in-house database searches with an m/z tolerance of 0.009 m/z or 10.0 ppm and a retention time tolerance of 0.2 min.
2.5. Statistical Analysis
3. Results
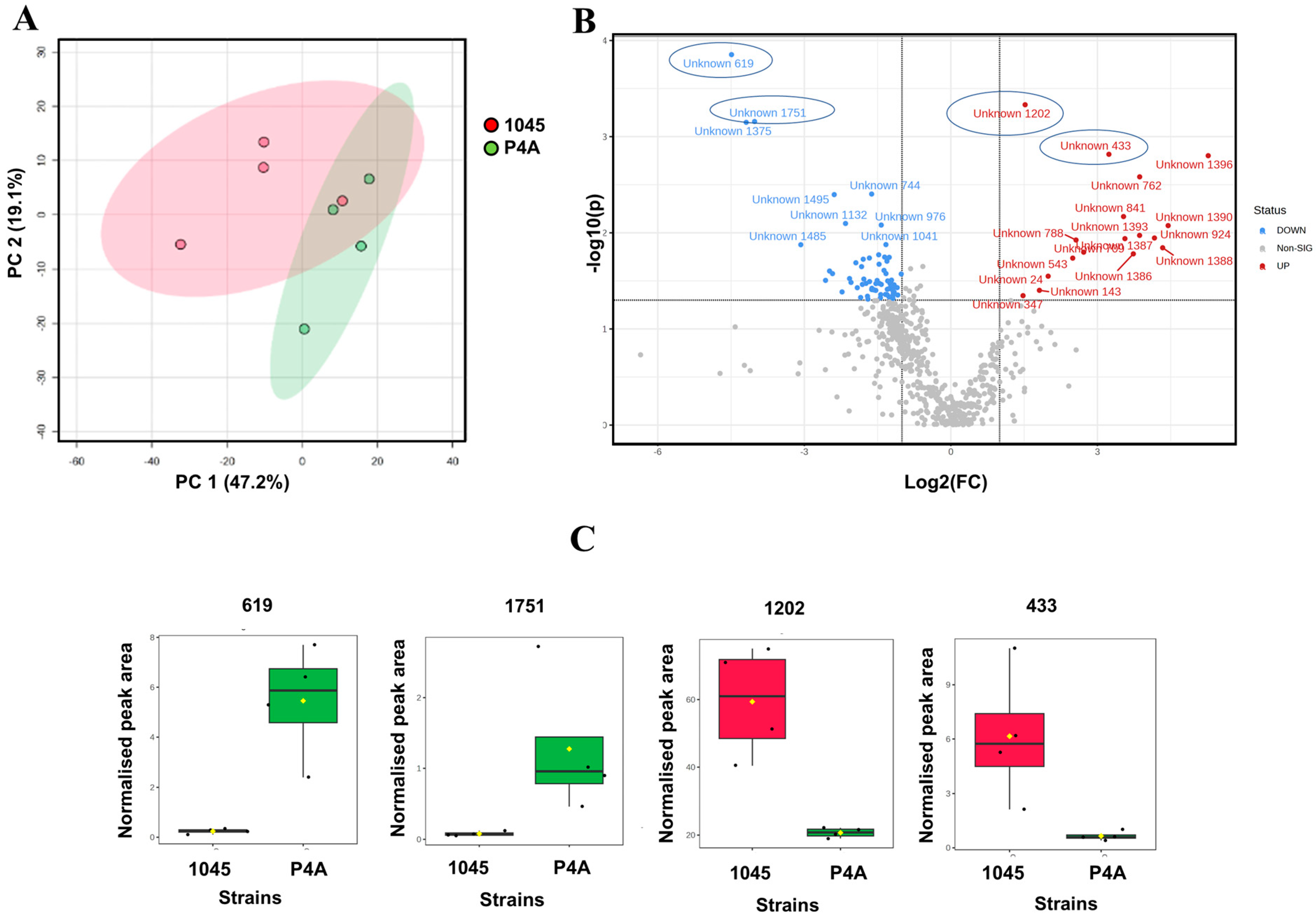
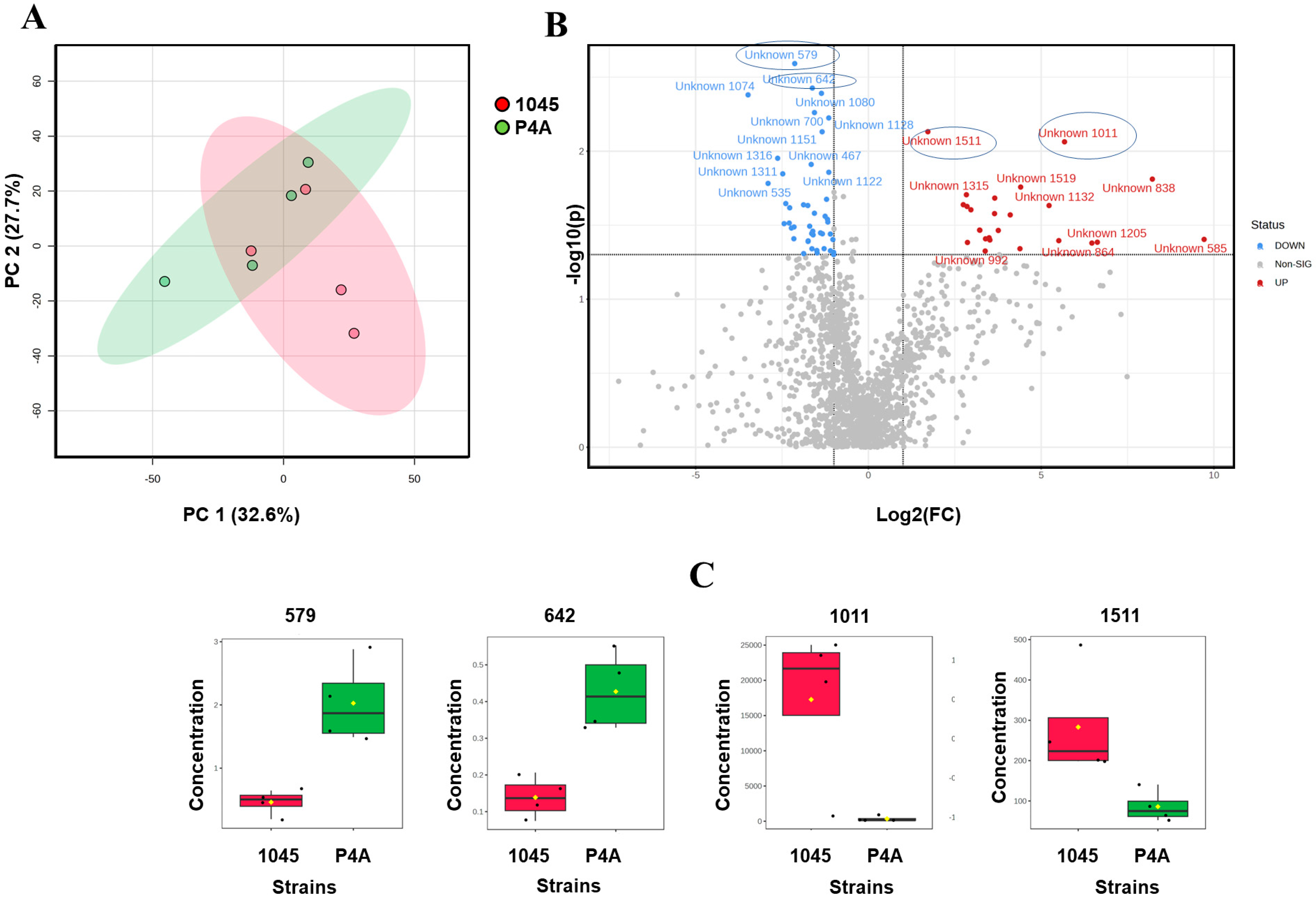
3.1. Lipid and Metabolite Profiles in Untreated P. oleovorans P4A and P. oleovorans 1045
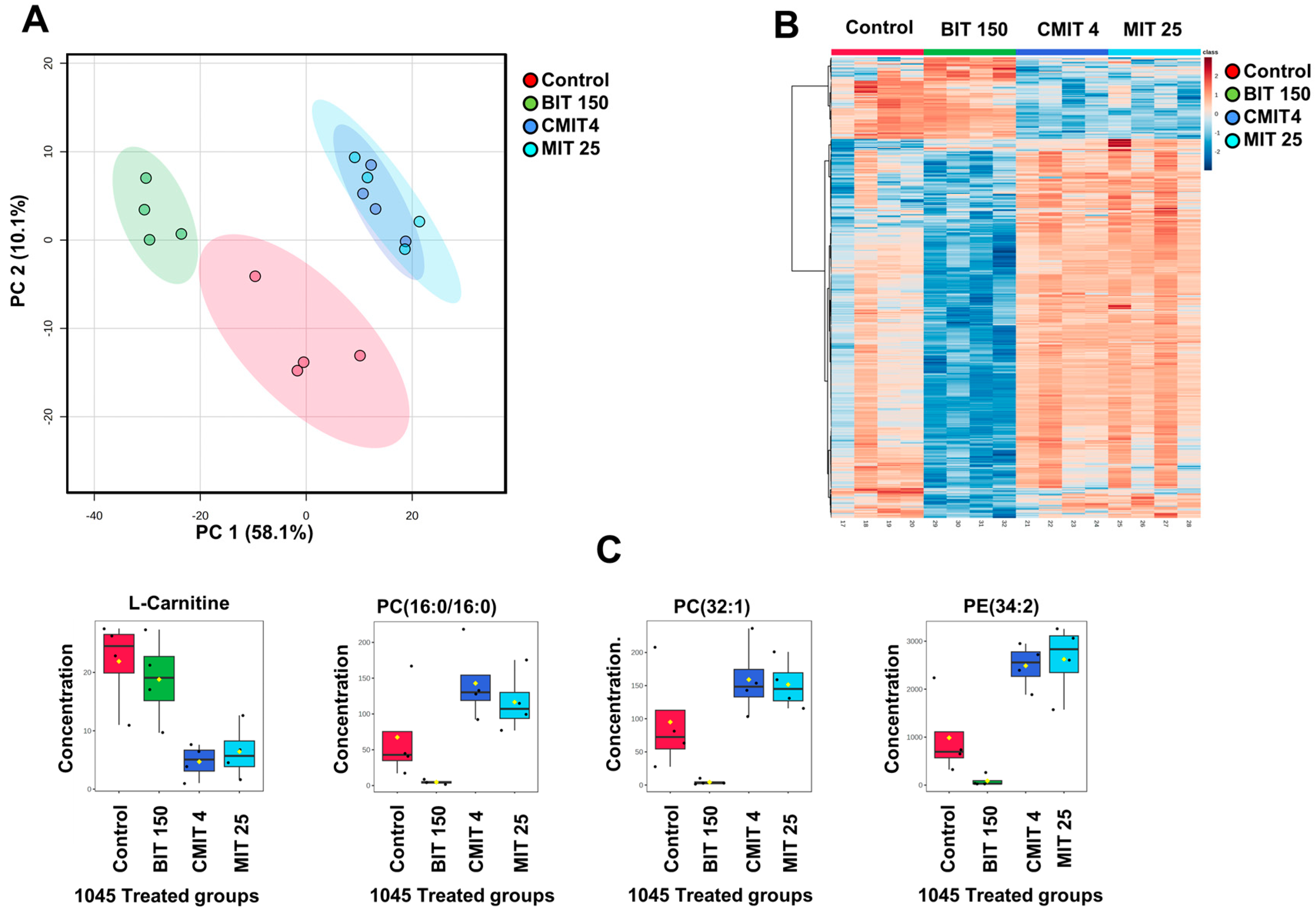
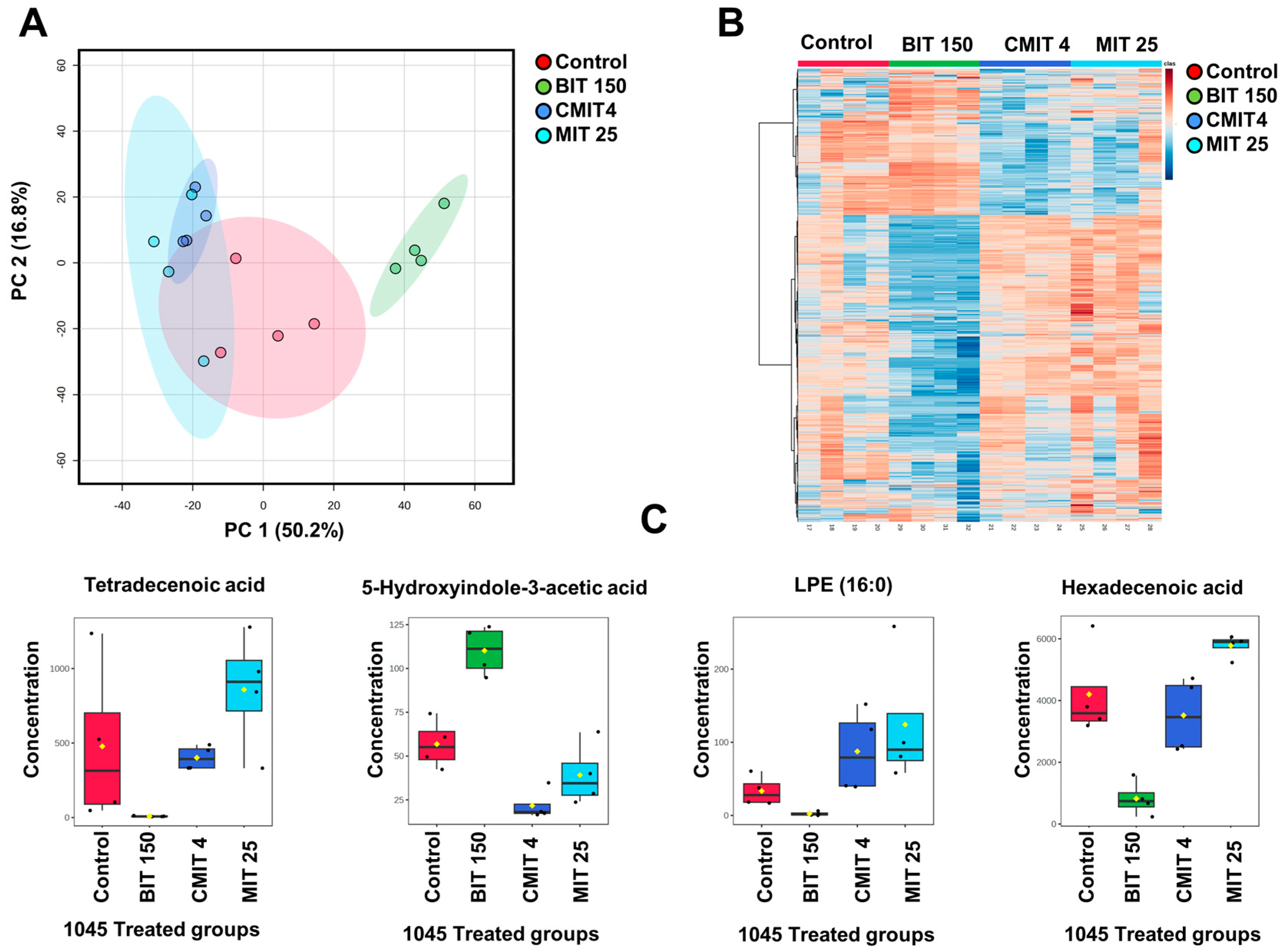
3.2. Lipid and Metabolite Profiles in Biocide-Treated P. oleovorans 1045 Reference Strains
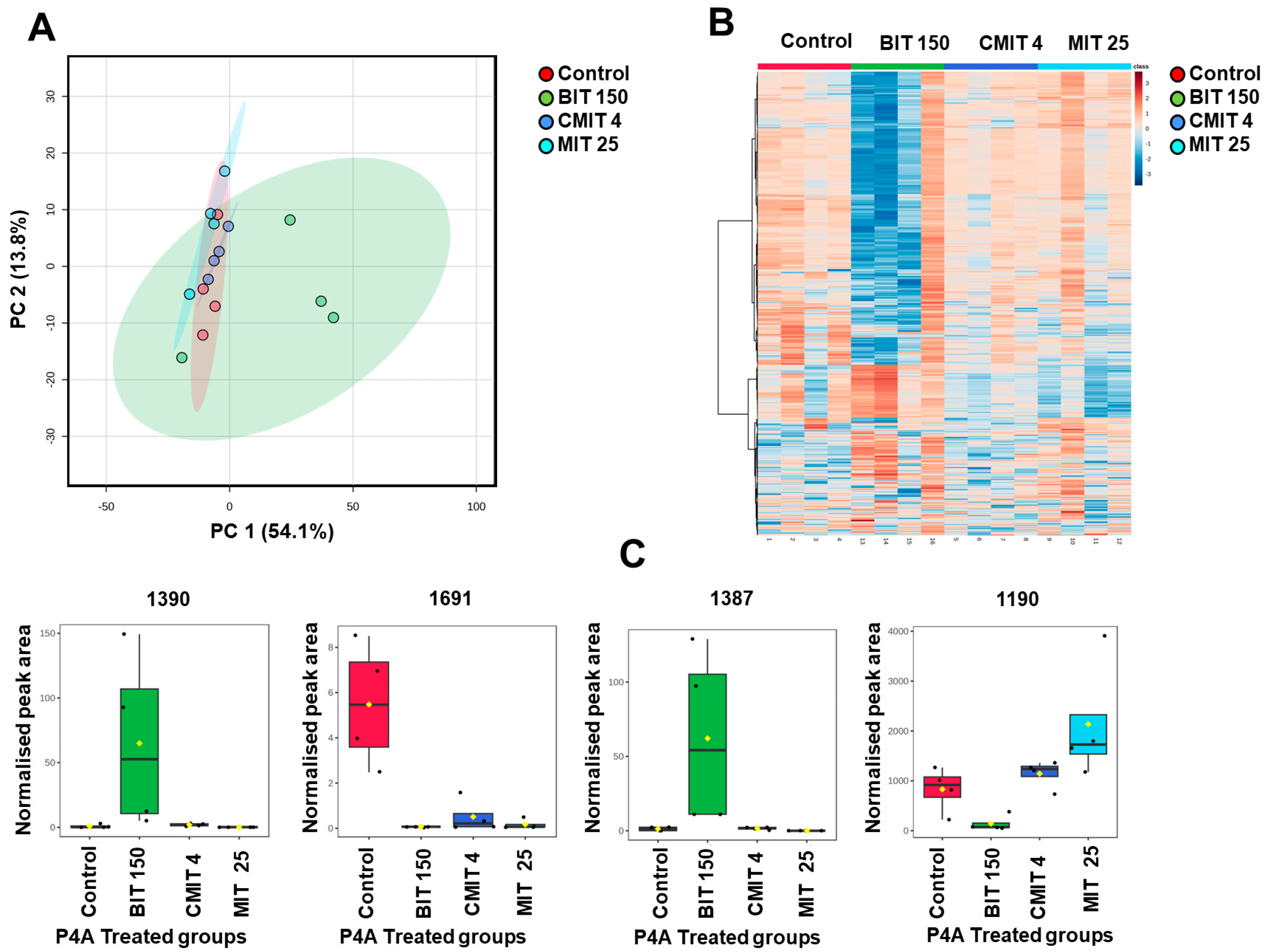
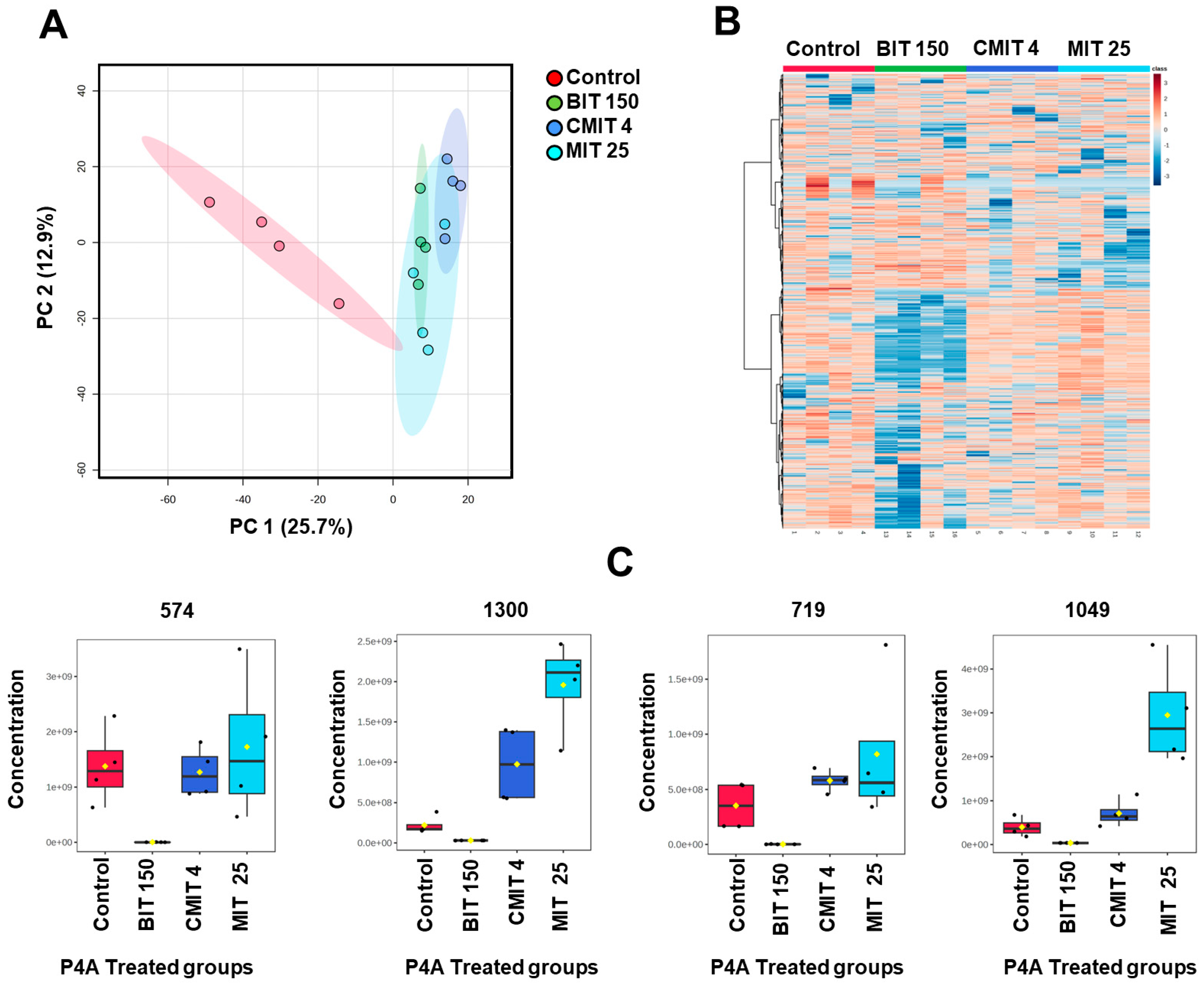
3.3. Lipids and Metabolites Profiles of Biocide-Treated P. oleovorans P4A Strain
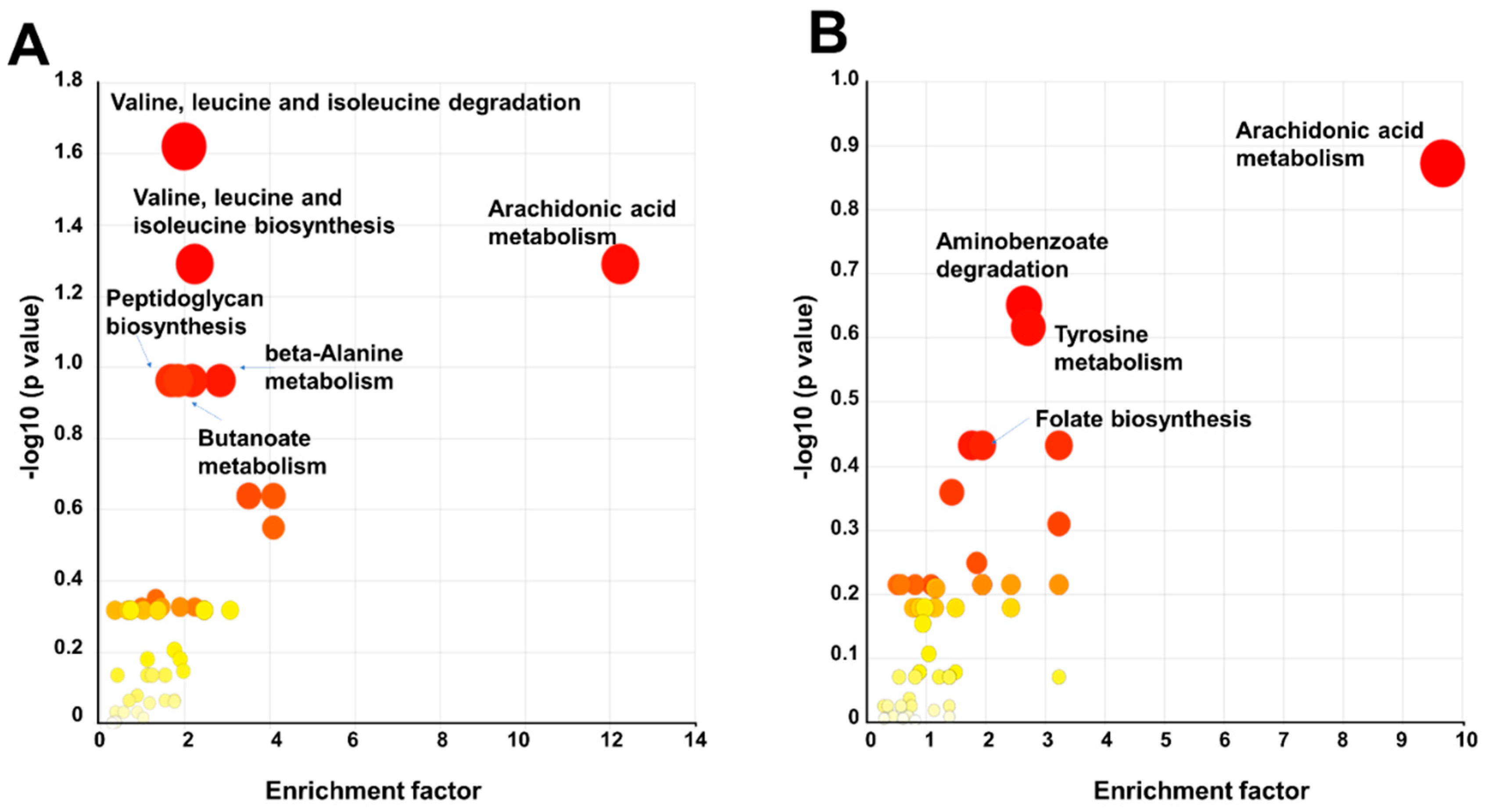
3.4. Metabolic Pathway Analysis of BIT-Exposed P. oleovorans Strains
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Williams, T.M. The Mechanism of Action of Isothiazolone Biocide. In Proceedings of the CORROSION 2006, San Diego, CA, USA, 12–16 March 2006. [Google Scholar]
- Luz, G.V.S.; Sousa, B.A.S.M.; Guedes, A.V.; Barreto, C.C.; Brasil, L.M. Biocides Used as Additives to Biodiesels and Their Risks to the Environment and Public Health: A Review. Molecules 2018, 23, 2698. [Google Scholar] [CrossRef]
- Kim, J.; Choi, J. Trans- and Multigenerational Effects of Isothiazolinone Biocide CMIT/MIT on Genotoxicity and Epigenotoxicity in Daphnia magna. Toxics 2023, 11, 388. [Google Scholar] [CrossRef]
- Lundov, M.D.; Krongaard, T.; Menne, T.L.; Johansen, J.D. Methylisothiazolinone contact allergy: A review. Br. J. Dermatol. 2011, 165, 1178–1182. [Google Scholar] [CrossRef]
- Scherrer, M.A.; Rocha, V.B.; Andrade, A.R. Contact dermatitis to methylisothiazolinone. An. Bras. Dermatol. 2015, 90, 912–914. [Google Scholar] [CrossRef]
- Basketter, D.; Casati, S. Dermal Toxicity: Skin Sensitization. In In Vitro Toxicology Systems; Bal-Price, A., Jennings, P., Eds.; Springer: New York, NY, USA, 2014; pp. 225–239. [Google Scholar]
- Martin, L.W.; Robson, C.L.; Watts, A.M.; Gray, A.R.; Wainwright, C.E.; Bell, S.C.; Ramsay, K.A.; Kidd, T.J.; Reid, D.W.; Brockway, B.; et al. Expression of Pseudomonas aeruginosa Antibiotic Resistance Genes Varies Greatly during Infections in Cystic Fibrosis Patients. Antimicrob. Agents Chemother. 2018, 62, 10–1128. [Google Scholar] [CrossRef]
- Pandey, P.; Kiran, U.V. Degradation of paints and its microbial effect on health and environment. J. Crit. Rev. 2020, 7, 4879–4884. [Google Scholar]
- Li, Z.J.; Wang, L.; Yuan, L.Y.; Xiao, C.L.; Mei, L.; Zheng, L.R.; Zhang, J.; Yang, J.H.; Zhao, Y.L.; Zhu, Z.T.; et al. Efficient removal of uranium from aqueous solution by zero-valent iron nanoparticle and its graphene composite. J. Hazard. Mater. 2015, 290, 26–33. [Google Scholar] [CrossRef]
- Coombs, K.; Rodriguez-Quijada, C.; Clevenger, J.O.; Sauer-Budge, A.F. Current Understanding of Potential Linkages between Biocide Tolerance and Antibiotic Cross-Resistance. Microorganisms 2023, 11, 2000. [Google Scholar] [CrossRef]
- Leahy, J.G.; Colwell, R.R. Microbial degradation of hydrocarbons in the environment. Microbiol. Rev. 1990, 54, 305–315. [Google Scholar] [CrossRef]
- Palleroni, N.J.; Pieper, D.H.; Moore, E.R.B. Microbiology of Hydrocarbon-Degrading Pseudomonas. In Handbook of Hydrocarbon and Lipid Microbiology; Timmis, K.N., Ed.; Springer: Berlin/Heidelberg, Germany, 2010; pp. 1787–1798. [Google Scholar]
- Mozejko-Ciesielska, J.; Szacherska, K.; Marciniak, P. Pseudomonas Species as Producers of Eco-friendly Polyhydroxyalkanoates. J. Polym. Environ. 2019, 27, 1151–1166. [Google Scholar] [CrossRef]
- Das, N.; Chandran, P. Microbial degradation of petroleum hydrocarbon contaminants: An overview. Biotechnol. Res. Int. 2011, 2011, 941810. [Google Scholar] [CrossRef]
- Yesankar, P.J.; Patil, A.; Kapley, A.; Qureshi, A. Catalytic resilience of multicomponent aromatic ring-hydroxylating dioxygenases in Pseudomonas for degradation of polycyclic aromatic hydrocarbons. World J. Microbiol. Biotechnol. 2023, 39, 166. [Google Scholar] [CrossRef]
- Medic, A.B.; Karadzic, I.M. Pseudomonas in environmental bioremediation of hydrocarbons and phenolic compounds-key catabolic degradation enzymes and new analytical platforms for comprehensive investigation. World J. Microbiol. Biotechnol. 2022, 38, 165. [Google Scholar] [CrossRef]
- Talukder, A.; Rahman, M.M.; Chowdhury, M.M.H.; Mobashshera, T.A.; Islam, N.N. Plasmid profiling of multiple antibiotic-resistant Pseudomonas aeruginosa isolated from soil of the industrial area in Chittagong, Bangladesh. Beni-Suef Univ. J. Basic Appl. Sci. 2021, 10, 1–7. [Google Scholar] [CrossRef]
- Chapman, J.S. Biocide resistance mechanisms. Int. Biodeterior. Biodegrad. 2003, 51, 133–138. [Google Scholar] [CrossRef]
- Fahmy, A.; Srinivasan, A.; Webber, M.A. The Relationship Between Bacterial Multidrug Efflux Pumps and Biofilm Formation. In Efflux-Mediated Antimicrobial Resistance in Bacteria; Li, X.-Z., Elkins, C.A., Zgurskaya, H.I., Eds.; Springer International Publishing: Berlin/Heidelberg, Germany, 2016; pp. 651–663. [Google Scholar]
- Ugwuanyi, F.C.; Ajayi, A.; Ojo, D.A.; Adeleye, A.I.; Smith, S.I. Evaluation of efflux pump activity and biofilm formation in multidrug resistant clinical isolates of Pseudomonas aeruginosa isolated from a Federal Medical Center in Nigeria. Ann. Clin. Microbiol. Antimicrob. 2021, 20, 11. [Google Scholar] [CrossRef]
- Anari, R.K.; Nikkhahi, F.; Javadi, A.; Bakht, M.; Rostamani, M.; Kelishomi, F.Z.; Alizadeh, S.A. Evaluation of antibacterial activity of five biocides and the synergistic effect of biocide/EDTA combinations on biofilm-producing and non-producing Stenotrophomonas maltophilia strains isolated from clinical specimens in Iran. BMC Microbiol. 2022, 22, 257. [Google Scholar] [CrossRef]
- Maillard, J.Y. Bacterial target sites for biocide action. J. Appl. Microbiol. 2002, 92, 16S–27S. [Google Scholar] [CrossRef]
- Dumas, J.L.; van Delden, C.; Perron, K.; Kohler, T. Analysis of antibiotic resistance gene expression in Pseudomonas aeruginosa by quantitative real-time-PCR. FEMS Microbiol. Lett. 2006, 254, 217–225. [Google Scholar] [CrossRef]
- Costerton, J.W.; Stewart, P.S.; Greenberg, E.P. Bacterial biofilms: A common cause of persistent infections. Science 1999, 284, 1318–1322. [Google Scholar] [CrossRef]
- Kamali, E.; Jamali, A.; Ardebili, A.; Ezadi, F.; Mohebbi, A. Evaluation of antimicrobial resistance, biofilm forming potential, and the presence of biofilm-related genes among clinical isolates of Pseudomonas aeruginosa. BMC Res. Notes 2020, 13, 27. [Google Scholar] [CrossRef]
- Sunil, B.; Ashraf, A. (Eds.) An Overview of Lipid Metabolism. In Pediatric Dyslipidemia; Springer International Publishing: Berlin/Heidelberg, Germany, 2023; pp. 9–14. [Google Scholar]
- Constantino-Teles, P.; Jouault, A.; Touqui, L.; Saliba, A.M. Role of Host and Bacterial Lipids in Pseudomonas aeruginosa Respiratory Infections. Front. Immunol. 2022, 13, 931027. [Google Scholar] [CrossRef]
- Russell, A.D. Similarities and differences in the responses of microorganisms to biocides. J. Antimicrob. Chemother. 2003, 52, 750–763. [Google Scholar] [CrossRef]
- Brügger, B. From lipidomics to cellular functions: Lipids as modulators of protein activity. FASEB J. 2015, 29, 492.1. [Google Scholar] [CrossRef]
- Nozawa, Y. Adaptive regulation of membrane lipids and fluidity during thermal acclimation in Tetrahymena. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2011, 87, 450–462. [Google Scholar] [CrossRef]
- Murinova, S.; Dercova, K. Response mechanisms of bacterial degraders to environmental contaminants on the level of cell walls and cytoplasmic membrane. Int. J. Microbiol. 2014, 2014, 873081. [Google Scholar] [CrossRef]
- Schuler, A.J.; Jenkins, J.L. Bacterial physiology of polyhydroxyalkanoates. In Advances in Microbial Physiology; Academic Press: Cambridge, MA, USA, 2001. [Google Scholar]
- Hewelt-Belka, W.; Kot-Wasik, A. Analytical Strategies and Applications in Lipidomics. In Handbook of Bioanalytics; Buszewski, B., Baranowska, I., Eds.; Springer International Publishing: Berlin/Heidelberg, Germany, 2022; pp. 1–26. [Google Scholar]
- Zullig, T.; Trotzmuller, M.; Kofeler, H.C. Lipidomics from sample preparation to data analysis: A primer. Anal. Bioanal. Chem. 2020, 412, 2191–2209. [Google Scholar] [CrossRef]
- Xie, Y.; Wu, Z.; Qin, Z.; Wu, B.; Lv, X.; Wei, F.; Chen, H. Methods of Lipidomic Analysis: Extraction, Derivatization, Separation, and Identification of Lipids. In Cancer Metabolomics: Methods and Applications; Hu, S., Ed.; Springer International Publishing: Berlin/Heidelberg, Germany, 2021; pp. 173–187. [Google Scholar]
- Jimenez-Diaz, L.; Caballero, A.; Segura, A. Pathways for the Degradation of Fatty Acids in Bacteria. In Aerobic Utilization of Hydrocarbons, Oils and Lipids; Rojo, F., Ed.; Springer International Publishing: Berlin/Heidelberg, Germany, 2017; pp. 1–23. [Google Scholar]
- Saha, R.; Sproer, C.; Beck, B.; Bagley, S. Pseudomonas oleovorans subsp. lubricantis subsp. nov., and reclassification of Pseudomonas pseudoalcaligenes ATCC 17440T as later synonym of Pseudomonas oleovorans ATCC 8062 T. Curr. Microbiol. 2010, 60, 294–300. [Google Scholar] [CrossRef]
- Folch, J.; Lees, M.; Sloane Stanley, G.H. A simple method for the isolation and purification of total lipides from animal tissues. J. Biol. Chem. 1957, 226, 497–509. [Google Scholar] [CrossRef]
- Nygren, H.; Seppanen-Laakso, T.; Castillo, S.; Hyotylainen, T.; Oresic, M. Liquid chromatography-mass spectrometry (LC-MS)-based lipidomics for studies of body fluids and tissues. Methods Mol. Biol. 2011, 708, 247–257. [Google Scholar]
- Pluskal, T.; Castillo, S.; Villar-Briones, A.; Oresic, M. MZmine 2: Modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinform. 2010, 11, 395. [Google Scholar] [CrossRef]
- Chong, J.; Soufan, O.; Li, C.; Caraus, I.; Li, S.; Bourque, G.; Wishart, D.S.; Xia, J. MetaboAnalyst 4.0: Towards more transparent and integrative metabolomics analysis. Nucleic Acids Res. 2018, 46, W486–W494. [Google Scholar] [CrossRef]
- Pang, Z.; Chong, J.; Zhou, G.; de Lima Morais, D.A.; Chang, L.; Barrette, M.; Gauthier, C.; Jacques, P.E.; Li, S.; Xia, J. MetaboAnalyst 5.0: Narrowing the gap between raw spectra and functional insights. Nucleic Acids Res. 2021, 49, W388–W396. [Google Scholar] [CrossRef]
- Li, S.; Park, Y.; Duraisingham, S.; Strobel, F.H.; Khan, N.; Soltow, Q.A.; Jones, D.P.; Pulendran, B. Predicting network activity from high throughput metabolomics. PLoS Comput. Biol. 2013, 9, e1003123. [Google Scholar] [CrossRef]
- Bitzenhofer Nora, L.; Kruse, L.; Thies, S.; Wynands, B.; Lechtenberg, T.; Rönitz, J.; Kozaeva, E.; Wirth Nicolas, T.; Eberlein, C.; Jaeger, K.-E.; et al. Towards robust Pseudomonas cell factories to harbour novel biosynthetic pathways. Essays Biochem. 2021, 65, 319–336. [Google Scholar]
- Han, M.L.; Zhu, Y.; Creek, D.J.; Lin, Y.W.; Gutu, A.D.; Hertzog, P.; Purcell, T.; Shen, H.H.; Moskowitz, S.M.; Velkov, T.; et al. Comparative Metabolomics and Transcriptomics Reveal Multiple Pathways Associated with Polymyxin Killing in Pseudomonas aeruginosa. mSystems 2019, 4, 10–1128. [Google Scholar] [CrossRef]
- Su, Y.B.; Tang, X.K.; Zhu, L.P.; Yang, K.X.; Pan, L.; Li, H.; Chen, Z.G. Enhanced Biosynthesis of Fatty Acids Contributes to Ciprofloxacin Resistance in Pseudomonas aeruginosa. Front. Microbiol. 2022, 13, 845173. [Google Scholar] [CrossRef]
- Hoppel, C. The role of carnitine in normal and altered fatty acid metabolism. Am. J. Kidney Dis. 2003, 41 (Suppl. 4), S4–S12. [Google Scholar] [CrossRef]
- Vaz, F.M.; Wanders, R.J. Carnitine biosynthesis in mammals. Biochem. J. 2002, 361 Pt 3, 417–429. [Google Scholar] [CrossRef]
- Maillard, J.Y. Antimicrobial biocides in the healthcare environment: Efficacy, usage, policies, and perceived problems. Ther. Clin. Risk Manag. 2005, 1, 307–320. [Google Scholar]
- Sáez, L.P.; Rodríguez-Caballero, G.; Olaya-Abril, A.; Cabello, P.; Moreno-Vivián, C.; Roldán, M.D.; Luque-Almagro, V.M. Genomic Insights into Cyanide Biodegradation in the Pseudomonas Genus. Int. J. Mol. Sci. 2024, 25, 4456. [Google Scholar] [CrossRef]
- Longo, N.; Frigeni, M.; Pasquali, M. Carnitine transport and fatty acid oxidation. Biochim. Biophys. Acta 2016, 1863, 2422–2435. [Google Scholar] [CrossRef]
- Meadows, J.A.; Wargo, M.J. Carnitine in bacterial physiology and metabolism. Microbiology 2015, 161, 1161–1174. [Google Scholar] [CrossRef]
- Sohlenkamp, C.; Geiger, O. Bacterial membrane lipids: Diversity in structures and pathways. FEMS Microbiol. Rev. 2016, 40, 133–159. [Google Scholar] [CrossRef]
- Vance, J.E.; Tasseva, G. Formation and function of phosphatidylserine and phosphatidylethanolamine in mammalian cells. Biochim. Biophys. Acta 2013, 1831, 543–554. [Google Scholar] [CrossRef]
- Sohlenkamp, C.; Lopez-Lara, I.M.; Geiger, O. Biosynthesis of phosphatidylcholine in bacteria. Prog. Lipid Res. 2003, 42, 115–162. [Google Scholar] [CrossRef]
- Rahman, S.; Kraljevic Pavelic, S.; Markova-Car, E. Circadian (De)regulation in Head and Neck Squamous Cell Carcinoma. Int. J. Mol. Sci. 2019, 20, 2662. [Google Scholar] [CrossRef]
- Li, A.; Schertzer, J.W.; Yong, X. Molecular dynamics modeling of Pseudomonas aeruginosa outer membranes. Phys. Chem. Chem. Phys. 2018, 20, 23635–23648. [Google Scholar] [CrossRef]
- Wilderman, P.J.; Vasil, A.I.; Martin, W.E.; Murphy, R.C.; Vasil, M.L. Pseudomonas aeruginosa synthesizes phosphatidylcholine by use of the phosphatidylcholine synthase pathway. J. Bacteriol. 2002, 184, 4792–4799. [Google Scholar] [CrossRef]
- Davies, D.G.; Marques, C.N. A fatty acid messenger is responsible for inducing dispersion in microbial biofilms. J. Bacteriol. 2009, 191, 1393–1403. [Google Scholar] [CrossRef]
- de Carvalho, C.; Caramujo, M.J. The Various Roles of Fatty Acids. Molecules 2018, 23, 2583. [Google Scholar] [CrossRef]
- Tang, J.; Li, Y.; Zhang, L.; Mu, J.; Jiang, Y.; Fu, H.; Zhang, Y.; Cui, H.; Yu, X.; Ye, Z. Biosynthetic Pathways and Functions of Indole-3-Acetic Acid in Microorganisms. Microorganisms 2023, 11, 2077. [Google Scholar] [CrossRef]
- Spaepen, S.; Vanderleyden, J. Auxin and plant-microbe interactions. Cold Spring Harb. Perspect. Biol. 2011, 3, a001438. [Google Scholar] [CrossRef]
- Tsouka, S.; Masoodi, M. Metabolic Pathway Analysis: Advantages and Pitfalls for the Functional Interpretation of Metabolomics and Lipidomics Data. Biomolecules 2023, 13, 244. [Google Scholar] [CrossRef]
- Yuan, Y.; Leeds, J.A.; Meredith, T.C. Pseudomonas aeruginosa directly shunts β-oxidation degradation intermediates into de novo fatty acid biosynthesis. J. Bacteriol. 2012, 194, 5185. [Google Scholar] [CrossRef]
- Bernal, P.; Muñoz-Rojas, J.; Hurtado, A.; Ramos, J.L.; Segura, A. A Pseudomonas putida cardiolipin synthesis mutant exhibits increased sensitivity to drugs related to transport functionality. Environ. Microbiol. 2007, 9, 1135–1145. [Google Scholar] [CrossRef]
- Ramos, J.-L.; Sol Cuenca, M.; Molina-Santiago, C.; Segura, A.; Duque, E.; Gómez-García, M.R.; Udaondo, Z.; Roca, A. Mechanisms of solvent resistance mediated by interplay of cellular factors in Pseudomonas putida. FEMS Microbiol. Rev. 2015, 39, 555–566. [Google Scholar] [CrossRef]
- Pallotti, F.; Bergamini, C.; Lamperti, C.; Fato, R. The Roles of Coenzyme Q in Disease: Direct and Indirect Involvement in Cellular Functions. Int. J. Mol. Sci. 2021, 23, 128. [Google Scholar] [CrossRef]
- Rudney, H. The biosynthesis of terpenoid quinones. Biochem. J. 1969, 113, 21P–23P. [Google Scholar] [CrossRef]
- Subramanian, M.; Srinivasan, T.; Sudarsanam, D. Examining the Gm18 and m(1)G Modification Positions in tRNA Sequences. Genom. Inform. 2014, 12, 71–75. [Google Scholar] [CrossRef]
- Nadal-Jimenez, P.; Koch, G.; Reis, C.R.; Muntendam, R.; Raj, H.; Jeronimus-Stratingh, C.M.; Cool, R.H.; Quax, W.J. PvdP is a tyrosinase that drives maturation of the pyoverdine chromophore in Pseudomonas aeruginosa. J. Bacteriol. 2014, 196, 2681–2690. [Google Scholar] [CrossRef]
- Carmona, M.; Zamarro, M.T.; Blazquez, B.; Durante-Rodriguez, G.; Juarez, J.F.; Valderrama, J.A.; Barragan, M.J.; Garcia, J.L.; Diaz, E. Anaerobic catabolism of aromatic compounds: A genetic and genomic view. Microbiol. Mol. Biol. Rev. 2009, 73, 71–133. [Google Scholar] [CrossRef]
- Nogales, J.; García, J.L.; Díaz, E. Degradation of Aromatic Compounds in Pseudomonas: A Systems Biology View. In Aerobic Utilization of Hydrocarbons, Oils and Lipids; Rojo, F., Ed.; Springer International Publishing: Berlin/Heidelberg, Germany, 2017; pp. 1–49. [Google Scholar]
- Valderrama, J.A.; Durante-Rodriguez, G.; Blazquez, B.; Garcia, J.L.; Carmona, M.; Diaz, E. Bacterial degradation of benzoate: Cross-regulation between aerobic and anaerobic pathways. J. Biol. Chem. 2012, 287, 10494–10508. [Google Scholar] [CrossRef]
- Zaar, A.; Eisenreich, W.; Bacher, A.; Fuchs, G. A novel pathway of aerobic benzoate catabolism in the bacteria Azoarcus evansii and Bacillus stearothermophilus. J. Biol. Chem. 2001, 276, 24997–25004. [Google Scholar] [CrossRef]
| Conditions | Polar/Semipolar Compounds | Lipidomics |
|---|---|---|
| Injection volume | 10 µL | 1 µL |
| Column | C18 precolumn (Waters Corporation, Wexford, Ireland) and an inline filter, pore size 0.2 µm (Waters Corporation, Wexford, Ireland). + ACQUITY UPLC® BEH C18 column (2.1 mm × 100 mm, particle size 1.7 µm) by Waters (Milford, MA, USA) | C18 precolumn (Waters Corporation, Wexford, Ireland) and an inline filter, pore size 0.2 µm (Waters Corporation, Wexford, Ireland). + ACQUITY UPLC® BEH C18 column (2.1 mm × 100 mm, particle size 1.7 µm) by Waters (Milford, MA, USA) |
| Mobile phases | A H2O:MeOH (v/v 70:30) with 2 mM ammonium acetate B MeOH with containing 2 mM ammonium acetate | A 10 mM ammonium acetate and 0.1% Formic Acid in H2O B Acetonitrile:Isopropanol (v/v 1:1) with 0.1% Formic Acid and 10 mM ammonium acetate |
| Gradient |
|
|
| Flow rate | 0.4 mL min−1 | 0.4 mL min−1 |
| MS conditions | Dual ESI ionization source with capillary voltage 4.5 kV, nozzle voltage 1500 V, N2 pressure in the nebulized was 21 psi and the N2 flow rate and temperature as sheath gas was 11 L min−1 and 379 °C, respectively. The drying gas flow was set to 10 L min−1 and the temperature to 150 °C. m/z range 100–1700 in negative ion mode. | Dual ESI ionization source with capillary voltage 3.64 kV, nozzle voltage 1500 V, N2 pressure in the nebulized was 21 psi and the N2 flow rate and temperature as sheath gas was 11 L min−1 and 379 °C, respectively. The drying gas flow was set to 10 L min−1 and temperature to 193 °C. m/z range 100–1700 in positive ion mode |
| Pathway Analysis for 1045 | Pathway Total | Hits. Total | Hits. Sig | P. Fisher | P. EASE | P. Gamma |
|---|---|---|---|---|---|---|
| Valine, leucine, and isoleucine degradation | 31 | 5 | 5 | 0.0239 | 0.1531 | 0.0052 |
| Valine, leucine, and isoleucine biosynthesis | 22 | 4 | 4 | 0.0512 | 0.2724 | 0.0071 |
| Arachidonic acid metabolism | 4 | 4 | 4 | 0.0512 | 0.2724 | 0.0071 |
| beta-Alanine metabolism | 13 | 3 | 3 | 0.1088 | 0.4613 | 0.0121 |
| Peptidoglycan biosynthesis | 17 | 3 | 3 | 0.1088 | 0.4613 | 0.0121 |
| Butanoate metabolism | 22 | 3 | 3 | 0.1088 | 0.4613 | 0.0121 |
| Pantothenate and CoA biosynthesis | 20 | 3 | 3 | 0.1088 | 0.4613 | 0.0121 |
| Ubiquinone and other terpenoid-quinone biosynthesis | 9 | 4 | 3 | 0.2816 | 0.6503 | 0.0220 |
| Porphyrin and chlorophyll metabolism | 56 | 11 | 6 | 0.4463 | 0.6711 | 0.0237 |
| Pathway Analysis for P4A | Pathway Total | Hits. Total | Hits. Sig | P. Fisher | P. EASE | P. Gamma |
|---|---|---|---|---|---|---|
| Arachidonic acid metabolism | 4 | 4 | 4 | 0.1341 | 0.4786 | 0.0158 |
| Tyrosine metabolism | 25 | 9 | 7 | 0.2417 | 0.4876 | 0.0162 |
| Aminobenzoate degradation | 11 | 3 | 3 | 0.2230 | 0.6527 | 0.0261 |
| Folate biosynthesis | 34 | 7 | 5 | 0.4371 | 0.7191 | 0.0323 |
| Ubiquinone and other terpenoid-quinone biosynthesis | 9 | 4 | 3 | 0.4898 | 0.8267 | 0.0485 |
| Aminoacyl–tRNA biosynthesis | 21 | 6 | 4 | 0.5632 | 0.8289 | 0.0489 |
| Porphyrin and chlorophyll metabolism | 56 | 11 | 6 | 0.7807 | 0.9118 | 0.0738 |
| Valine, leucine, and isoleucine degradation | 31 | 5 | 3 | 0.7004 | 0.9184 | 0.0769 |
| Biotin metabolism | 6 | 2 | 2 | 0.3693 | 0.8436 | 0.0521 |
| Peptidoglycan biosynthesis | 17 | 3 | 2 | 0.6619 | 0.9391 | 0.0885 |
| Butanoate metabolism | 22 | 3 | 2 | 0.6619 | 0.9391 | 0.0885 |
| Pantothenate and CoA biosynthesis | 20 | 3 | 2 | 0.6619 | 0.9391 | 0.0885 |
| Valine, leucine, and isoleucine biosynthesis | 22 | 4 | 2 | 0.8340 | 0.9765 | 0.1271 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ali, M.L.; Ferrieres, L.; Jass, J.; Hyötyläinen, T. Metabolic Changes in Pseudomonas oleovorans Isolated from Contaminated Construction Material Exposed to Varied Biocide Treatments. Metabolites 2024, 14, 326. https://doi.org/10.3390/metabo14060326
Ali ML, Ferrieres L, Jass J, Hyötyläinen T. Metabolic Changes in Pseudomonas oleovorans Isolated from Contaminated Construction Material Exposed to Varied Biocide Treatments. Metabolites. 2024; 14(6):326. https://doi.org/10.3390/metabo14060326
Chicago/Turabian StyleAli, Muatasem Latif, Lionel Ferrieres, Jana Jass, and Tuulia Hyötyläinen. 2024. "Metabolic Changes in Pseudomonas oleovorans Isolated from Contaminated Construction Material Exposed to Varied Biocide Treatments" Metabolites 14, no. 6: 326. https://doi.org/10.3390/metabo14060326
APA StyleAli, M. L., Ferrieres, L., Jass, J., & Hyötyläinen, T. (2024). Metabolic Changes in Pseudomonas oleovorans Isolated from Contaminated Construction Material Exposed to Varied Biocide Treatments. Metabolites, 14(6), 326. https://doi.org/10.3390/metabo14060326







