1H NMR Serum Metabolomic Change of Trimethylamine N-oxide (TMAO) Is Associated with Alcoholic Liver Disease Progression
Abstract
1. Introduction
2. Materials and Methods
2.1. Patient Information and Data Collection
2.2. 1H Nuclear Magnetic Resonance (1H NMR)
2.3. 1H NMR Data Processing and Multivariate Statistical Analysis
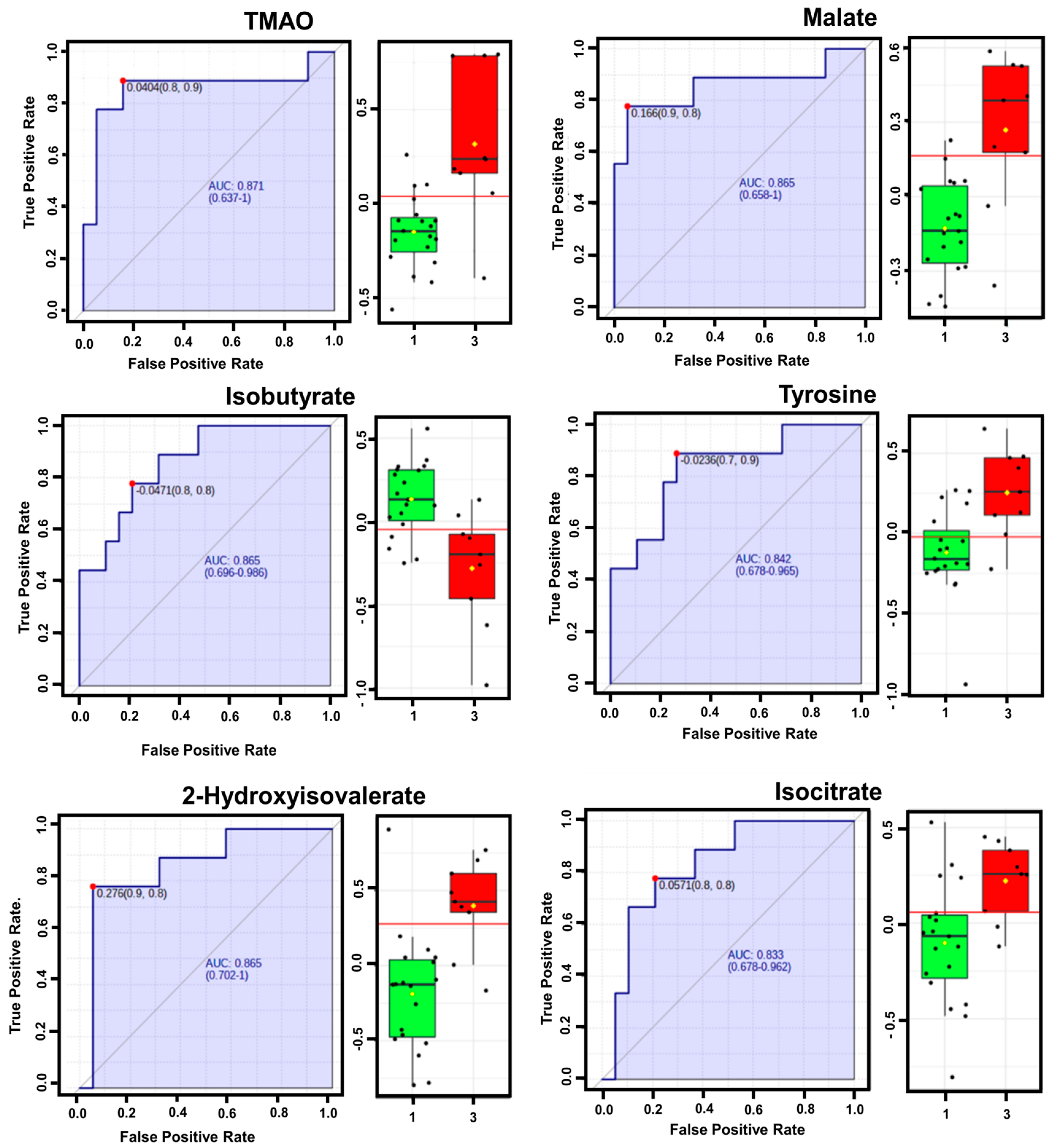
2.4. Receiver Operating Characteristic (ROC) Curve and Heatmap Analyses
3. Results
3.1. Clinical Characteristics of the Study Population
| CTP-A (n = 19) | CTP-B (n = 10) | CTP-C (n = 9) | p-Value | |
|---|---|---|---|---|
| Age (years) | 55.5 (45–73) | 51 (34–66) | 51 (41–68) | 0.36500 |
| Gender (Male) | 17 (89.5) | 9 (80.0) | 6 (66.7) | 0.34363 |
| Hypertension | 7 (36.8) | 3 (30.0) | 2 (22.2) | 0.73352 |
| Diabetes Mellitus | 4 (21.1) | 1 (9.1) | 2 (5.3) | 0.72398 |
| Hyperlipidemia | 3 (15.8) | 1 (10.0) | 1 (11.1) | 0.88895 |
| BMI (kg/m2) | 23.6 (16.8–29.7) | 21.9 (16.5–32) | 23.1 (18.7–30.1) | 0.64900 |
| Stiffness (kPa) | 11.0 (4.2–75) | 48.4 (5.6–75) | 44.6 (17.6–67.8) | 0.06324 |
| Total bilirubin (mg/dL) | 1.0 (0.4–4.0) | 2.95 (0.8–20.6) | 5.4 (1.2–38.1) | 0.00017 |
| Albumin (mg/dL) | 3.6 (3.1–4.6) | 2.95 (2.0–4.7) | 2.5 (1.8–3.4) | 0.00004 |
| AST (IU/dL) | 55 (14–3387) | 76 (18–789) | 89 (31–331) | 0.57530 |
| ALT (IU/dL) | 46 (8–2835) | 46.5 (10–2080) | 26 (11–66) | 0.62040 |
| GGT (mg/dL) | 218 (22–2760) | 138 (22–884) | 146 (16–477) | 0.57350 |
| Platelet (103/μL) | 182 (54–372) | 110 (55–247) | 62 (20–139) | 0.00203 |
| INR | 1.05 (0.89–1.55) | 1.37 (0.98–1.91) | 1.55 (1.31–2.26) | 0.00005 |
| Creatinine | 0.92 (0.16–4.67) | 0.72 (0.32–6.22) | 0.77 (0.39–4.41) | 0.55880 |
3.2. Identification of Serum Metabolites in ALD Patients by 1H-NMR
3.3. Multivariate Analyses of Serum Metabolite Data from ALD Patients According to CTP Classes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Asrani, S.K.; Devarbhavi, H.; Eaton, J.; Kamath, P.S. Burden of Liver Diseases in the World. J. Hepatol. 2019, 70, 151–171. [Google Scholar] [CrossRef] [PubMed]
- Seitz, H.K.; Bataller, R.; Cortez-Pinto, H.; Gao, B.; Gual, A.; Lackner, C.; Mathurin, P.; Mueller, S.; Szabo, G.; Tsukamoto, H. Alcoholic Liver Disease. Nat. Rev. Dis. Primers 2018, 4, 1–22. [Google Scholar] [CrossRef] [PubMed]
- Pan, H.-C.; Jenq, C.-C.; Lee, W.-C.; Tsai, M.-H.; Fan, P.-C.; Chang, C.-H.; Chang, M.-Y.; Tian, Y.-C.; Hung, C.-C.; Fang, J.-T.; et al. Scoring Systems for Predicting Mortality after Liver Transplantation. PLoS ONE 2014, 9, e107138. [Google Scholar] [CrossRef] [PubMed]
- Shanmugham, M.; Bellanger, S.; Leo, C.H. Gut-Derived Metabolite, Trimethylamine-N-Oxide (TMAO) in Cardio-Metabolic Diseases: Detection, Mechanism, and Potential Therapeutics. Pharmaceuticals 2023, 16, 504. [Google Scholar] [CrossRef] [PubMed]
- Sharon, G.; Garg, N.; Debelius, J.; Knight, R.; Dorrestein, P.C.; Mazmanian, S.K. Specialized Metabolites from the Microbiome in Health and Disease. Cell Metab. 2014, 20, 719–730. [Google Scholar] [CrossRef] [PubMed]
- Janeiro, M.H.; Ramírez, M.J.; Milagro, F.I.; Martínez, J.A.; Solas, M. Implication of Trimethylamine N-Oxide (TMAO) in Disease: Potential Biomarker or New Therapeutic Target. Nutrients 2018, 10, 1398. [Google Scholar] [CrossRef] [PubMed]
- Francque, S.M.; Van Der Graaff, D.; Kwanten, W.J. Non-Alcoholic Fatty Liver Disease and Cardiovascular Risk: Pathophysiological Mechanisms and Implications. J. Hepatol. 2016, 65, 425–443. [Google Scholar] [CrossRef]
- Fan, Y.; Pedersen, O. Gut Microbiota in Human Metabolic Health, and Disease. Nat. Rev. Microbiol. 2021, 19, 55–71. [Google Scholar] [CrossRef]
- Theofilis, P.; Vordoni, A.; Kalaitzidis, R.G. Trimethylamine N-Oxide Levels in Non-Alcoholic Fatty Liver Disease: A Systematic Review and Meta-Analysis. Metabolites 2022, 12, 1243. [Google Scholar] [CrossRef]
- Canfora, E.E.; Meex, R.C.R.; Venema, K.; Blaak, E.E. Gut Microbial Metabolites in Obesity, NAFLD and T2DM. Nat. Rev. Endocrinol. 2019, 15, 261–273. [Google Scholar] [CrossRef]
- Bajaj, J.S. Alcohol, Liver Disease, and the Gut Microbiota. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 235–246. [Google Scholar] [CrossRef] [PubMed]
- Crabb, D.W.; Bataller, R.; Chalasani, N.P.; Kamath, P.S.; Lucey, M.; Mathurin, P.; McClain, C.; McCullough, A.; Mitchell, M.C.; Morgan, T.R.; et al. Standard Definitions and Common Data Elements for Clinical Trials in Patients with Alcoholic Hepatitis: Recommendation from the NIAAA Alcoholic Hepatitis Consortia. Gastroenterology 2016, 150, 785–790. [Google Scholar] [CrossRef] [PubMed]
- Albers, I.; Hartmann, H.; Bircher, J.; Creutzfeldt, W. Superiority of the Child-Pugh Classification to Quantitative Liver Function Tests for Assessing Prognosis of Liver Cirrhosis. Scand. J. Gastroenterol. 1989, 24, 269–276. [Google Scholar] [CrossRef] [PubMed]
- Infante-Rivard, C.; Esnaola, S.; Villeneuve, J.-P. Clinical and statistical validity of conventional prognostic factors in predicting short-term survival among cirrhotics. Hepatology 1987, 7, 660–664. [Google Scholar] [CrossRef] [PubMed]
- Llorente, C.; Schnabl, B. The Gut Microbiota and Liver Disease. Cell. Mol. Gastroenterol. Hepatol. 2015, 1, 275–284. [Google Scholar] [CrossRef] [PubMed]
- Dubinkina, V.B.; Tyakht, A.V.; Odintsova, V.Y.; Yarygin, K.S.; Kovarsky, B.A.; Pavlenko, A.V.; Ischenko, D.S.; Popenko, A.S.; Alexeev, D.G.; Taraskina, A.Y.; et al. Links of Gut Microbiota Composition with Alcohol Dependence Syndrome and Alcoholic Liver Disease. Microbiome 2017, 5, 141. [Google Scholar] [CrossRef] [PubMed]
- Schwenger, K.J.; Clermont-Dejean, N.; Allard, J.P. The Role of the Gut Microbiome in Chronic Liver Disease: Clinical Evidence Revised. JHEP Rep. 2019, 1, 214–226. [Google Scholar] [CrossRef]
- León-Mimila, P.; Villamil-Ramírez, H.; Li, X.S.; Shih, D.M.; Hui, S.T.; Ocampo-Medina, E.; López-Contreras, B.; Morán-Ramos, S.; Olivares-Arevalo, M.; Grandini-Rosales, P.; et al. Trimethylamine N-Oxide Levels Are Associated with NASH in Obese Subjects with Type 2 Diabetes. Diabetes Metab. 2021, 47, 101183. [Google Scholar] [CrossRef]
- Chen, Y.; Liu, Y.; Zhou, R.; Chen, X.; Wang, C.; Tan, X.; Wang, L.; Zheng, R.; Zhang, H.; Ling, W.; et al. Associations of Gut-Flora-Dependent Metabolite Trimethylamine-N-Oxide, Betaine and Choline with Non-Alcoholic Fatty Liver Disease in Adults. Sci. Rep. 2016, 6, 19076. [Google Scholar] [CrossRef]
- Dumas, M.-E.; Barton, R.H.; Toye, A.; Cloarec, O.; Blancher, C.; Rothwell, A.; Fearnside, J.; Tatoud, R.; Blanc, V.; Lindon, J.C.; et al. Metabolic Profiling Reveals a Contribution of Gut Microbiota to Fatty Liver Phenotype in Insulin-Resistant Mice. Proc. Natl. Acad. Sci. USA 2006, 103, 12511–12516. [Google Scholar] [CrossRef]
- Zhou, D.; Fan, J.-G. Microbial Metabolites in Non-Alcoholic Fatty Liver Disease. World J. Gastroenterol. 2019, 25, 2019–2028. [Google Scholar] [CrossRef] [PubMed]
- Chambers, E.S.; Viardot, A.; Psichas, A.; Morrison, D.J.; Murphy, K.G.; Zac-Varghese, S.E.K.; MacDougall, K.; Preston, T.; Tedford, C.; Finlayson, G.S.; et al. Effects of Targeted Delivery of Propionate to the Human Colon on Appetite Regulation, Body Weight Maintenance and Adiposity in Overweight Adults. Gut 2015, 64, 1744–1754. [Google Scholar] [CrossRef] [PubMed]
- Zhou, D.; Pan, Q.; Liu, X.-L.; Yang, R.-X.; Chen, Y.-W.; Liu, C.; Fan, J.-G. Clostridium Butyricum B1 Alleviates High-Fat Diet-Induced Steatohepatitis in Mice via Enterohepatic Immunoregulation. J. Gastroenterol. Hepatol. 2017, 32, 1640–1648. [Google Scholar] [CrossRef] [PubMed]
- Singh, V.; Yeoh, B.S.; Chassaing, B.; Xiao, X.; Saha, P.; Aguilera Olvera, R.; Lapek, J.D.; Zhang, L.; Wang, W.-B.; Hao, S.; et al. Dysregulated Microbial Fermentation of Soluble Fiber Induces Cholestatic Liver Cancer. Cell 2018, 175, 679–694. [Google Scholar] [CrossRef] [PubMed]
- Corbin, K.D.; Zeisel, S.H. Choline Metabolism Provides Novel Insights into Nonalcoholic Fatty Liver Disease, and Its Progression. Curr. Opin. Gastroenterol. 2012, 28, 159–165. [Google Scholar] [CrossRef]
- Ueland, P.M. Choline and Betaine in Health and Disease. J. Inherit. Metab. Dis. 2011, 34, 3–15. [Google Scholar] [CrossRef]
- Arias, N.; Arboleya, S.; Allison, J.; Kaliszewska, A.; Higarza, S.G.; Gueimonde, M.; Arias, J.L. The Relationship between Choline Bioavailability from Diet, Intestinal Microbiota Composition, and Its Modulation of Human Diseases. Nutrients 2020, 12, 2340. [Google Scholar] [CrossRef]
- Michálková, L.; Horník, Š.; Sýkora, J.; Habartová, L.; Setnička, V. Diagnosis of Pancreatic Cancer via 1 H NMR Metabolomics of Human Plasma. Analyst 2018, 143, 5974–5978. [Google Scholar] [CrossRef]
- Holeček, M. Branched-Chain Amino Acids in Health and Disease: Metabolism, Alterations in Blood Plasma, and as Supplements. Nutr. Metab. 2018, 15, 33. [Google Scholar] [CrossRef]


| HMDB Card | Metabolites | Chemical Shifts (Multiplicities) (ppm) | CTP-A Mean ± SD | CTA-B Mean ± SD | CTA-C Mean ± SD |
|---|---|---|---|---|---|
| HMDB0000042 | Acetate | 1.9 (s) | 0.056 ± 0.019 | 0.051 ± 0.028 | 0.057 ± 0.026 |
| HMDB0001310 | Alanine | 1.46 (q) | 0.131 ± 0.033 | 0.137 ± 0.031 | 0.109 ± 0.025 |
| HMDB0000517 | Arginine | 1.86 (m) | 0.085 ± 0.027 | 0.070 ± 0.027 | 0.111 ± 0.057 |
| HMDB0000168 | Asparagine | 2.86–2.94 (t) | 0.043 ± 0.013 | 0.039 ± 0.013 | 0.041 ± 0.012 |
| HMDB0000191 | Aspartate | 2.78–2.82 (dd) | 0.041 ± 0.011 | 0.035 ± 0.010 | 0.038 ± 0.012 |
| HMDB0000043 | Betaine | 3.26 (s), 3.90 (s) | 0.527 ± 0.117 | 0.607 ± 0.174 | 0.714 ± 0.259 |
| HMDB0000062 | Carnitine | 2.42–2.46 (m), 3.22 (s) | 0.524 ± 0.092 | 0.579 ± 0.161 | 0.603 ± 0.227 |
| HMDB0000619 | Cholate | 0.70 (s) | 0.017 ± 0.007 | 0.015 ± 0.004 | 0.019 ± 0.007 |
| HMDB0000097 | Choline | 3.18 (s), 3.50 (dd) | 0.317 ± 0.089 | 0.355 ± 0.121 | 0.380 ± 0.169 |
| HMDB0000134 | Fumarate | 6.50 (s) | 0.012 ± 0.005 | 0.013 ± 0.003 | 0.013 ± 0.005 |
| HMDB0000122 | Glucose | 3.38 (dd), 3.50 (dd), 3.70–3.72 (m), 3.82–3.86 (m) | 2.024 ± 0.627 | 2.245 ± 0.781 | 2.501 ± 1.179 |
| HMDB0003339 | Glutamate | 2.02–2.06 (m), 2.34 (m) | 0.100 ± 0.033 | 0.087 ± 0.027 | 0.095 ± 0.047 |
| HMDB0000641 | Glutamine | 2.14–2.18 (m), 2.42 (m) | 0.188 ± 0.037 | 0.191 ± 0.048 | 0.199 ± 0.058 |
| HMDB0000131 | Glycerol | 3.54 (dd), 3.66 (dd), 3.78 (m) | 15.206 ± 2.789 | 14.336 ± 2.420 | 14.926 ± 2.274 |
| HMDB0000123 | Glycine | 3.54 (s) | 5.149 ± 0.690 | 4.906 ± 0.664 | 5.067 ± 0.487 |
| HMDB0000138 | Glycocholate | 0.74 (s), 0.90(s) | 0.072 ± 0.019 | 0.063 ± 0.016 | 0.075 ± 0.016 |
| HMDB0000128 | Guanidoacetate | 3.78 (s) | 4.355 ± 1.105 | 3.960 ± 0.748 | 4.179 ± 0.663 |
| HMDB0000177 | Histidine | 7.22 (s), 8.3 (s) | 0.031 ± 0.009 | 0.030 ± 0.005 | 0.029 ± 0.007 |
| HMDB0000011 | 3-Hydroxybutyrate | 1.18–1.22 (d), 2.3 (dd) | 0.132 ± 0.100 | 0.079 ± 0.035 | 0.183 ± 0.198 |
| HMDB0000407 | 2-Hydroxyisovalerate | 0.82 (d) | 0.020 ± 0.013 | 0.030 ± 0.017 a | 0.036 ± 0.012 a |
| HMDB0011631 | 3-Hydroxykynurenine | 7.42–7.48 (dd) | 0.030 ± 0.007 | 0.028 ± 0.008 | 0.037 ± 0.012 |
| HMDB0001873 | Isobutyrate | 1.02–1.04 (d) | 0.085 ± 0.021 | 0.061 ± 0.015 a | 0.066 ± 0.018 a |
| HMDB0000193 | Isocitrate | 2.50–2.54 (dd), 2.96–3.06 (m) | 0.084 ± 0.012 | 0.088 ± 0.028 | 0.110 ± 0.016 |
| HMDB0000172 | Isoleucine | 0.90–0.93 (t), 1.26 (m) | 0.074 ± 0.022 | 0.062 ± 0.015 | 0.074 ± 0.016 |
| HMDB0000190 | Lactate | 1.30 (d), 4.10 (q) | 0.565 ± 0.129 | 0.710 ± 0.229 | 0.622 ± 0.204 |
| HMDB0000687 | Leucine | 0.94 (m), 1.66–170 (m) | 0.126 ± 0.042 | 0.103 ± 0.031 | 0.108 ± 0.038 |
| HMDB0000182 | Lysine | 1.7 (m), 1.86–1.94 (m), 3.02 (t) | 0.211 ± 0.066 | 0.191 ± 0.064 | 0.254 ± 0.067 |
| HMDB0000156 | Malate | 2.66 (dd) | 0.038 ± 0.008 | 0.041 ± 0.009 a | 0.051 ± 0.008 a |
| HMDB0001875 | Methanol | 3.34 (s) | 0.062 ± 0.018 | 0.058 ± 0.022 | 0.064 ± 0.015 |
| HMDB0000001 | 1-Methylhistidine | 3.10 (dd), 7.78 (s) | 0.045 ± 0.014 | 0.053 ± 0.015 | 0.056 ± 0.007 |
| HMDB0000208 | 2-Oxoglutatate | 2.43–2.46 (t), 2.98–3.0 (t) | 0.082 ± 0.019 | 0.081 ± 0.017 | 0.078 ± 0.019 |
| HMDB0000786 | Oxypurinol | 8.26(s) | 0.016 ± 0.006 | 0.014 ± 0.004 | 0.016 ± 0.003 |
| HMDB0000159 | Phenylalanine | 7.3 (m), 7.42 (m) | 0.054 ± 0.010 | 0.055 ± 0.013 | 0.063 ± 0.014 |
| HMDB0000162 | Proline | 2.20 (m) | 0.103 ± 0.030 | 0.089 ± 0.021 | 0.118 ± 0.036 |
| HMDB0000243 | Pyruvate | 2.38 (s) | 0.042 ± 0.015 | 0.034 ± 0.008 | 0.049 ± 0.031 |
| HMDB0000187 | Serine | 3.94–3.98 (dd) | 0.078 ± 0.037 | 0.081 ± 0.027 | 0.083 ± 0.039 |
| HMDB0000254 | Succinate | 2.39–2.42 (t) | 0.077 ± 0.013 | 0.076 ± 0.020 | 0.081 ± 0.021 |
| HMDB0000251 | Taurine | 3.26 (t), 3.42 (t) | 0.582 ± 0.133 | 0.682 ± 0.207 | 0.822 ± 0.352 |
| HMDB0000906 | TMA | 2.90 (s) | 0.025 ± 0.007 | 0.022 ± 0.009 | 0.025 ± 0.007 |
| HMDB0000925 | TMAO | 3.26 (s) | 0.201 ± 0.035 | 0.246 ± 0.064 a | 0.316 ± 0.095 a |
| HMDB0000158 | Tyrosine | 6.90 (ddd), 7.18 (ddd) | 0.053 ± 0.009 | 0.061 ± 0.024 a | 0.073 ± 0.016 a |
| HMDB0000883 | Valine | 0.98 (m), 0.99–1.02 (dd), 2.26 (m) | 0.235 ± 0.076 | 0.163 ± 0.032 a | 0.190 ± 0.093 a |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Oh, J.; Kim, J.; Lee, S.; Park, G.; Baritugo, K.-A.G.; Han, K.J.; Lee, S.; Sung, G.-H. 1H NMR Serum Metabolomic Change of Trimethylamine N-oxide (TMAO) Is Associated with Alcoholic Liver Disease Progression. Metabolites 2024, 14, 39. https://doi.org/10.3390/metabo14010039
Oh J, Kim J, Lee S, Park G, Baritugo K-AG, Han KJ, Lee S, Sung G-H. 1H NMR Serum Metabolomic Change of Trimethylamine N-oxide (TMAO) Is Associated with Alcoholic Liver Disease Progression. Metabolites. 2024; 14(1):39. https://doi.org/10.3390/metabo14010039
Chicago/Turabian StyleOh, Junsang, Jayoung Kim, Sanghak Lee, Gyubin Park, Kei-Anne Garcia Baritugo, Ki Jun Han, Sangheun Lee, and Gi-Ho Sung. 2024. "1H NMR Serum Metabolomic Change of Trimethylamine N-oxide (TMAO) Is Associated with Alcoholic Liver Disease Progression" Metabolites 14, no. 1: 39. https://doi.org/10.3390/metabo14010039
APA StyleOh, J., Kim, J., Lee, S., Park, G., Baritugo, K.-A. G., Han, K. J., Lee, S., & Sung, G.-H. (2024). 1H NMR Serum Metabolomic Change of Trimethylamine N-oxide (TMAO) Is Associated with Alcoholic Liver Disease Progression. Metabolites, 14(1), 39. https://doi.org/10.3390/metabo14010039










