Non-Targeted Metabolomic Analysis of Arabidopsis thaliana (L.) Heynh: Metabolic Adaptive Responses to Stress Caused by N Starvation
Abstract
:1. Introduction
2. Materials and Methods
2.1. Biological Material
2.2. Disinfection of Arabidopsis Thaliana Seeds
2.3. Preparation of Murashige and Skoog (MS) Medium for Hydroponic Cultures
2.4. Construction of Hydroponic Chambers
2.5. Growth Conditions and Times-Sampling
2.6. Quantification of Phenotypic Traits
2.7. Methanolic Extracts
2.8. Non-Targeted Metabolomic Analysis Using Ultra-High Performance Liquid Chromatography and Accurate Mass Spectrometry
2.9. Statistical, Diversity, and Chemometric Analyses
2.9.1. Identification of Paired Contrasts and Differential “Metabolites” Represented by Mass-to-Charge Ratio (m/z) Values Detected at a Specific Retention Time (rt)
2.9.2. Metabolic Pathway Reconstruction, Functional Enrichment Analysis, and Presumptive Annotation
2.9.3. Tentative Identification of Differential Metabolites (Chemical Markers) between Binary Classes
3. Results
3.1. Establishment of Hydroponic Cultures in a Time-Course Experiment Using the WT Genotype and Loss-of-Function Double Mutant (nia1/nia2) of A. thaliana
3.2. Effect of Stress Induced by Nitrate Deficiency on the Growth of the WT Genotype and Loss-of-Function Double Mutant (nia1/nia2) of A. thaliana
3.3. Evaluation of the Effect of Nutritional Stress Induced by Nitrate Deficiency on the Growth of the Rosettes of WT and nia1/nia2 Genotypes
3.4. Evaluation of the Effect of Nutritional Stress Induced by Nitrate Deficiency on the Growth of the Root System of WT and nia1/nia2 Genotypes
3.5. Metabolomic Compositional Analysis as a Function of α Diversity, Richness, and Equity of Metabolites (m/z_rt)
3.5.1. Acquisition of Spectrometric Signals
3.5.2. Suspected Metabolite (m/z_rt) Diversity Analysis by Species Accumulation Curves (SACs)
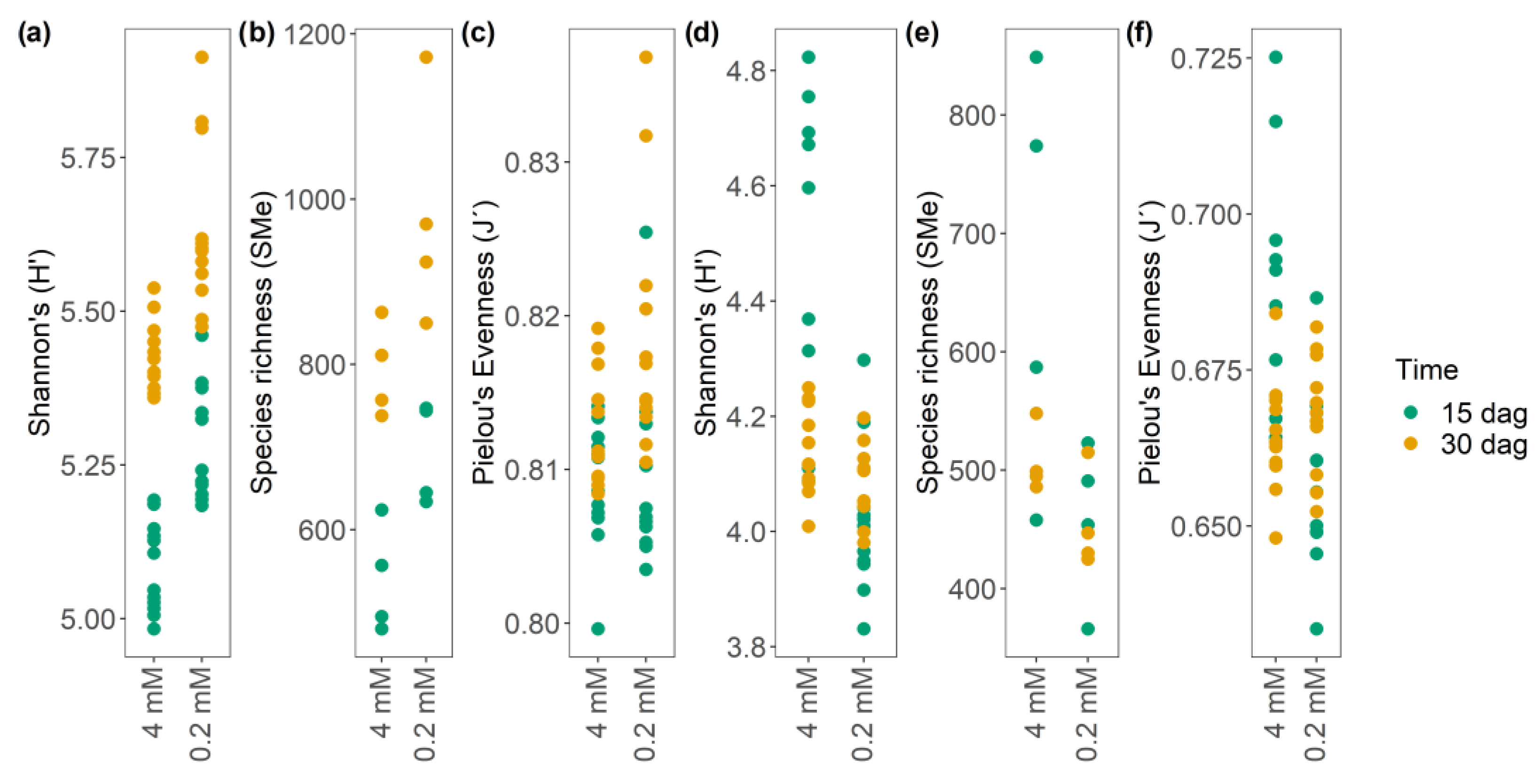
3.5.3. Nitrate Starvation as a Modulator of the α-Diversity, Richness, and Evenness of m/z_rt Pairs in the Metabolome of A. thaliana
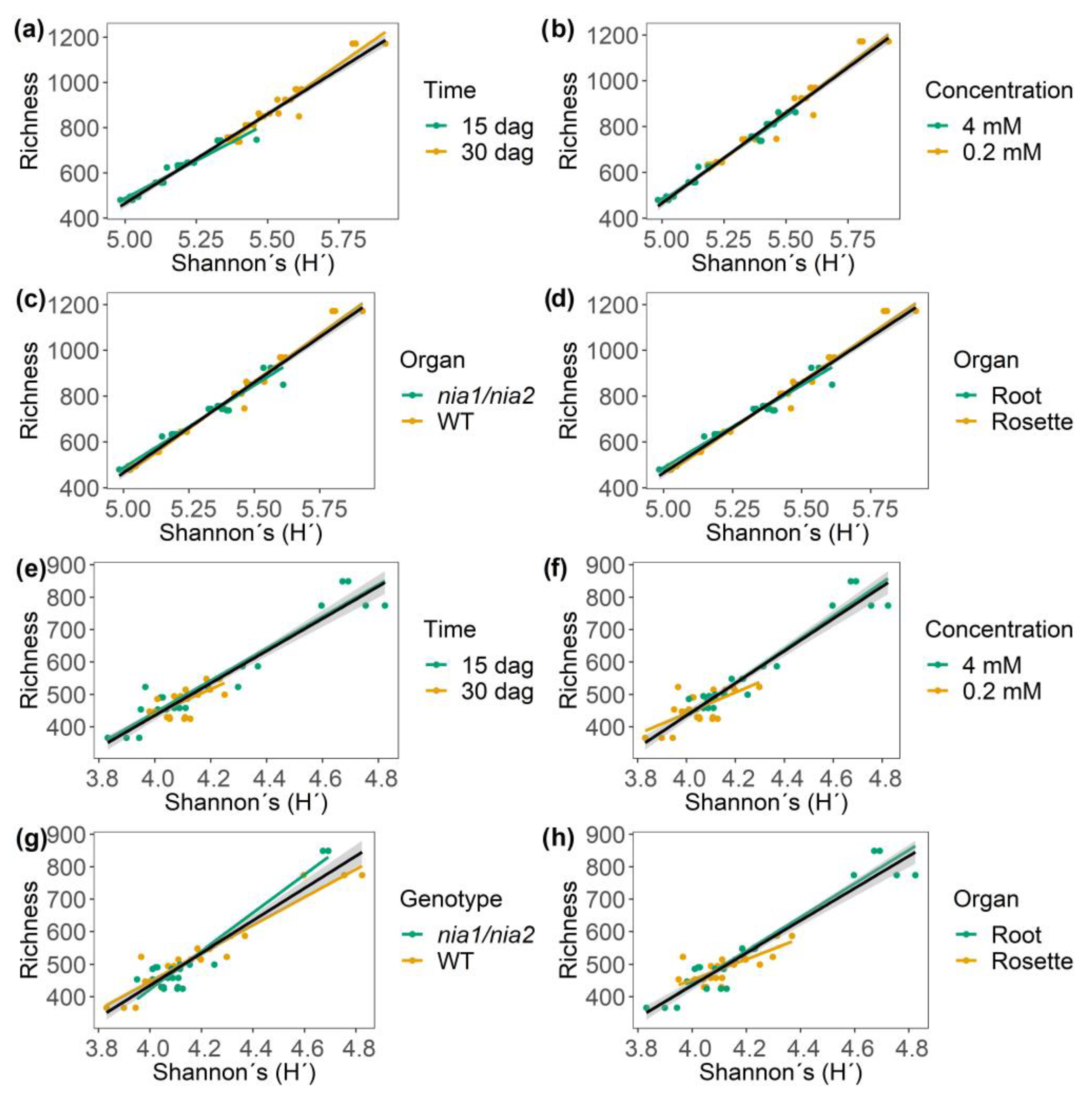
3.5.4. Identification of Correlation Patterns between α Diversity and Richness Equity (J’) of m/z_rt over Time and Across Nitrate Concentrations
3.6. Chemometric Analyses to Identify Discriminant Groups in the Metabolome of A. thaliana
3.6.1. Two-Dimensional Principal Component Analysis (2D PCA) for the Identification of Statistically Relevant Binary Classes
3.6.2. Identification of Differential m/z_rt (Chemical Markers) by Fold-Change (FC) Analysis
3.6.3. Reconstruction of Metabolic Pathways, Functional Enrichment Analysis, and Presumptive Annotation of Differentially Synthesized/Accumulated m/z_rt in the Metabolomes of WT and Loss-of-Function nia1/nia2 Double-Mutant Genotypes
WT Genotype
Loss-of-Function nia1/nia2 Double Mutant
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Des Marais, D.L.; Juenger, T.E. Pleiotropy, plasticity, and the evolution of plant abiotic stress tolerance. Ann. N. Y. Acad. Sci. 2010, 1206, 56–79. [Google Scholar] [CrossRef]
- Dar, F.A.; Mushtaq, N.U.; Saleem, S.; Rehman, R.U.; Dar, T.U.H.; Hakeem, K.R. Role of epigenetics in modulating phenotypic plasticity against abiotic stresses in plants. Int. J. Genom. 2022, 2022, 1092894. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Forde, B.G. An Arabidopsis MADS box gene that controls nutrient-induced changes in root architecture. Science 1998, 279, 407–409. [Google Scholar] [CrossRef]
- Bi, Y.M.; Zhang, Y.; Signorelli, T.; Zhao, R.; Zhu, T.; Rothstein, S. Genetic analysis of Arabidopsis GATA transcription factor gene family reveals a nitrate-inducible member important for chlorophyll synthesis and glucose sensitivity. Plant J. 2005, 44, 680–692. [Google Scholar] [CrossRef] [PubMed]
- Xu, G.; Fan, X.; Miller, A.J. Plant nitrogen assimilation and use efficiency. Annu. Rev. Plant Biol. 2012, 63, 153–182. [Google Scholar] [CrossRef]
- Pandey, P.K.; Yu, J.; Omranian, N.; Alseekh, S.; Vaid, N.; Fernie, A.R.; Nikoloski, Z.; Laitinen, R.A. Plasticity in metabolism underpins local responses to nitrogen in Arabidopsis thaliana populations. Plant Direct. 2019, 3, e00186. [Google Scholar] [CrossRef]
- Shaar-Moshe, L.; Hayouka, R.; Roessner, U.; Peleg, Z. Phenotypic and metabolic plasticity shapes life-history strategies under combinations of abiotic stresses. Plant Direct. 2019, 3, e00113. [Google Scholar] [CrossRef]
- Sack, L.; Buckley, T.N. Trait multi-functionality in plant stress response. Integr. Comp. Biol. 2020, 60, 98–112. [Google Scholar] [CrossRef] [PubMed]
- Fàbregas, N.; Fernie, A.R. The metabolic response to drought. J. Exp. Bot. 2019, 70, 1077–1085. [Google Scholar] [CrossRef]
- Kollist, H.; Zandalinas, S.I.; Sengupta, S.; Nuhkat, M.; Kangasjärvi, J.; Mittler, R. Rapid responses to abiotic stress: Priming the landscape for the signal transduction network. Trends Plant Sci. 2019, 24, 25–37. [Google Scholar] [CrossRef]
- Varoquaux, N.; Cole, B.; Gao, C.; Pierroz, G.; Baker, C.R.; Patel, D.; Madera, M.; Jeffers, T.; Hollingsworth, J.; Sievert, J.; et al. Transcriptomic analysis of field-droughted sorghum from seedling to maturity reveals biotic and metabolic responses. Proc. Natl. Acad. Sci. USA 2019, 116, 27124–27132. [Google Scholar] [CrossRef]
- Xu, Y.; Fu, X. Reprogramming of plant central metabolism in response to abiotic stresses: A metabolomics view. Int. J. Mol. Sci. 2022, 23, 5716. [Google Scholar] [CrossRef] [PubMed]
- Yeshi, K.; Crayn, D.; Ritmejerytė, E.; Wangchuk, P. Plant secondary metabolites produced in response to abiotic stresses has potential application in pharmaceutical product development. Molecules 2022, 27, 313. [Google Scholar] [CrossRef] [PubMed]
- Allwood, J.W.; Williams, A.; Uthe, H.; van Dam, N.M.; Mur, L.A.; Grant, M.R.; Pétriacq, P. Unravelling plant responses to stress—The importance of targeted and untargeted metabolomics. Metabolites 2021, 11, 558. [Google Scholar] [CrossRef] [PubMed]
- Lephatsi, M.M.; Meyer, V.; Piater, L.A.; Dubery, I.A.; Tugizimana, F. Plant responses to abiotic stresses and rhizobacterial biostimulants: Metabolomics and epigenetics perspectives. Metabolites 2021, 11, 457. [Google Scholar] [CrossRef] [PubMed]
- Tinte, M.M.; Chele, K.H.; van der Hooft, J.J.J.; Tugizimana, F. Metabolomics-guided elucidation of plant abiotic stress responses in the 4IR era: An Overview. Metabolites 2021, 11, 445. [Google Scholar] [CrossRef]
- Wilkinson, J.Q.; Crawford, N.M. Identification and characterization of a chlorate-resistant mutant of Arabidopsis thaliana with mutations in both nitrate reductase structural genes NIA1 and NIA2. Mol. Gen. Genet. 1993, 239, 289–297. [Google Scholar] [CrossRef]
- Classic Murashige, T.; Skoog, F. A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physio plant. 1962, 15, 473–497. [Google Scholar] [CrossRef]
- Alatorre-Cobos, F.; Calderón-Vázquez, C.; Ibarra-Laclette, E.; Yong-Villalobos, L.; Pérez-Torres, C.-A.; Oropeza-Aburto, A.; Méndez-Bravo, A.; González-Morales, S.-I.; Gutiérrez-Alanís, D.; Chacón-López, A.; et al. An improved, low-cost, hydroponic system for growing Arabidopsis and other plant species under aseptic conditions. BMC Plant Biol. 2014, 14, 69. [Google Scholar] [CrossRef]
- Mareya, C.R.; Tugizimana, F.; Piater, L.A.; Madala, N.E.; Steenkamp, P.A.; Dubery, I.A. Untargeted metabolomics reveal defensome-related metabolic reprogramming in Sorghum bicolor against infection by Burkholderia andropogonis. Metabolites 2019, 9, 8. [Google Scholar] [CrossRef]
- Tugizimana, F.; Djami-Tchatchou, A.T.; Steenkamp, P.A.; Piater, L.A.; Dubery, I.A. Metabolomic analysis of defense-related reprogramming in Sorghum bicolor in response to Colletotrichum sublineolum infection reveals a functional metabolic web of phenylpropanoid and flavonoid pathways. Front. Plant Sci. 2018, 9, 1840. [Google Scholar] [CrossRef] [PubMed]
- Monribot-Villanueva, J.L.; Elizalde-Contreras, J.M.; Aluja, M.; Segura-Cabrera, A.; Birke, A.; Guerrero-Analco, J.A.; Ruiz-May, E. Endorsing and extending the repertory of nutraceutical and antioxidant sources in mangoes during postharvest shelf life. Food Chem. 2019, 285, 119–129. [Google Scholar] [CrossRef] [PubMed]
- Cadena-Zamudio, J.D.; Nicasio-Torres, P.; Monribot-Villanueva, J.L.; Guerrero-Analco, J.A.; Ibarra-Laclette, E. Integrated analysis of the transcriptome and metabolome of Cecropia obtusifolia: A plant with high chlorogenic acid content traditionally used to treat diabetes Mellitus. Int. J. Mol. Sci. 2020, 21, 7572. [Google Scholar] [CrossRef]
- Cadena-Zamudio, J.D.; Monribot-Villanueva, J.L.; Pérez-Torres, C.-A.; Alatorre-Cobos, F.; Jiménez-Moraila, B.; Guerrero-Analco, J.A.; Ibarra-Laclette, E.J.F.; Genomics, I. The use of ecological analytical tools as an unconventional approach for untargeted metabolomics data analysis: The case of Cecropia obtusifolia and its adaptive responses to nitrate starvation. Funct. Integr. Genom. 2022, 22, 1467–1493. [Google Scholar] [CrossRef] [PubMed]
- Beattie, J.R.; Esmonde-White, F.W.L. Exploration of principal component analysis: Deriving principal component analysis visually using spectra. Appl. Spectrosc. 2021, 75, 361–375. [Google Scholar] [CrossRef]
- Chen, R.-C.; Dewi, C.; Huang, S.-W.; Caraka, R.E. Selecting critical features for data classification based on machine learning methods. J. Big Data 2020, 7, 52. [Google Scholar] [CrossRef]
- Martey, E.N.; Ahmed, L.; Attoh-Okine, N. Track geometry big data analysis: A machine learning approach. In Proceedings of the 2017 IEEE International Conference on Big Data (Big Data), Boston, MA, USA, 11–14 December 2017; pp. 3800–3809. [Google Scholar]
- Mishra, S.; Mallick, P.K.; Tripathy, H.K.; Bhoi, A.K.; González-Briones, A. Performance evaluation of a proposed machine learning model for chronic disease datasets using an integrated attribute evaluator and an improved decision tree classifier. Appl. Sci. 2020, 10, 8137. [Google Scholar] [CrossRef]
- Yu, H.; Xing, S.; Nierves, L.; Lange, P.F.; Huan, T. Fold-change compression: An unexplored but correctable quantitative bias caused by nonlinear electrospray ionization responses in untargeted metabolomics. Anal. Chem. 2020, 92, 7011–7019. [Google Scholar] [CrossRef]
- Shen, H.; Zamboni, N.; Heinonen, M.; Rousu, J. Metabolite identification through machine learning—Tackling CASMI challenge using FingerID. Metabolites 2013, 3, 484–505. [Google Scholar] [CrossRef]
- Hoffmann, M.A.; Kretschmer, F.; Ludwig, M.; Böcker, S. MAD HATTER correctly annotates 98% of small molecule tandem mass spectra searching in PubChem. Metabolites 2023, 13, 314. [Google Scholar] [CrossRef]
- Cran.r-project. Available CRAN Packages by Name. Available online: https://cran.r-project.org/web/packages/available_packages_by_name.html (accessed on 24 August 2020).
- Xia, J.; Psychogios, N.; Young, N.; Wishart, D.S. MetaboAnalyst: A web server for metabolomic data analysis and interpretation. Nucleic Acids Res. 2009, 37, W652–W660. [Google Scholar] [CrossRef]
- Chong, J.; Wishart, D.S.; Xia, J. Using MetaboAnalyst 4.0 for comprehensive and integrative metabolomics data analysis. Curr. Protoc. Bioinform. 2019, 68, e86. [Google Scholar] [CrossRef] [PubMed]
- Liigand, P.; Kaupmees, K.; Haav, K.; Liigand, J.; Leito, I.; Girod, M.; Antoine, R.; Kruve, A. Think negative: Finding the best electrospray ionization/MS mode for your analyte. Anal. Chem. 2017, 89, 5665–5668. [Google Scholar] [CrossRef]
- Zhao, H.; Ni, S.; Cai, S.; Zhang, G. Comprehensive dissection of primary metabolites in response to diverse abiotic stress in barley at seedling stage. Plant Physiol. Biochem. 2021, 161, 54–64. [Google Scholar] [CrossRef]
- Krapp, A.; Berthomé, R.; Orsel, M.; Mercey-Boutet, S.; Yu, A.; Castaings, L.; Elftieh, S.; Major, H.; Renou, J.-P.; Daniel-Vedele, F. Arabidopsis roots and shoots show distinct temporal adaptation patterns toward nitrogen starvation. Plant Physiol. 2011, 157, 1255–1282. [Google Scholar] [CrossRef] [PubMed]
- Karpievitch, Y.V.; Taverner, T.; Adkins, J.N.; Callister, S.J.; Anderson, G.A.; Smith, R.D.; Dabney, A.R. Normalization of peak intensities in bottom-up MS-based proteomics using singular value decomposition. Bioinformatics 2009, 25, 2573–2580. [Google Scholar] [CrossRef] [PubMed]
- Changyong, F.; Hongyue, W.; Naiji, L.; Tian, C.; Hua, H.; Ying, L. Log-transformation and its implications for data analysis. Shanghai Arch. Psychiatry 2014, 26, 105. [Google Scholar] [CrossRef]
- Li, S.; Park, Y.; Duraisingham, S.; Strobel, F.H.; Khan, N.; Soltow, Q.A.; Jones, D.P.; Pulendran, B. Predicting network activity from high throughput metabolomics. PLoS Comput. Biol. 2013, 9, e1003123. [Google Scholar] [CrossRef]
- Johnson, C.H.; Ivanisevic, J.; Benton, H.P.; Siuzdak, G. Bioinformatics: The next frontier of metabolomics. Anal. Chem. 2015, 87, 147–156. [Google Scholar] [CrossRef]
- Liebal, U.W.; Phan, A.N.T.; Sudhakar, M.; Raman, K.; Blank, L.M. Machine learning applications for mass spectrometry-based metabolomics. Metabolites 2020, 10, 243. [Google Scholar] [CrossRef]
- Karnovsky, A.; Li, S. Pathway analysis for targeted and untargeted metabolomics. Methods Mol. Biol. 2020, 2104, 387–400. [Google Scholar] [CrossRef]
- Blancaflor, E.B.; Kilaru, A.; Keereetaweep, J.; Khan, B.R.; Faure, L.; Chapman, K.D. N-Acylethanolamines: Lipid metabolites with functions in plant growth and development. Plant J. 2014, 79, 568–583. [Google Scholar] [CrossRef] [PubMed]
- Pichersky, E.; Raguso, R.A.J.N.P. Why do plants produce so many terpenoid compounds? New Phytol. 2018, 220, 692–702. [Google Scholar] [CrossRef] [PubMed]
- Nagegowda, D.A.; Gupta, P. Advances in biosynthesis, regulation, and metabolic engineering of plant specialized terpenoids. Plant Sci. 2020, 294, 110457. [Google Scholar] [CrossRef]
- Sharma, A.; Shahzad, B.; Rehman, A.; Bhardwaj, R.; Landi, M.; Zheng, B. Response of phenylpropanoid pathway and the role of polyphenols in plants under abiotic stress. Molecules 2019, 24, 2452. [Google Scholar] [CrossRef] [PubMed]
- Shah, A.; Smith, D.L. Flavonoids in agriculture: Chemistry and roles in, biotic and abiotic stress responses, and microbial associations. Agronomy 2020, 10, 1209. [Google Scholar] [CrossRef]
- Šamec, D.; Karalija, E.; Šola, I.; Vujčić Bok, V.; Salopek-Sondi, B. The role of polyphenols in abiotic stress response: The influence of molecular structure. Plants 2021, 10, 118. [Google Scholar] [CrossRef]
- Jia, Q.; Kong, D.; Li, Q.; Sun, S.; Song, J.; Zhu, Y.; Liang, K.; Ke, Q.; Lin, W.; Huang, J. The function of inositol phosphatases in plant tolerance to abiotic stress. Int. J. Mol. Sci. 2019, 20, 3999. [Google Scholar] [CrossRef]
- Basak, P.; Majumder, A.L. Regulation of stress-induced inositol metabolism in plants: A phylogenetic search for conserved cis elements. J. Plant Biochem. Biotechnol. 2021, 30, 756–778. [Google Scholar] [CrossRef]
- Hu, D.; Wei, L.; Liao, W. Brassinosteroids in plants: Crosstalk with small-molecule compounds. Biomolecules 2021, 11, 1800. [Google Scholar] [CrossRef]
- Nolan, T.M.; Vukasinovic, N.; Liu, D.; Russinova, E.; Yin, Y. Brassinosteroids: Multidimensional regulators of plant growth, development, and stress responses. Plant Cell 2020, 32, 295–318. [Google Scholar] [CrossRef]
- Vukašinović, N.; Wang, Y.; Vanhoutte, I.; Fendrych, M.; Guo, B.; Kvasnica, M.; Jiroutová, P.; Oklestkova, J.; Strnad, M.; Russinova, E. Local brassinosteroid biosynthesis enables optimal root growth. Nat. Plants 2021, 7, 619–632. [Google Scholar] [CrossRef] [PubMed]
- Chapman, J.M.; Muday, G.K. Flavonols modulate lateral root emergence by scavenging reactive oxygen species in Arabidopsis thaliana. J. Biol. Chem. 2021, 296, 100222. [Google Scholar] [CrossRef] [PubMed]
- Richmond, B.L.; Coelho, C.L.; Wilkinson, H.; McKenna, J.; Ratchinski, P.; Schwarze, M.; Frost, M.; Lagunas, B.; Gifford, M.L. Elucidating connections between the strigolactone biosynthesis pathway, flavonoid production and root system architecture in Arabidopsis thaliana. Physiol. Plant. 2022, 174, e13681. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Xiong, F.; Nong, S.; Liao, J.; Xing, A.; Shen, Q.; Ma, Y.; Fang, W.; Zhu, X. Effects of nitric oxide on the GABA, polyamines, and proline in tea (Camellia sinensis) roots under cold stress. Sci. Rep. 2020, 10, 12240. [Google Scholar] [CrossRef]
- Sun, C.; Zhang, Y.; Liu, L.; Liu, X.; Li, B.; Jin, C.; Lin, X. Molecular functions of nitric oxide and its potential applications in horticultural crops. Hortic. Res. 2021, 8, 71. [Google Scholar] [CrossRef]
- Tan, P.; Zeng, C.; Wan, C.; Liu, Z.; Dong, X.; Peng, J.; Lin, H.; Li, M.; Liu, Z.; Yan, M. Metabolic profiles of Brassica juncea roots in response to cadmium stress. Metabolites 2021, 11, 383. [Google Scholar] [CrossRef]
- Zi, X.; Zhou, S.; Wu, B.J.M. Alpha-linolenic acid mediates diverse drought responses in maize (Zea mays L.) at seedling and flowering stages. Molecules 2022, 27, 771. [Google Scholar] [CrossRef]
- Emamverdian, A.; Ding, Y.; Mokhberdoran, F. The role of salicylic acid and gibberellin signaling in plant responses to abiotic stress with an emphasis on heavy metals. Plant Signal. Behav. 2020, 15, 1777372. [Google Scholar] [CrossRef]
- López-Ruiz, B.A.; Zluhan-Martínez, E.; Sánchez, M.d.l.P.; Álvarez-Buylla, E.R.; Garay-Arroyo, A. Interplay between hormones and several abiotic stress conditions on Arabidopsis thaliana primary root development. Cells 2020, 9, 2576. [Google Scholar] [CrossRef]
- Touraine, B. Nitrate Uptake by Roots-Transporters and Root Development. In Nitrogen Acquisition and Assimilation in Higher Plants; Springer: Dordrecht, The Netherlands, 2004; pp. 1–34. [Google Scholar]
- Sachdev, S.; Ansari, S.A.; Ansari, M.I.; Fujita, M.; Hasanuzzaman, M. Abiotic stress and reactive oxygen species: Generation, signaling, and defense mechanisms. Antioxidants 2021, 10, 277. [Google Scholar] [CrossRef]
- D’Auria, J.C.; Pichersky, E.; Schaub, A.; Hansel, A.; Gershenzon, J. Characterization of a BAHD acyltransferase responsible for producing the green leaf volatile (Z)-3-hexen-1-yl acetate in Arabidopsis thaliana. Plant J. 2007, 49, 194–207. [Google Scholar] [CrossRef]
- Wakai, J.; Kusama, S.; Nakajima, K.; Kawai, S.; Okumura, Y.; Shiojiri, K. Effects of trans-2-hexenal and cis-3-hexenal on post-harvest strawberry. Sci. Rep. 2019, 9, 10112. [Google Scholar] [CrossRef]
- Shomali, A.; Das, S.; Arif, N.; Sarraf, M.; Zahra, N.; Yadav, V.; Aliniaeifard, S.; Chauhan, D.K.; Hasanuzzaman, M. Diverse physiological roles of flavonoids in plant environmental stress responses and tolerance. Plants 2022, 11, 3158. [Google Scholar] [CrossRef] [PubMed]
- Ye, H.; Liu, S.; Tang, B.; Chen, J.; Xie, Z.; Nolan, T.M.; Jiang, H.; Guo, H.; Lin, H.-Y.; Li, L. RD26 mediates crosstalk between drought and brassinosteroid signalling pathways. Nat. Commun. 2017, 8, 14573. [Google Scholar] [CrossRef] [PubMed]
- Chaudhuri, A.; Halder, K.; Abdin, M.Z.; Majee, M.; Datta, A. Abiotic stress tolerance in plants: Brassinosteroids navigate competently. Int. J. Mol. Sci. 2022, 23, 14577. [Google Scholar] [CrossRef] [PubMed]
- Colebrook, E.H.; Thomas, S.G.; Phillips, A.L.; Hedden, P. The role of gibberellin signalling in plant responses to abiotic stress. J. Exp. Biol. 2014, 217, 67–75. [Google Scholar] [CrossRef]
- Forghani, A.; Almodares, A.; Ehsanpour, A. The role of gibberellic acid and paclobutrazol on oxidative stress responses induced by in vitro salt stress in sweet sorghum. Russ. J. Plant Physiol. 2020, 67, 555–563. [Google Scholar] [CrossRef]
- Hao, R.; Shan, S.; Yang, D.; Zhang, H.; Sun, Y.; Li, Z. Peonidin-3-O-glucoside from purple corncob ameliorates nonalcoholic fatty liver disease by regulating mitochondrial and lysosome functions to reduce oxidative stress and inflammation. Nutrients 2023, 15, 372. [Google Scholar] [CrossRef]
- Clouse, S.D. Arabidopsis mutants reveal multiple roles for sterols in plant development. Plant Cell 2002, 14, 1995–2000. [Google Scholar] [CrossRef]
- Du, Y.; Fu, X.; Chu, Y.; Wu, P.; Liu, Y.; Ma, L.; Tian, H.; Zhu, B. Biosynthesis and the roles of plant sterols in development and stress responses. Int. J. Mol. Sci. 2022, 23, 2332. [Google Scholar] [CrossRef] [PubMed]
- Gorelova, V.; Ambach, L.; Rébeillé, F.; Stove, C.; Van Der Straeten, D. Folates in plants: Research advances and progress in crop biofortification. Front. Chem. 2017, 5, 21. [Google Scholar] [CrossRef] [PubMed]
- Kołton, A.; Długosz-Grochowska, O.; Wojciechowska, R.; Czaja, M. Biosynthesis regulation of folates and phenols in plants. Sci. Hortic. 2022, 291, 110561. [Google Scholar] [CrossRef]
- Xuan, W.; Beeckman, T.; Xu, G. Plant nitrogen nutrition: Sensing and signaling. Curr. Opin. Plant Biol. 2017, 39, 57–65. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Hu, B.; Chu, C. Towards understanding the hierarchical nitrogen signalling network in plants. Curr. Opin. Plant Biol. 2020, 55, 60–65. [Google Scholar] [CrossRef]
- Modolo, L.V.; Augusto, O.; Almeida, I.M.; Pinto-Maglio, C.A.; Oliveira, H.C.; Seligman, K.; Salgado, I. Decreased arginine and nitrite levels in nitrate reductase-deficient Arabidopsis thaliana plants impair nitric oxide synthesis and the hypersensitive response to Pseudomonas syringae. Plant Sci. 2006, 171, 34–40. [Google Scholar] [CrossRef]
- Medici, A.; Krouk, G. The primary nitrate response: A multifaceted signalling pathway. J. Exp. Bot. 2014, 65, 5567–5576. [Google Scholar] [CrossRef]
- Xiao, C.; Fang, Y.; Wang, S.; He, K. The alleviation of ammonium toxicity in plants. J. Integr. Plant Biol. 2023, 65, 1362–1368. [Google Scholar] [CrossRef]
- Mayer, M.; Ludewig, U. Role of AMT1;1 in NH4+ acquisition in Arabidopsis thaliana. Plant Biol. (Stuttg) 2006, 8, 522–528. [Google Scholar] [CrossRef]
- Koltun, A.; Maniero, R.A.; Vitti, M.; de Setta, N.; Giehl, R.F.H.; Lima, J.E.; Figueira, A. Functional characterization of the sugarcane (Saccharum spp.) ammonium transporter AMT2;1 suggests a role in ammonium root-to-shoot translocation. Front. Plant Sci. 2022, 13, 1039041. [Google Scholar] [CrossRef]
- Huang, L.; Li, J.; Zhang, B.; Hao, Y.; Ma, F.J.H. Genome-wide identification and expression analysis of AMT gene family in apple (Malus domestica Borkh.). Horticulturae 2022, 8, 457. [Google Scholar] [CrossRef]
- Shin, S.Y.; Jeong, J.S.; Lim, J.Y.; Kim, T.; Park, J.H.; Kim, J.K.; Shin, C. Transcriptomic analyses of rice (Oryza sativa) genes and non-coding RNAs under nitrogen starvation using multiple omics technologies. BMC Genom. 2018, 19, 532. [Google Scholar] [CrossRef] [PubMed]
- Gazzarrini, S.; Lejay, L.; Gojon, A.; Ninnemann, O.; Frommer, W.B.; von Wirén, N. Three functional transporters for constitutive, diurnally regulated, and starvation-induced uptake of ammonium into Arabidopsis roots. Plant Cell 1999, 11, 937–947. [Google Scholar] [CrossRef]
- Von Wirén, N.; Lauter, F.R.; Ninnemann, O.; Gillissen, B.; Walch-Liu, P.; Engels, C.; Jost, W.; Frommer, W.B. Differential regulation of three functional ammonium transporter genes by nitrogen in root hairs and by light in leaves of tomato. Plant J. 2000, 21, 167–175. [Google Scholar] [CrossRef]
- Sonoda, Y.; Ikeda, A.; Saiki, S.; Yamaya, T.; Yamaguchi, J. Feedback regulation of the ammonium transporter gene family AMT1 by glutamine in rice. Plant Cell Physiol. 2003, 44, 1396–1402. [Google Scholar] [CrossRef] [PubMed]
- Suenaga, A.; Moriya, K.; Sonoda, Y.; Ikeda, A.; Von Wirén, N.; Hayakawa, T.; Yamaguchi, J.; Yamaya, T. Constitutive expression of a novel-type ammonium transporter OsAMT2 in rice plants. Plant Cell Physiol. 2003, 44, 206–211. [Google Scholar] [CrossRef] [PubMed]
- Guiboileau, A.; Yoshimoto, K.; Soulay, F.; Bataillé, M.P.; Avice, J.C.; Masclaux-Daubresse, C. Autophagy machinery controls nitrogen remobilization at the whole-plant level under both limiting and ample nitrate conditions in Arabidopsis. New Phytol. 2012, 194, 732–740. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Chung, T.; Pennington, J.G.; Federico, M.L.; Kaeppler, H.F.; Kaeppler, S.M.; Otegui, M.S.; Vierstra, R.D. Autophagic recycling plays a central role in maize nitrogen remobilization. Plant Cell 2015, 27, 1389–1408. [Google Scholar] [CrossRef]
- Naik, B.; Kumar, V.; Rizwanuddin, S.; Chauhan, M.; Choudhary, M.; Gupta, A.K.; Kumar, P.; Kumar, V.; Saris, P.E.J.; Rather, M.A.; et al. Genomics, proteomics, and metabolomics approaches to improve abiotic stress tolerance in tomato plant. Int. J. Mol. Sci. 2023, 24, 3025. [Google Scholar] [CrossRef]
- Hwang, J.-H.; Yu, S.-I.; Lee, B.-H.; Lee, D.-H. Modulation of energy metabolism is important for low-oxygen stress adaptation in Brassicaceae species. Int. J. Mol. Sci. 2020, 21, 1787. [Google Scholar] [CrossRef]
- Smolko, A.; Bauer, N.; Pavlović, I.; Pěnčík, A.; Novák, O.; Salopek-Sondi, B. Altered root growth, auxin metabolism and distribution in Arabidopsis thaliana exposed to salt and osmotic stress. Int. J. Mol. Sci. 2021, 22, 7993. [Google Scholar] [CrossRef]
- Schneider, G.F.; Salazar, D.; Hildreth, S.B.; Helm, R.F.; Whitehead, S.R. Comparative metabolomics of fruits and leaves in a hyperdiverse lineage suggests fruits are a key incubator of phytochemical diversification. Front. Plant Sci. 2021, 12, 693739. [Google Scholar] [CrossRef]
- Zhou, Y.; Xu, X.; Chen, Y.; Gao, J.; Shi, Q.; Tian, L.; Cao, L. Combined metabolome and transcriptome analyses reveal the flavonoids changes and biosynthesis mechanisms in different organs of Hibiseu manihot L. Front. Plant Sci. 2022, 13, 817378. [Google Scholar] [CrossRef]
- Beckles, D.M.; Roessner, U. Plant Metabolomics: Applications and Opportunities for Agricultural Biotechnology. In Plant Biotechnology and Agriculture; Academic Press: London, UK, 2012; pp. 67–81. [Google Scholar]
- Savchenko, T.; Walley, J.W.; Chehab, E.W.; Xiao, Y.; Kaspi, R.; Pye, M.F.; Mohamed, M.E.; Lazarus, C.M.; Bostock, R.M.; Dehesh, K. Arachidonic acid: An evolutionarily conserved signaling molecule modulates plant stress signaling networks. Plant Cell 2010, 22, 3193–3205. [Google Scholar] [CrossRef]
- Knight, V.I.; Wang, H.; Lincoln, J.E.; Lulai, E.C.; Gilchrist, D.G.; Bostock, R.M. Hydroperoxides of fatty acids induce programmed cell death in tomato protoplasts. Physiol. Mol. Plant Pathol. 2001, 59, 277–286. [Google Scholar] [CrossRef]
- Choi, D.; Bostock, R.M.; Avdiushko, S.; Hildebrand, D.F. Lipid-derived signals that discriminate wound-and pathogen-responsive isoprenoid pathways in plants: Methyl jasmonate and the fungal elicitor arachidonic acid induce different 3-hydroxy-3-methylglutaryl-coenzyme A reductase genes and antimicrobial isoprenoids in Solanum tuberosum L. Proc. Natl. Acad. Sci. USA 1994, 91, 2329–2333. [Google Scholar] [CrossRef] [PubMed]
- Tjamos, E.C.; Kucacute, J.A. Inhibition of steroid glycoalkaloid accumulation by arachidonic and eicosapentaenoic acids in potato. Science 1982, 217, 542–544. [Google Scholar] [CrossRef]
- Bryant, J.P.; Chapin, F.S., III; Klein, D.R. Carbon/nutrient balance of boreal plants in relation to vertebrate herbivory. Oikos 1983, 40, 357–368. [Google Scholar] [CrossRef]
- Massad, T.J.; Dyer, L.A.; Vega, C.G. Costs of defense and a test of the carbon-nutrient balance and growth-differentiation balance hypotheses for two co-occurring classes of plant defense. PLoS ONE 2012, 7, e47554. [Google Scholar] [CrossRef]
- Bostock, R.M.; Kuc, J.A.; Laine, R.A. Eicosapentaenoic and arachidonic acids from Phytophthora infestans elicit fungitoxic sesquiterpenes in the potato. Science 1981, 212, 67–69. [Google Scholar] [CrossRef] [PubMed]
- Laha, N.P.; Dhir, Y.W.; Giehl, R.F.; Schäfer, E.M.; Gaugler, P.; Shishavan, Z.H.; Gulabani, H.; Mao, H.; Zheng, N.; von Wirén, N. ITPK1-dependent inositol polyphosphates regulate auxin responses in Arabidopsis thaliana. Biorxiv 2020, 1–29. [Google Scholar] [CrossRef]
- Kong, X.; Zhang, C.; Zheng, H.; Sun, M.; Zhang, F.; Zhang, M.; Cui, F.; Lv, D.; Liu, L.; Guo, S. Antagonistic interaction between auxin and SA signaling pathways regulates bacterial infection through lateral root in Arabidopsis. Cell Rep. 2020, 32, 108060. [Google Scholar] [CrossRef]
- Zaninotto, F.; Camera, S.L.; Polverari, A.; Delledonne, M. Cross talk between reactive nitrogen and oxygen species during the hypersensitive disease resistance response. Plant Physiol. 2006, 141, 379–383. [Google Scholar] [CrossRef]
- Laha, D.; Riemer, E.; Pullagurla, N.J.; Yadav, R.; Hansjosten, L.; Priyanshi, R.; Jork, N.; Furkert, D.; Fiedler, D.; Jessen, H.J. Regulation of plant biotic interactions and abiotic stress responses by inositol polyphosphates. Front. Plant Sci. 2022, 13, 944515. [Google Scholar] [CrossRef]
- Schwechheimer, C. Gibberellin signaling in plants–the extended version. Front. Plant Sci. 2012, 2, 107. [Google Scholar] [CrossRef]
- Tu-Sekine, B.; Kim, S.F. The inositol phosphate system—A coordinator of metabolic adaptability. Int. J. Mol. Sci. 2022, 23, 6747. [Google Scholar] [CrossRef] [PubMed]
- Wei, Z.; Li, J. Brassinosteroids regulate root growth, development, and symbiosis. Mol. Plant 2016, 9, 86–100. [Google Scholar] [CrossRef]
- Phua, S.Y.; De Smet, B.; Remacle, C.; Chan, K.X.; Van Breusegem, F. Reactive oxygen species and organellar signaling. J. Exp. Bot. 2021, 72, 5807–5824. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Mubeen, B.; Hasnain, A.; Rizwan, M.; Adrees, M.; Naqvi, S.A.H.; Iqbal, S.; Kamran, M.; El-Sabrout, A.M.; Elansary, H.O. Role of promising secondary metabolites to confer resistance against environmental stresses in crop plants: Current scenario and future perspectives. Front. Plant Sci. 2022, 13, 881032. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.Y.; Zheng, W. Effect of plant growth temperature on antioxidant capacity in strawberry. J. Agric. Food Chem. 2001, 49, 4977–4982. [Google Scholar] [CrossRef]
- Munné-Bosch, S.; Alegre, L. The function of tocopherols and tocotrienols in plants. Crit. Rev. Plant Sci. 2002, 21, 31–57. [Google Scholar] [CrossRef]
- Zhang, M.; Swarts, S.G.; Yin, L.; Liu, C.; Tian, Y.; Cao, Y.; Swarts, M.; Yang, S.; Zhang, S.B.; Zhang, K. Antioxidant Properties of Quercetin. In Oxygen Transport to Tissue XXXII, 2011; Advances in Experimental Medicine and Biology, Volume 701; Springer: Boston, MA, USA; pp. 283–289. [CrossRef]
- García-Pérez, E.; Gutiérrez-Uribe, J.A.; García-Lara, S.J.P.C. Luteolin content and antioxidant activity in micropropagated plants of Poliomintha glabrescens (Gray). Plant Cell Tissue Organ Cult. (PCTOC) 2012, 108, 521–527. [Google Scholar] [CrossRef]
- Saleem, H.; Htar, T.T.; Naidu, R.; Nawawi, N.S.; Ahmad, I.; Ashraf, M.; Ahemad, N. Biological, chemical and toxicological perspectives on aerial and roots of Filago germanica (L.) huds: Functional approaches for novel phyto-pharmaceuticals. Food Chem. Toxicol. 2019, 123, 363–373. [Google Scholar] [CrossRef] [PubMed]
- Masferrer, A.; Arró, M.; Manzano, D.; Schaller, H.; Fernández-Busquets, X.; Moncaleán, P.; Fernández, B.; Cunillera, N.; Boronat, A.; Ferrer, A. Overexpression of Arabidopsis thaliana farnesyl diphosphate synthase (FPS1S) in transgenic Arabidopsis induces a cell death/senescence-like response and reduced cytokinin levels. Plant J. 2002, 30, 123–132. [Google Scholar] [CrossRef] [PubMed]
- Manzano, D.; Andrade, P.; Caudepón, D.; Altabella, T.; Arró, M.; Ferrer, A. Suppressing farnesyl diphosphate synthase alters chloroplast development and triggers sterol-dependent induction of jasmonate-and Fe-related responses. Plant Physiol. 2016, 172, 93–117. [Google Scholar] [CrossRef]
- Del Río, L.A. ROS and RNS in plant physiology: An overview. J. Exp. Bot. 2015, 66, 2827–2837. [Google Scholar] [CrossRef]
- Youssef, M.H.; Raafat, A.; El-Yazied, A.A.; Selim, S.; Azab, E.; Khojah, E.; El Nahhas, N.; Ibrahim, M.F. Exogenous application of alpha-Lipoic acid mitigates salt-induced oxidative damage in sorghum plants through regulation growth, leaf pigments, ionic homeostasis, antioxidant enzymes, and expression of salt stress responsive genes. Plants 2021, 10, 2519. [Google Scholar] [CrossRef]
- Gaude, N.; Bréhélin, C.; Tischendorf, G.; Kessler, F.; Dörmann, P. Nitrogen deficiency in Arabidopsis affects galactolipid composition and gene expression and results in accumulation of fatty acid phytyl esters. Plant J. 2007, 49, 729–739. [Google Scholar] [CrossRef]
- Liang, J.; He, J. Protective role of anthocyanins in plants under low nitrogen stress. Biochem. Biophys. Res. Commun. 2018, 498, 946–953. [Google Scholar] [CrossRef]
- Reuveny, Z.; Dougall, D.K.; Trinity, P.M. Regulatory coupling of nitrate and sulfate assimilation pathways in cultured tobacco cells. Proc. Natl. Acad. Sci. USA 1980, 77, 6670–6672. [Google Scholar] [CrossRef]
- Prosser, I.M.; Purves, J.V.; Saker, L.R.; Clarkson, D.T. Rapid disruption of nitrogen metabolism and nitrate transport in spinach plants deprived of sulphate. J. Exp. Bot. 2001, 52, 113–121. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Hirai, M.Y.; Hayashi, H.; Chino, M.; Naito, S.; Fujiwara, T. Role of O-acetyl-L-serine in the coordinated regulation of the expression of a soybean seed storage-protein gene by sulfur and nitrogen nutrition. Planta 1999, 209, 282–289. [Google Scholar] [CrossRef] [PubMed]
- Hirai, M.Y.; Fujiwara, T.; Awazuhara, M.; Kimura, T.; Noji, M.; Saito, K. Global expression profiling of sulfur-starved Arabidopsis by DNA macroarray reveals the role of O-acetyl-l-serine as a general regulator of gene expression in response to sulfur nutrition. Plant J. 2003, 33, 651–663. [Google Scholar] [CrossRef]
- Hirai, M.Y.; Yano, M.; Goodenowe, D.B.; Kanaya, S.; Kimura, T.; Awazuhara, M.; Arita, M.; Fujiwara, T.; Saito, K. Integration of transcriptomics and metabolomics for understanding of global responses to nutritional stresses in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2004, 101, 10205–10210. [Google Scholar] [CrossRef] [PubMed]
- Nikiforova, V.; Freitag, J.; Kempa, S.; Adamik, M.; Hesse, H.; Hoefgen, R. Transcriptome analysis of sulfur depletion in Arabidopsis thaliana: Interlacing of biosynthetic pathways provides response specificity. Plant J. 2003, 33, 633–650. [Google Scholar] [CrossRef] [PubMed]
- Nikiforova, V.J.; Kopka, J.; Tolstikov, V.; Fiehn, O.; Hopkins, L.; Hawkesford, M.J.; Hesse, H.; Hoefgen, R. Systems rebalancing of metabolism in response to sulfur deprivation, as revealed by metabolome analysis of Arabidopsis plants. Plant Physiol. 2005, 138, 304–318. [Google Scholar] [CrossRef] [PubMed]
- Ohkama-Ohtsu, N.; Wasaki, J. Recent progress in plant nutrition research: Cross-talk between nutrients, plant physiology and soil microorganisms. Plant Cell Physiol. 2010, 51, 1255–1264. [Google Scholar] [CrossRef]












Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cadena-Zamudio, J.D.; Monribot-Villanueva, J.L.; Pérez-Torres, C.-A.; Alatorre-Cobos, F.; Guerrero-Analco, J.A.; Ibarra-Laclette, E. Non-Targeted Metabolomic Analysis of Arabidopsis thaliana (L.) Heynh: Metabolic Adaptive Responses to Stress Caused by N Starvation. Metabolites 2023, 13, 1021. https://doi.org/10.3390/metabo13091021
Cadena-Zamudio JD, Monribot-Villanueva JL, Pérez-Torres C-A, Alatorre-Cobos F, Guerrero-Analco JA, Ibarra-Laclette E. Non-Targeted Metabolomic Analysis of Arabidopsis thaliana (L.) Heynh: Metabolic Adaptive Responses to Stress Caused by N Starvation. Metabolites. 2023; 13(9):1021. https://doi.org/10.3390/metabo13091021
Chicago/Turabian StyleCadena-Zamudio, Jorge David, Juan Luis Monribot-Villanueva, Claudia-Anahí Pérez-Torres, Fulgencio Alatorre-Cobos, José Antonio Guerrero-Analco, and Enrique Ibarra-Laclette. 2023. "Non-Targeted Metabolomic Analysis of Arabidopsis thaliana (L.) Heynh: Metabolic Adaptive Responses to Stress Caused by N Starvation" Metabolites 13, no. 9: 1021. https://doi.org/10.3390/metabo13091021
APA StyleCadena-Zamudio, J. D., Monribot-Villanueva, J. L., Pérez-Torres, C.-A., Alatorre-Cobos, F., Guerrero-Analco, J. A., & Ibarra-Laclette, E. (2023). Non-Targeted Metabolomic Analysis of Arabidopsis thaliana (L.) Heynh: Metabolic Adaptive Responses to Stress Caused by N Starvation. Metabolites, 13(9), 1021. https://doi.org/10.3390/metabo13091021






