A Metabolomics-Based Study on the Discriminative Classification Models and Toxicological Mechanism of Estazolam Fatal Intoxication
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemicals and Reagents
2.2. Animals Diets and Grouping
2.3. Blood and Brainstem Tissues Sample Collection
2.4. Blood and Brainstem Tissue Metabolite Extraction
2.5. Data Acquisition with Full Scan-MS/MS Using LC-HR MS/MS
2.6. Data Processing and Discriminating Component Analysis
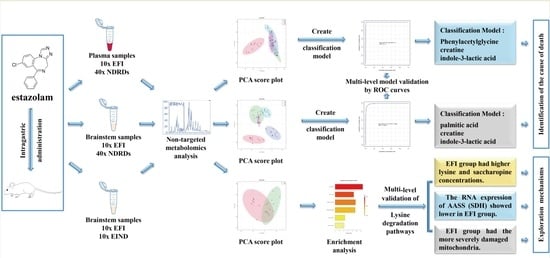
3. Results
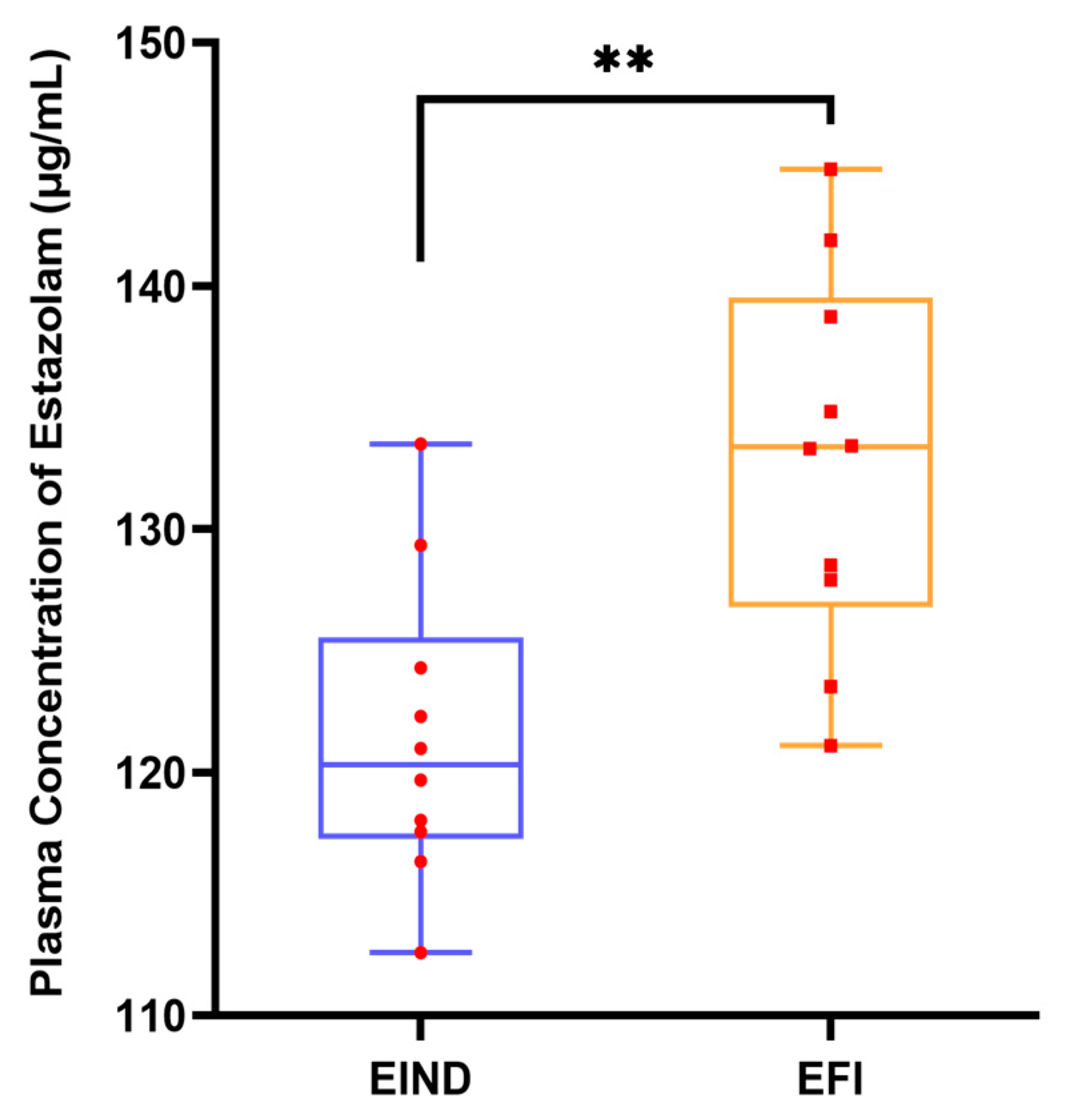
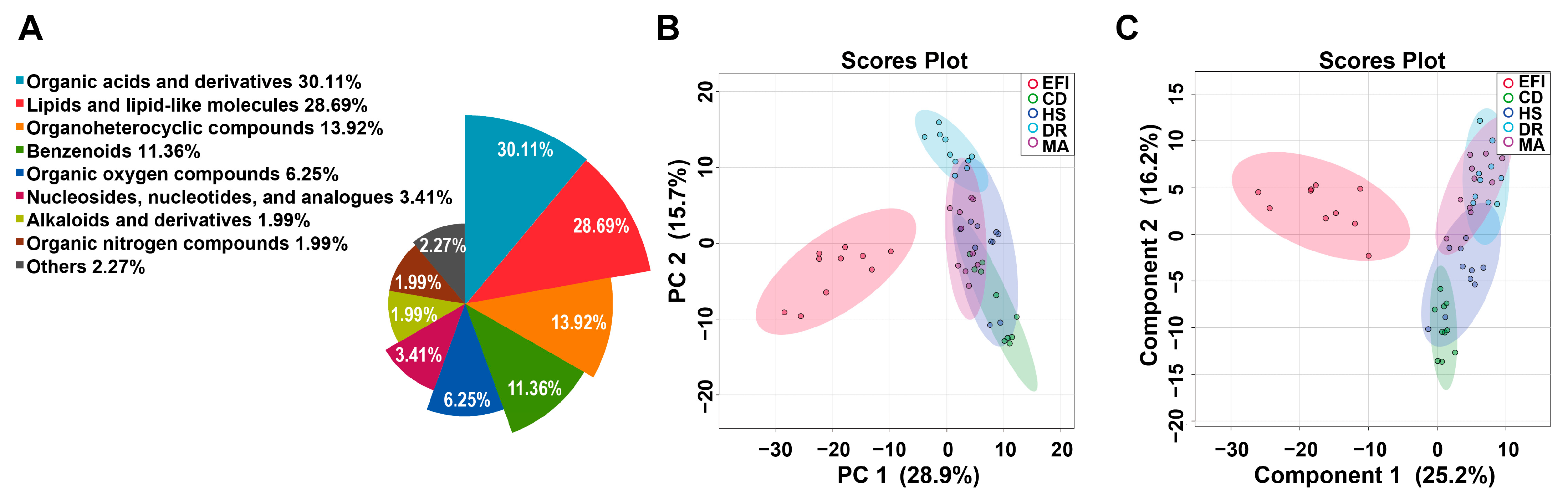
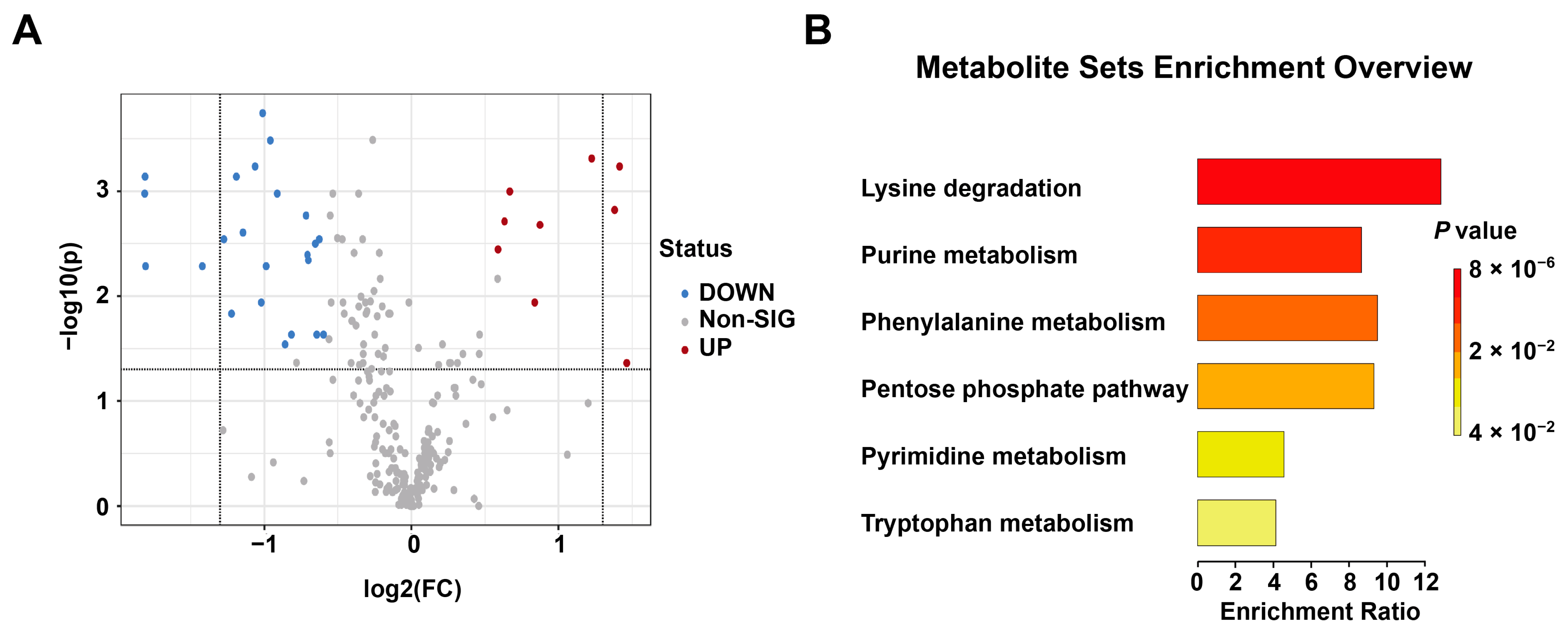
3.1. Metabolomics Profiling of Plasma Samples in the EFI and NDRDs Mice
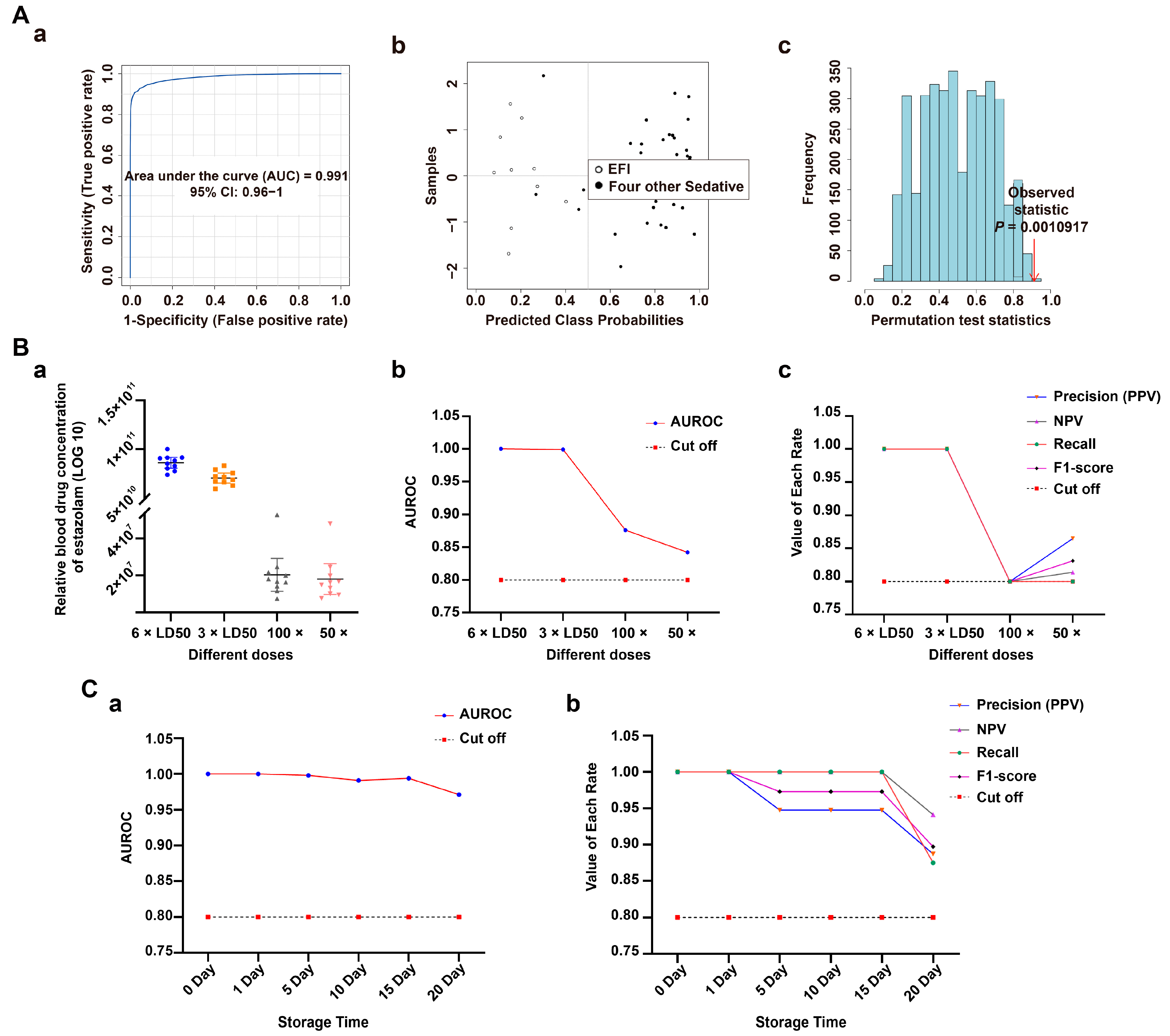
3.2. Classification Model Screening and Verification in EFI Plasma Samples Relative to NDRDs Mice
3.3. Metabolomics Profiling of Brainstem Tissue Samples in the EFI and NDRDs Mice
3.4. Classification Model Screening and Verification in EFI Brainstem Tissue Samples Relative to NDRDs Mice
3.5. New Toxicological Mechanism of Estazolam
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Gummin, D.D.; Mowry, J.B.; Spyker, D.A.; Brooks, D.E.; Fraser, M.O.; Banner, W. 2016 Annual Report of the American Association of Poison Control Centers’ National Poison Data System (NPDS): 34th Annual Report. Clin. Toxicol. 2017, 55, 1072–1252. [Google Scholar] [CrossRef] [PubMed]
- Gummin, D.D.; Mowry, J.B.; Spyker, D.A.; Brooks, D.E.; Osterthaler, K.M.; Banner, W. 2017 Annual Report of the American Association of Poison Control Centers’ National Poison Data System (NPDS): 35th Annual Report. Clin. Toxicol. 2018, 56, 1213–1415. [Google Scholar] [CrossRef] [PubMed]
- Gummin, D.D.; Mowry, J.B.; Spyker, D.A.; Brooks, D.E.; Beuhler, M.C.; Rivers, L.J.; Hashem, H.A.; Ryan, M.L. 2018 Annual Report of the American Association of Poison Control Centers’ National Poison Data System (NPDS): 36th Annual Report. Clin. Toxicol. 2019, 57, 1220–1413. [Google Scholar] [CrossRef]
- Gummin, D.D.; Mowry, J.B.; Beuhler, M.C.; Spyker, D.A.; Brooks, D.E.; Dibert, K.W.; Rivers, L.J.; Pham, N.P.T.; Ryan, M.L. 2019 Annual Report of the American Association of Poison Control Centers’ National Poison Data System (NPDS): 37th Annual Report. Clin. Toxicol. 2020, 58, 1360–1541. [Google Scholar] [CrossRef]
- Gummin, D.D.; Mowry, J.B.; Beuhler, M.C.; Spyker, D.A.; Bronstein, A.C.; Rivers, L.J.; Pham, N.P.T.; Weber, J. 2020 Annual Report of the American Association of Poison Control Centers’ National Poison Data System (NPDS): 38th Annual Report. Clin. Toxicol. 2021, 59, 1282–1501. [Google Scholar] [CrossRef]
- Nakano, H. SY01-5 current conditions and treatment of prescription drug abuse in Japan. Alcohol. Alcohol. 2014, 49, i3. [Google Scholar] [CrossRef]
- Värnik, A.; Sisask, M.; Värnik, P.; Wu, J.; Kõlves, K.; Arensman, E.; Maxwell, M.; Reisch, T.; Gusmão, R.; van Audenhove, C.; et al. Drug suicide: A sex-equal cause of death in 16 European countries. BMC Public. Health 2011, 11, 61. [Google Scholar] [CrossRef]
- Kuehn, B.M. Assessing a Veterinary Sedative’s Role in Drug Overdose Deaths. JAMA 2021, 326, 1573. [Google Scholar] [CrossRef]
- Kacinko, S.L.; Mohr, A.L.A.; Logan, B.K.; Barbieri, E.J. Xylazine: Pharmacology review and prevalence and drug combinations in forensic toxicology casework. J. Anal. Toxicol. 2022, 46, 911–917. [Google Scholar] [CrossRef]
- Dye, D.W.; McGwin, G.; Atherton, D.S.; McCleskey, B.; Davis, G.G. Correctly Identifying Deaths Due to Drug Toxicity without a Forensic Autopsy. Am. J. Forensic Med. Pathol. 2019, 40, 99–101. [Google Scholar] [CrossRef] [PubMed]
- Melo, P.; Costa, P.; Quintas, M.J.; Castro, A.; Tarelho, S.; Franco, J.M.; Teixeira, H.M. Pentobarbital in the context of possible suicides: Analysis of a Case. Forensic Sci. Int. 2017, 274, 109–112. [Google Scholar] [CrossRef]
- Ketola, R.A.; Ojanperä, I. Summary statistics for drug concentrations in post-mortem femoral blood representing all causes of death. Drug. Test. Anal. 2019, 11, 1326–1337. [Google Scholar] [CrossRef] [PubMed]
- Jönsson, A.K.; Söderberg, C.; Espnes, K.A.; Ahlner, J.; Eriksson, A.; Reis, M.; Druid, H. Sedative and hypnotic drugs--fatal and non-fatal reference blood concentrations. Forensic Sci. Int. 2014, 236, 138–145. [Google Scholar] [CrossRef]
- Mantinieks, D.; Gerostamoulos, D.; Glowacki, L.; Di Rago, M.; Schumann, J.; Woodford, N.W.; Drummer, O.H. Postmortem Drug Redistribution: A Compilation of Postmortem/Antemortem Drug Concentration Ratios. J. Anal. Toxicol. 2021, 45, 368–377. [Google Scholar] [CrossRef]
- Szeremeta, M.; Pietrowska, K.; Niemcunowicz-Janica, A.; Kretowski, A.; Ciborowski, M. Applications of Metabolomics in Forensic Toxicology and Forensic Medicine. Int. J. Mol. Sci. 2021, 22, 3010. [Google Scholar] [CrossRef]
- Dawidowska, J.; Krzyżanowska, M.; Markuszewski, M.J.; Kaliszan, M. The Application of Metabolomics in Forensic Science with Focus on Forensic Toxicology and Time-of-Death Estimation. Metabolites 2021, 11, 801. [Google Scholar] [CrossRef] [PubMed]
- Maeda, H.; Ishikawa, T.; Michiue, T. Forensic molecular pathology: Its impacts on routine work, education and training. Leg. Med. 2014, 16, 61–69. [Google Scholar] [CrossRef]
- Zhang, K.; Yan, H.; Liu, R.N.; Xiang, P.; Deng, K.F.; Zhang, J.; Tuo, Y.; Wang, Z.Y.; Huang, P. Exploring metabolic alterations associated with death from asphyxia and the differentiation of asphyxia from sudden cardiac death by GC-HRMS-based untargeted metabolomics. J. Chromatogr. B 2021, 1171, 122638. [Google Scholar] [CrossRef]
- Zhang, K.; Yan, H.; Liu, R.N.; Xiang, P.; Zhang, J.; Deng, K.F.; Huang, P.; Wang, Z.Y. The Use of Gas Chromatography Coupled with High-Resolution Mass Spectrometry-Based Untargeted Metabolomics to Discover Metabolic Changes and Help in the Determination of Complex Causes of Death: A Preliminary Study. ACS Omega 2021, 6, 2100–2109. [Google Scholar] [CrossRef]
- Bai, R.; Dai, X.H.; Miao, X.G.; Xie, B.; Yu, F.; Cong, B.; Wen, D.; Ma, C.L. Dynamic Changes in Plasma Metabolic Profiles Reveal a Potential Metabolite Panel for Interpretation of Fatal Intoxication by Chlorpromazine or Olanzapine in Mice. Metabolites 2022, 12, 1184. [Google Scholar] [CrossRef]
- Xu, J.N.; Chen, L.F.; Su, J.; Liu, Z.L.; Chen, J.; Lin, Q.F.; Mao, W.D.; Gao, Z.W.; Shen, D. The anxiolytic-like effects of estazolam on a PTSD animal model. Psychiatry Res. 2018, 269, 529–535. [Google Scholar] [CrossRef]
- Lou, G.; Yu, Z.; Chen, L.; Zhou, Y.; Zhang, L. Trends in Prescriptions for Insomnia in a Province in China Between 2015 and 2019. Front. Psychiatry 2022, 13, 915823. [Google Scholar] [CrossRef]
- Scotto di Tella, A.; Ricci, P.; Di Nunzio, C.; Cassandro, P. A new method for the determination in blood and urine of a novel triazolobenzodiazepine (estazolam) by HPLC. J. Anal. Toxicol. 1986, 10, 65–67. [Google Scholar] [CrossRef]
- Yu, S.; Na, J.Y.; Lee, Y.J.; Kim, K.T.; Park, J.T.; Kim, H.S. Forensic application of microRNA-706 as a biomarker for drowning pattern identification. Forensic Sci. Int. 2015, 255, 96–101. [Google Scholar] [CrossRef]
- Zeng, Y.; Lv, Y.; Tao, L.; Ma, J.; Zhang, H.; Xu, H.; Xiao, B.; Shi, Q.; Ma, K.; Chen, L. G6PC3, ALDOA and CS induction accompanies mir-122 down-regulation in the mechanical asphyxia and can serve as hypoxia biomarkers. Oncotarget 2016, 7, 74526–74536. [Google Scholar] [CrossRef] [PubMed]
- Tani, N.; Ikeda, T.; Aoki, Y.; Shida, A.; Oritani, S.; Ishikawa, T. Pathophysiological significance of clock genes BMAL1 and PER2 as erythropoietin-controlling factors in acute blood hemorrhage. Hum. Cell. 2019, 32, 275–284. [Google Scholar] [CrossRef]
- Wang, R.; Zhuang, C.; Su, J.; Qu, G.; Xu, S.; Pan, C.; Liu, L.; Liu, Q. A primary study on the influence of ethanol on the median lethal dose and metabolism in early stages of diazepam. Chin. J. Forens. Med. 2015, 30, 5–8. [Google Scholar] [CrossRef]
- Drugfuture. Chemical Identification: 2H-1,4-Benzodiazepin-2-one, 1,3-dihydro-7-nitro-5-phenyl-. Available online: https://www.drugfuture.com/toxic/q24-q496.html (accessed on 15 April 2023).
- Carmichael, E.B.; Johnson, W.H. The LD50 of pentobarbital sodium for both nursed and unnursed newborn rats. Anesthesiology 1951, 12, 340–343. [Google Scholar] [CrossRef]
- Wang, L. Study on the Forensic Toxicokinetics of Diazepam Poisoning Biomarkers. Ph.D. Thesis, Shanxi Medical University, Taiyuan, China, 2020. [Google Scholar]
- Gertsman, I.; Barshop, B.A. Promises and pitfalls of untargeted metabolomics. J. Inherit. Metab. Dis. 2018, 41, 355–366. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Sanderson, S.M.; Dai, Z.; Reid, M.A.; Cooper, D.E.; Lu, M.; Richie, J.P., Jr.; Ciccarella, A.; Calcagnotto, A.; Mikhael, P.G.; et al. Dietary methionine influences therapy in mouse cancer models and alters human metabolism. Nature 2019, 572, 397–401. [Google Scholar] [CrossRef]
- Huang, H.; Shi, L.Y.; Wei, L.L.; Han, Y.S.; Yi, W.J.; Pan, Z.W.; Jiang, T.T.; Chen, J.; Tu, H.H.; Li, Z.B.; et al. Plasma metabolites Xanthine, 4-Pyridoxate, and d-glutamic acid as novel potential biomarkers for pulmonary tuberculosis. Clin. Chim. Acta 2019, 498, 135–142. [Google Scholar] [CrossRef]
- Hemmer, S.; Manier, S.K.; Fischmann, S.; Westphal, F.; Wagmann, L.; Meyer, M.R. Comparison of Three Untargeted Data Processing Workflows for Evaluating LC-HRMS Metabolomics Data. Metabolites 2020, 10, 378. [Google Scholar] [CrossRef]
- Chong, J.; Wishart, D.S.; Xia, J. Using MetaboAnalyst 4.0 for Comprehensive and Integrative Metabolomics Data Analysis. Curr. Protoc. Bioinform. 2019, 68, e86. [Google Scholar] [CrossRef] [PubMed]
- Smith, A.M.; Neal, T.M.S. The distinction between discriminability and reliability in forensic science. Sci. Justice 2021, 61, 319–331. [Google Scholar] [CrossRef]
- Usman, M.; Gunjan, V.K.; Wajid, M.; Zubair, M.; Siddiquee, K.N. Speech as a Biomarker for COVID-19 Detection Using Machine Learning. Comput. Intell. Neurosci. 2022, 2022, 6093613. [Google Scholar] [CrossRef]
- Yu, Y.; Gao, Z.; Lou, J.; Mao, Z.; Li, K.; Chu, C.; Hu, L.; Li, Z.; Deng, C.; Fan, H.; et al. Identification of Serum-Based Metabolic Feature and Characteristic Metabolites in Paraquat Intoxicated Mouse Models. Front. Physiol. 2020, 11, 65. [Google Scholar] [CrossRef] [PubMed]
- Salman Khan, M.; Ullah, A.; Khan, K.N.; Riaz, H.; Yousafzai, Y.M.; Rahman, T.; Chowdhury, M.E.H.; Abul Kashem, S.B. Deep Learning Assisted Automated Assessment of Thalassaemia from Haemoglobin Electrophoresis Images. Diagnostics 2022, 12, 2405. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.; Mathelier, A. Intervene: A tool for intersection and visualization of multiple gene or genomic region sets. BMC Bioinform. 2017, 18, 287. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Xu, Z.; Lu, X.; Yang, X.; Yin, P.; Kong, H.; Yu, Y.; Xu, G. Comprehensive two-dimensional gas chromatography/time-of-flight mass spectrometry for metabonomics: Biomarker discovery for diabetes mellitus. Anal. Chim. Acta 2009, 633, 257–262. [Google Scholar] [CrossRef]
- Manna, S.K.; Patterson, A.D.; Yang, Q.; Krausz, K.W.; Li, H.; Idle, J.R.; Fornace, A.J., Jr.; Gonzalez, F.J. Identification of noninvasive biomarkers for alcohol-induced liver disease using urinary metabolomics and the Ppara-null mouse. J. Proteome Res. 2010, 9, 4176–4188. [Google Scholar] [CrossRef]
- Dieterle, F.; Ross, A.; Schlotterbeck, G.; Senn, H. Metabolite projection analysis for fast identification of metabolites in metabonomics. Application in an amiodarone study. Anal. Chem. 2006, 78, 3551–3561. [Google Scholar] [CrossRef] [PubMed]
- Cai, H.; Cao, T.; Li, N.; Fang, P.; Xu, P.; Wu, X.; Zhang, B.; Xiang, D. Quantitative monitoring of a panel of stress-induced biomarkers in human plasma by liquid chromatography-tandem mass spectrometry: An application in a comparative study between depressive patients and healthy subjects. Anal. Bioanal. Chem. 2019, 411, 5765–5777. [Google Scholar] [CrossRef] [PubMed]
- Fawcett, T. An introduction to ROC analysis. Pattern Recogn. Lett. 2006, 27, 861–874. [Google Scholar] [CrossRef]
- Deng, X.; Liu, Q.; Deng, Y.; Mahadevan, S. An improved method to construct basic probability assignment based on the confusion matrix for classification problem. Inform. Sci. 2016, 340, 250–261. [Google Scholar] [CrossRef]
- Kurian, B.; Jyothi, V.L. Comparative Analysis of Machine Learning Methods for Breast Cancer Classification in Genetic Sequences. J. Environ. Public. Health 2022, 2022, 7199290. [Google Scholar] [CrossRef]
- McCall, W.V.; Benca, R.M.; Rosenquist, P.B.; Riley, M.A.; McCloud, L.; Newman, J.C.; Case, D.; Rumble, M.; Krystal, A.D. Hypnotic medications and suicide: Risk, mechanisms, mitigation, and the FDA. Am. J. Psychiatry 2017, 174, 18–25. [Google Scholar] [CrossRef] [PubMed]
- Søreide, K.; Kørner, H.; Søreide, J.A. Diagnostic accuracy and receiver-operating characteristics curve analysis in surgical research and decision making. Ann. Surg. 2011, 253, 27–34. [Google Scholar] [CrossRef] [PubMed]
- Leandro, J.; Houten, S.M. Saccharopine, a lysine degradation intermediate, is a mitochondrial toxin. J. Cell. Biol. 2019, 218, 391–392. [Google Scholar] [CrossRef]
- Zhou, J.; Wang, X.; Wang, M.; Chang, Y.; Zhang, F.; Ban, Z.; Tang, R.; Gan, Q.; Wu, S.; Guo, Y.; et al. The lysine catabolite saccharopine impairs development by disrupting mitochondrial homeostasis. J. Cell. Biol. 2019, 218, 580–597. [Google Scholar] [CrossRef]


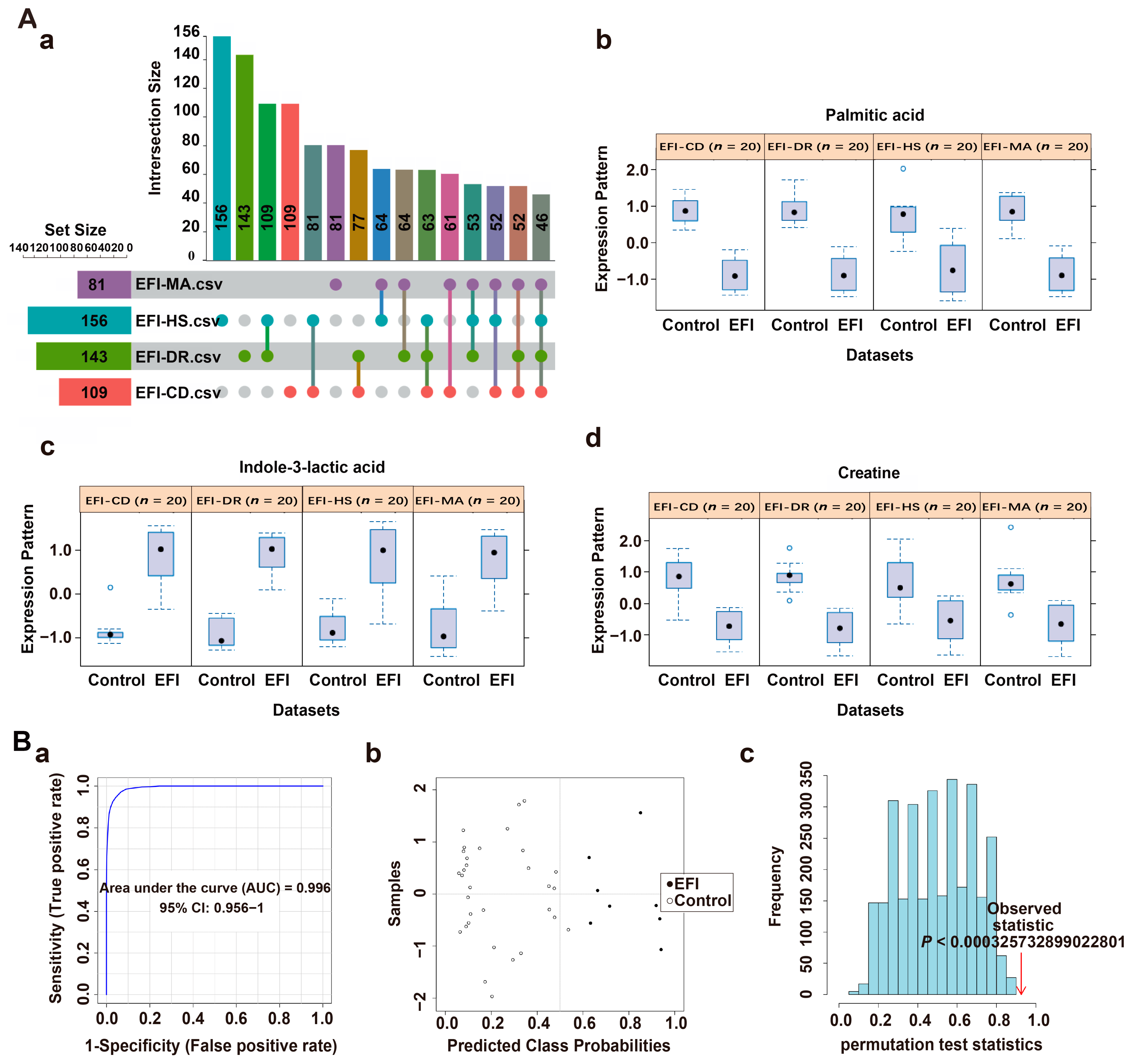


| Compound Name a | Metabolite Identification | p d | Combined Log FC e | Trend f | Combined VIP g | ||
|---|---|---|---|---|---|---|---|
| Accurate Mass b | Retention Time b | mzCloud Best Match c | |||||
| Phenylacetylglycine | 193.07388 | 6.414 | 90 | 1.52 × 10−33 | 1.822502209 | up | 1.27002 |
| 7-Methylguanine | 165.06493 | 1.767 | 87.1 | 1.26 × 10−32 | 1.812534031 | up | 1.03212 |
| Propionylcarnitine | 217.13116 | 2.578 | 92.9 | 9.31 × 10−32 | −1.80225988 | down | 1.09656 |
| Creatine | 131.06937 | 1.489 | 95.2 | 4.95 × 10−30 | 1.780471987 | up | 1.2083 |
| L-Ascorbic acid 2-sulfate | 255.98905 | 1.222 | 88 | 4.22 × 10−28 | 1.754421741 | up | 1.07764 |
| Methionine | 149.0509 | 1.67 | 91.8 | 3.98 × 10−25 | −1.706198279 | down | 1.27548 |
| Ascorbic acid | 176.03203 | 1.226 | 87.4 | 1.54 × 10−23 | 1.676402203 | up | 1.11066 |
| N-Isovalerylglycine | 159.08942 | 5.848 | 89.3 | 3.21 × 10−22 | 1.649229414 | up | 1.1655 |
| 4-Pyridoxic acid | 183.05305 | 3.268 | 94.8 | 1.91 × 10−21 | 1.632156605 | up | 1.16982 |
| Xanthurenic acid | 205.03731 | 5.402 | 91.2 | 6.65 × 10−21 | 1.619327608 | up | 1.25248 |
| Acetyl-L-carnitine | 203.11558 | 1.659 | 93.8 | 4.74 × 10−20 | −1.599262594 | down | 1.09352 |
| Indole-3-lactic acid | 205.07359 | 6.928 | 91.8 | 1.62 × 10−18 | 1.560302287 | up | 1.060518 |
| Valine | 117.0788 | 1.619 | 95.8 | 8.78 × 10−15 | −1.443708344 | down | 1.004272 |
| 3-Phenyllactic acid | 166.06286 | 6.78 | 91.1 | 5.48 × 10−7 | 1.036145258 | up | 1.18646 |
| Compound Name | AUC | 95%CI | p |
|---|---|---|---|
| Phenylacetylglycine | 1 | 1−1 | 11.3668 × 10−15 |
| Creatine | 0.9975 | 0.989−1 | 3.4137 × 10−15 |
| Indole-3-lactic acid | 0.96 | 0.894−1 | 1.5252 × 10−10 |
| Compound Name | Metabolite Identification | p | Combined Log FC | Trend | Combined VIP | ||
|---|---|---|---|---|---|---|---|
| Accurate Mass | Retention Time | mzCloud Best Match | |||||
| Palmitic acid | 256.23977 | 9.512 | 96.3 | 2.09 × 10−21 | −1.653094773 | down | 1.63824 |
| Prostaglandin D2 | 352.22435 | 8.491 | 93.1 | 3.22 × 10−21 | −1.648333391 | down | 1.8836 |
| Indole-3-lactic acid | 205.07668 | 6.625 | 89.6 | 7.08 × 10−21 | 1.640230298 | up | 1.72006 |
| Creatine | 131.06942 | 1.05 | 96.7 | 2.39 × 10−15 | −1.48559731 | down | 1.24968 |
| DL-Tryptophan | 204.08967 | 5.174 | 87.3 | 2.49 × 10−15 | 1.483820281 | up | 1.48568 |
| Indole-3-acrylic acid | 187.06325 | 5.176 | 93.4 | 3.98 × 10−15 | 1.476502537 | up | 1.48258 |
| Compound Name | AUC | 95%CI | p |
|---|---|---|---|
| Palmitic acid | 0.98 | 0.939−1 | 9.6901 × 10−11 |
| Indole-3-lactic acid | 0.973 | 0.914−1 | 1.6272 × 10−9 |
| Creatine | 0.958 | 0.896−0.992 | 3.0926 × 10−7 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dai, X.; Bai, R.; Xie, B.; Xiang, J.; Miao, X.; Shi, Y.; Yu, F.; Cong, B.; Wen, D.; Ma, C. A Metabolomics-Based Study on the Discriminative Classification Models and Toxicological Mechanism of Estazolam Fatal Intoxication. Metabolites 2023, 13, 567. https://doi.org/10.3390/metabo13040567
Dai X, Bai R, Xie B, Xiang J, Miao X, Shi Y, Yu F, Cong B, Wen D, Ma C. A Metabolomics-Based Study on the Discriminative Classification Models and Toxicological Mechanism of Estazolam Fatal Intoxication. Metabolites. 2023; 13(4):567. https://doi.org/10.3390/metabo13040567
Chicago/Turabian StyleDai, Xiaohui, Rui Bai, Bing Xie, Jiahong Xiang, Xingang Miao, Yan Shi, Feng Yu, Bin Cong, Di Wen, and Chunling Ma. 2023. "A Metabolomics-Based Study on the Discriminative Classification Models and Toxicological Mechanism of Estazolam Fatal Intoxication" Metabolites 13, no. 4: 567. https://doi.org/10.3390/metabo13040567
APA StyleDai, X., Bai, R., Xie, B., Xiang, J., Miao, X., Shi, Y., Yu, F., Cong, B., Wen, D., & Ma, C. (2023). A Metabolomics-Based Study on the Discriminative Classification Models and Toxicological Mechanism of Estazolam Fatal Intoxication. Metabolites, 13(4), 567. https://doi.org/10.3390/metabo13040567