A Novel Approach on the Use of Samples from Faecal Occult Blood Screening Kits for Metabolomics Analysis: Application in Colorectal Cancer Population
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemicals
2.2. Clinical Samples and Study Population
2.3. Sample Collection and Metabolite Extraction
2.4. UHPLC-MS Metabolic Profiling
2.5. Data Pre-Processing
2.6. Data Normalization and Quality Control
2.7. Statistical Analysis
3. Results
3.1. Reproducibility of Metabolite Extraction Procedure (Batch 1)
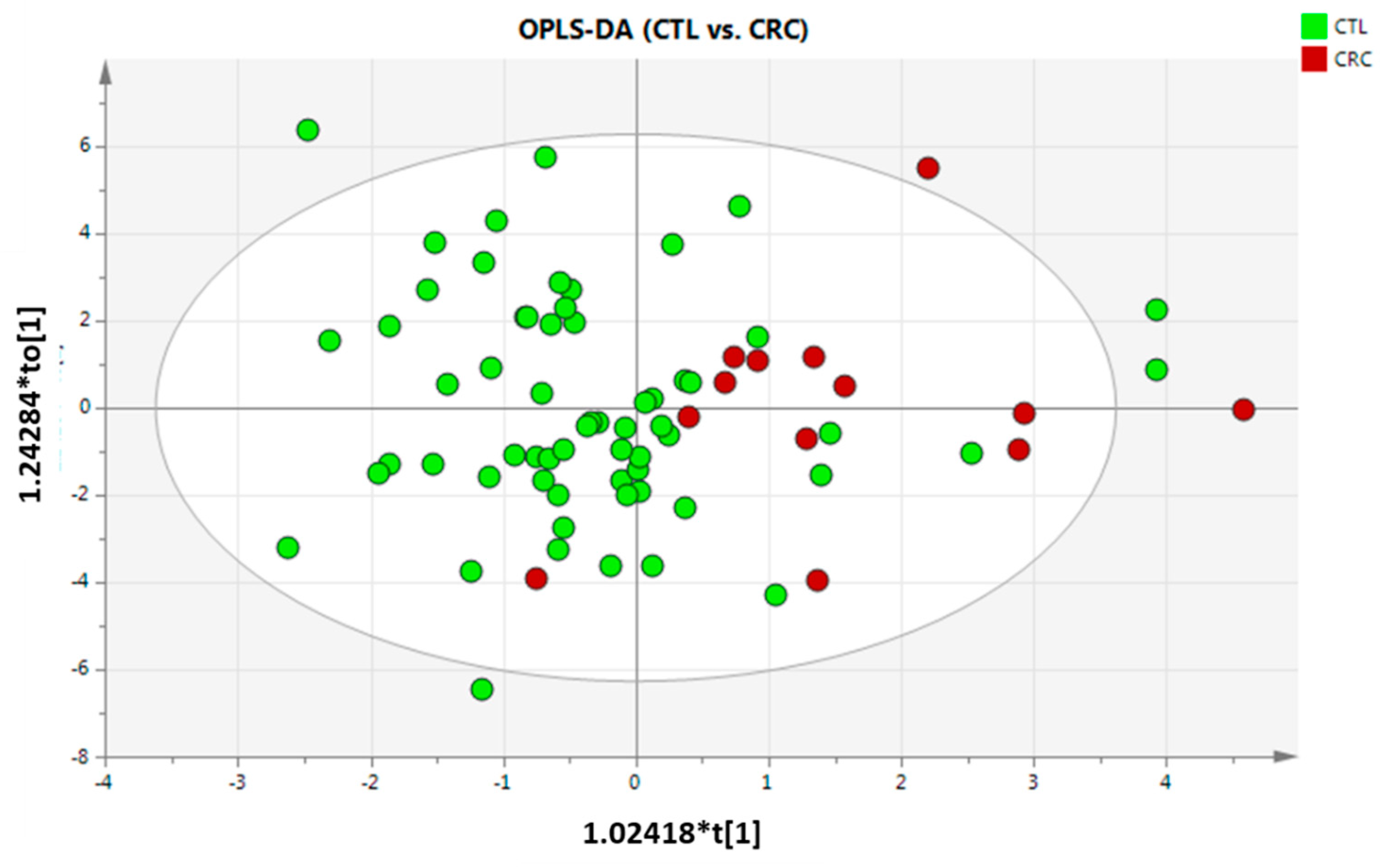
3.2. Metabolic Differences per Group
3.3. Fusion of Independent Studies
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Amir Hashim, N.A.; Ab-Rahim, S.; Wan Ngah, W.Z.; Nathan, S.; Ab Mutalib, N.S.; Sagap, I.; Jamal, A.R.A.; Mazlan, M. Global Metabolomics Profiling of Colorectal Cancer in Malaysian Patients. Bioimpacts 2021, 11, 33–43. [Google Scholar] [CrossRef] [PubMed]
- Fearon, E.R.; Vogelstein, B. A Genetic Model for Colorectal Tumorigenesis. Cell 1990, 61, 759–767. [Google Scholar] [CrossRef] [PubMed]
- Jones, S.; Chen, W.-D.; Parmigiani, G.; Diehl, F.; Beerenwinkel, N.; Antal, T.; Traulsen, A.; Nowak, M.A.; Siegel, C.; Velculescu, V.E.; et al. Comparative Lesion Sequencing Provides Insights into Tumor Evolution. Proc. Natl. Acad. Sci. USA 2008, 105, 4283–4288. [Google Scholar] [CrossRef] [PubMed]
- Erben, V.; Bhardwaj, M.; Schrotz-King, P.; Brenner, H. Metabolomics Biomarkers for Detection of Colorectal Neoplasms: A Systematic Review. Cancers 2018, 10, 246. [Google Scholar] [CrossRef] [PubMed]
- Hashim, N.A.A.; Ab-Rahim, S.; Suddin, L.S.; Saman, M.S.A.; Mazlan, M. Global Serum Metabolomics Profiling of Colorectal Cancer. Mol. Clin. Oncol. 2019, 11, 3–14. [Google Scholar] [CrossRef]
- Zhang, F.; Zhang, Y.; Zhao, W.; Deng, K.; Wang, Z.; Yang, C.; Ma, L.; Openkova, M.S.; Hou, Y.; Li, K. Metabolomics for Biomarker Discovery in the Diagnosis, Prognosis, Survival and Recurrence of Colorectal Cancer: A Systematic Review. Oncotarget 2017, 8, 35460–35472. [Google Scholar] [CrossRef] [PubMed]
- Ni, Y.; Xie, G.; Jia, W. Metabonomics of Human Colorectal Cancer: New Approaches for Early Diagnosis and Biomarker Discovery. J. Proteome Res. 2014, 13, 3857–3870. [Google Scholar] [CrossRef]
- Gold, A.; Choueiry, F.; Jin, N.; Mo, X.; Zhu, J. The Application of Metabolomics in Recent Colorectal Cancer Studies: A State-of-the-Art Review. Cancers 2022, 14, 725. [Google Scholar] [CrossRef]
- Yusof, H.M.; Ab-Rahim, S.; Suddin, L.S.; Saman, M.S.A.; Mazlan, M. Metabolomics Profiling on Different Stages of Colorectal Cancer: A Systematic Review. Malays. J. Med. Sci. 2018, 25, 16–34. [Google Scholar] [CrossRef]
- Clos-Garcia, M.; Garcia, K.; Alonso, C.; Iruarrizaga-Lejarreta, M.; D’Amato, M.; Crespo, A.; Iglesias, A.; Cubiella, J.; Bujanda, L.; Falcón-Pérez, J.M. Integrative Analysis of Fecal Metagenomics and Metabolomics in Colorectal Cancer. Cancers 2020, 12, 1142. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Etxebarria, K.; Clos-Garcia, M.; Telleria, O.; Nafría, B.; Alonso, C.; Iruarrizaga-Lejarreta, M.; Franke, A.; Crespo, A.; Iglesias, A.; Cubiella, J.; et al. Interplay between Genome, Metabolome and Microbiome in Colorectal Cancer. Cancers 2021, 13, 6216. [Google Scholar] [CrossRef] [PubMed]
- Phua, L.C.; Chue, X.P.; Koh, P.K.; Cheah, P.Y.; Ho, H.K.; Chan, E.C.Y. Non-Invasive Fecal Metabonomic Detection of Colorectal Cancer. Cancer Biol. Ther. 2014, 15, 389–397. [Google Scholar] [CrossRef] [PubMed]
- Cubiella, J.; Clos-Garcia, M.; Alonso, C.; Martinez-Arranz, I.; Perez-Cormenzana, M.; Barrenetxea, Z.; Berganza, J.; Rodríguez-Llopis, I.; D’Amato, M.; Bujanda, L.; et al. Targeted UPLC-MS Metabolic Analysis of Human Faeces Reveals Novel Low-Invasive Candidate Markers for Colorectal Cancer. Cancers 2018, 10, 300. [Google Scholar] [CrossRef]
- Song, E.M.; Byeon, J.-S.; Lee, S.M.; Yoo, H.J.; Kim, S.J.; Lee, S.-H.; Chang, K.; Hwang, S.W.; Yang, D.-H.; Jeong, J.-Y. Fecal Fatty Acid Profiling as a Potential New Screening Biomarker in Patients with Colorectal Cancer. Dig. Dis. Sci. 2018, 63, 1229–1236. [Google Scholar] [CrossRef] [PubMed]
- Goedert, J.J.; Sampson, J.N.; Moore, S.C.; Xiao, Q.; Xiong, X.; Hayes, R.B.; Ahn, J.; Shi, J.; Sinha, R. Fecal Metabolomics: Assay Performance and Association with Colorectal Cancer. Carcinogenesis 2014, 35, 2089–2096. [Google Scholar] [CrossRef]
- Brown, D.G.; Rao, S.; Weir, T.L.; O’Malia, J.; Bazan, M.; Brown, R.J.; Ryan, E.P. Metabolomics and Metabolic Pathway Networks from Human Colorectal Cancers, Adjacent Mucosa, and Stool. Cancer Metab. 2016, 4, 11. [Google Scholar] [CrossRef]
- Sinha, R.; Ahn, J.; Sampson, J.N.; Shi, J.; Yu, G.; Xiong, X.; Hayes, R.B.; Goedert, J.J. Fecal Microbiota, Fecal Metabolome, and Colorectal Cancer Interrelations. PLoS ONE 2016, 11, e0152126. [Google Scholar] [CrossRef]
- Yang, Y.; Misra, B.B.; Liang, L.; Bi, D.; Weng, W.; Wu, W.; Cai, S.; Qin, H.; Goel, A.; Li, X.; et al. Integrated Microbiome and Metabolome Analysis Reveals a Novel Interplay between Commensal Bacteria and Metabolites in Colorectal Cancer. Theranostics 2019, 9, 4101–4114. [Google Scholar] [CrossRef]
- Weir, T.L.; Manter, D.K.; Sheflin, A.M.; Barnett, B.A.; Heuberger, A.L.; Ryan, E.P. Stool Microbiome and Metabolome Differences between Colorectal Cancer Patients and Healthy Adults. PLoS ONE 2013, 8, e70803. [Google Scholar] [CrossRef]
- Kim, M.; Vogtmann, E.; Ahlquist, D.A.; Devens, M.E.; Kisiel, J.B.; Taylor, W.R.; White, B.A.; Hale, V.L.; Sung, J.; Chia, N.; et al. Fecal Metabolomic Signatures in Colorectal Adenoma Patients Are Associated with Gut Microbiota and Early Events of Colorectal Cancer Pathogenesis. mBio 2020, 11, e03186-19. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.J.; Yang, J.; Seo, H.; Lee, W.H.; Lee, D.H.; Kym, S.; Park, Y.S.; Kim, J.G.; Jang, I.-J.; Kim, Y.-K.; et al. Colorectal Cancer Diagnostic Model Utilizing Metagenomic and Metabolomic Data of Stool Microbial Extracellular Vesicles. Sci. Rep. 2020, 10, 2860. [Google Scholar] [CrossRef] [PubMed]
- Cubiella, J.; Vega, P.; Salve, M.; Díaz-Ondina, M.; Alves, M.T.; Quintero, E.; Álvarez-Sánchez, V.; Fernández-Bañares, F.; Boadas, J.; Campo, R.; et al. Development and External Validation of a Faecal Immunochemical Test-Based Prediction Model for Colorectal Cancer Detection in Symptomatic Patients. BMC Med. 2016, 14, 128. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Arranz, I.; Mayo, R.; Pérez-Cormenzana, M.; Mincholé, I.; Salazar, L.; Alonso, C.; Mato, J.M. Enhancing Metabolomics Research through Data Mining. J. Proteom. 2015, 127, 275–288. [Google Scholar] [CrossRef] [PubMed]
- Schiffman, C.; Petrick, L.; Perttula, K.; Yano, Y.; Carlsson, H.; Whitehead, T.; Metayer, C.; Hayes, J.; Rappaport, S.; Dudoit, S. Filtering Procedures for Untargeted LC-MS Metabolomics Data. BMC Bioinform. 2019, 20, 334. [Google Scholar] [CrossRef] [PubMed]
- Want, E.J.; Masson, P.; Michopoulos, F.; Wilson, I.D.; Theodoridis, G.; Plumb, R.S.; Shockcor, J.; Loftus, N.; Holmes, E.; Nicholson, J.K. Global Metabolic Profiling of Animal and Human Tissues via UPLC-MS. Nat. Protoc. 2013, 8, 17–32. [Google Scholar] [CrossRef] [PubMed]
- Yan, G.; Li, L.; Zhu, B.; Li, Y. Lipidome in Colorectal Cancer. Oncotarget 2016, 7, 33429–33439. [Google Scholar] [CrossRef]
- Mika, A.; Kobiela, J.; Pakiet, A.; Czumaj, A.; Sokołowska, E.; Makarewicz, W.; Chmielewski, M.; Stepnowski, P.; Marino-Gammazza, A.; Sledzinski, T. Preferential Uptake of Polyunsaturated Fatty Acids by Colorectal Cancer Cells. Sci. Rep. 2020, 10, 1954. [Google Scholar] [CrossRef]
- Czumaj, A.; Zabielska, J.; Pakiet, A.; Mika, A.; Rostkowska, O.; Makarewicz, W.; Kobiela, J.; Sledzinski, T.; Stelmanska, E. In Vivo Effectiveness of Orlistat in the Suppression of Human Colorectal Cancer Cell Proliferation. Anticancer Res. 2019, 39, 3815–3822. [Google Scholar] [CrossRef]
- Roynette, C.E.; Calder, P.C.; Dupertuis, Y.M.; Pichard, C. N-3 Polyunsaturated Fatty Acids and Colon Cancer Prevention. Clin. Nutr. 2004, 23, 139–151. [Google Scholar] [CrossRef]
- Kim, S.H.; Roh, K.H.; Park, J.-S.; Kim, K.-S.; Kim, H.U.; Lee, K.-R.; Kang, H.-C.; Kim, J.-B. Heterologous Reconstitution of Omega-3 Polyunsaturated Fatty Acids in Arabidopsis. BioMed Res. Int. 2015, 2015, 768478. [Google Scholar] [CrossRef] [PubMed]
- Park, W.J.; Kothapalli, K.S.D.; Lawrence, P.; Tyburczy, C.; Brenna, J.T. An Alternate Pathway to Long-Chain Polyunsaturates: The FADS2 Gene Product Δ8-Desaturates 20:2n-6 and 20:3n-3. J. Lipid Res. 2009, 50, 1195–1202. [Google Scholar] [CrossRef] [PubMed]
- Pickens, C.A.; Albuquerque Pereira, M.d.F.; Fenton, J.I. Long-Chain ω-6 Plasma Phospholipid Polyunsaturated Fatty Acids and Association with Colon Adenomas in Adult Men: A Cross-Sectional Study. Eur. J. Cancer Prev. 2017, 26, 497–505. [Google Scholar] [CrossRef] [PubMed]
- McEntee, M.F.; Whelan, J. Dietary Polyunsaturated Fatty Acids and Colorectal Neoplasia. Biomed. Pharmacother. 2002, 56, 380–387. [Google Scholar] [CrossRef]
- Xie, C.; Woollett, L.A.; Turley, S.D.; Dietschy, J.M. Fatty Acids Differentially Regulate Hepatic Cholesteryl Ester Formation and Incorporation into Lipoproteins in the Liver of the Mouse. J. Lipid Res. 2002, 43, 1508–1519. [Google Scholar] [CrossRef]
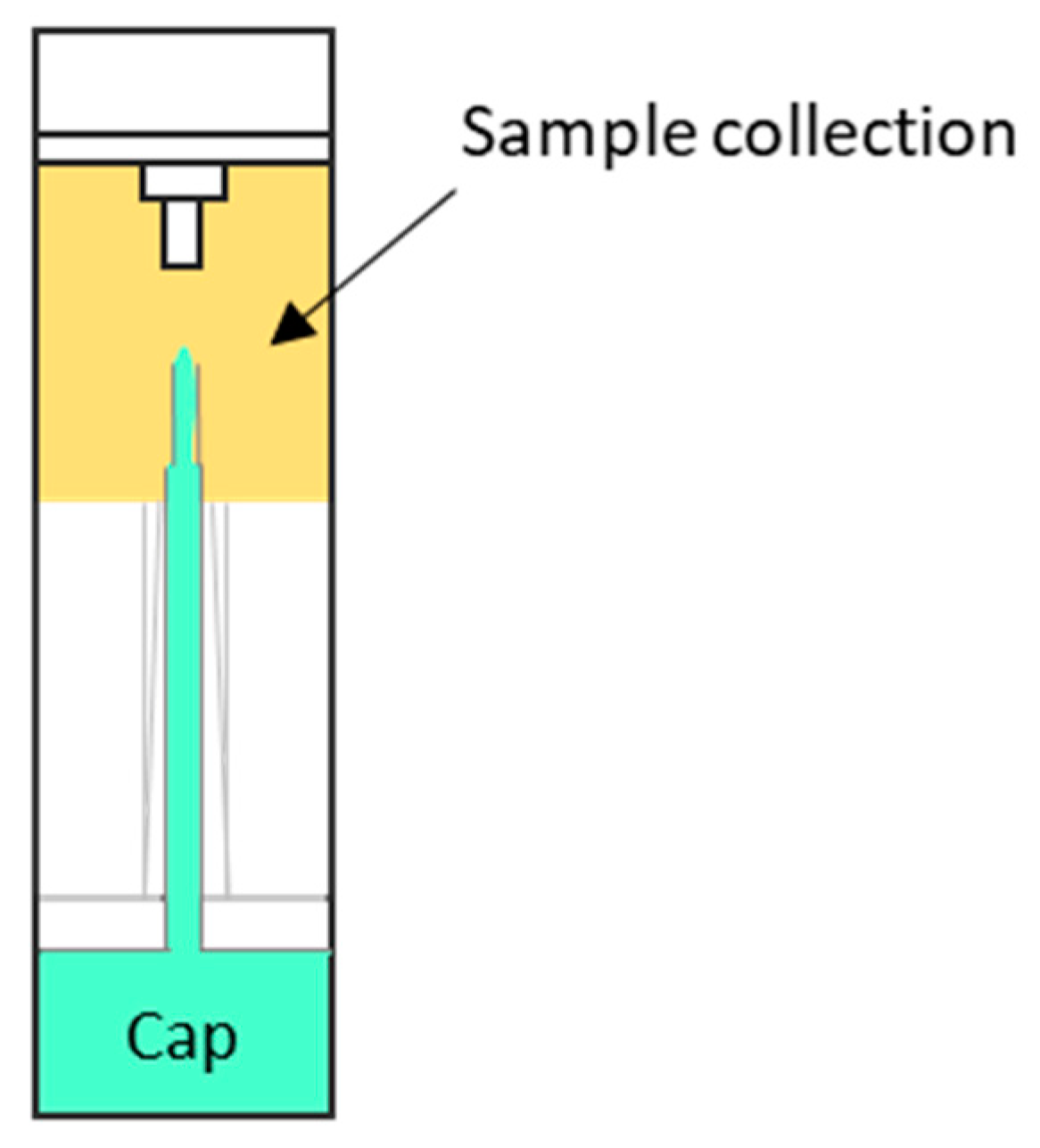
| Batch | Experimental Group | Group Code | Number of Samples | Gender (% Women) | Age, Median |
|---|---|---|---|---|---|
| Batch 1 | Control | CTL | 3 | NA 1 | NA 1 |
| Adenoma | AD | 3 | NA 1 | NA 1 | |
| Colorectal cancer | CRC | 3 | NA 1 | NA 1 | |
| Batch 2 | Control | CTL | 11 | 54 | 61.5 (24–70) 2 |
| Adenoma | AD | 11 | 18 | 60 (53–64) 2 | |
| Colorectal cancer | CRC | 11 | 45 | 62 (50–69) 2 | |
| Batch 3 | Control | CTL | 51 | 55 | 61 (51–70) 2 |
| Adenoma | AD | 45 | 47 | 63 (50–71) 2 | |
| Colorectal cancer | CRC | 2 | 50 | 70 (69–71) 2 | |
| Batch 2 + 3 | Control | CTL | 62 | 41 | 61 (24–70) 2 |
| Adenoma | AD | 56 | 56 | 63 (50–71) 2 | |
| Colorectal cancer | CRC | 13 | 46 | 65 (50–71) 2 |
| Adenoma vs. Colorectal Cancer (AD vs. CTL) | Colorectal Cancer vs. Control (CRC vs. CTL) | Colorectal Cancer vs. Adenoma (CRC vs. AD) | ||
|---|---|---|---|---|
| FOB | q-value | 1 | 0.0012 | 0.0186 |
| log2(FC) | −0.6748 | 1.6339 | 2.3088 | |
| ChoE (16:0) | q-value | 1 | 1 | 1 |
| log2(FC) | 0.0791 | −0.0182 | −0.0973 | |
| ChoE (18:1) | q-value | 0.3990 | 0.1419 | 1 |
| log2(FC) | −0.1491 | −0.1936 | −0.04453 | |
| ChoE (18:2) | q-value | 1 | 0.3042 | 0.0385 |
| log2(FC) | −0.7916 | 0.3736 | 1.1653 | |
| ChoE (18:3) | q-value | 1 | 1 | 1 |
| log2(FC) | −0.2334 | −0.1391 | 0.09433 | |
| ChoE (20:2) | q-value | 1 | 1 | 1 |
| log2(FC) | 0.6879 | −0.1814 | −0.8693 | |
| ChoE (20:4) | q-value | 1 | 0.0473 | 0.0473 |
| log2(FC) | −0.7378 | 1.3763 | 2.1141 | |
| ChoE (20:5) | q-value | 1 | 1 | 1 |
| log2(FC) | 0.3711 | 2.2539 | 1.8828 | |
| ChoE (22:4) | q-value | 1 | 1 | 0.6508 |
| log2(FC) | −0.3206 | −0.6638 | −0.3432 | |
| ChoE (22:5) | q-value | 1 | 1 | 1 |
| log2(FC) | −0.5777 | 1.2950 | 1.8732 | |
| ChoE (22:6) | q-value | 1 | 0.8109 | 1 |
| log2(FC) | 1.1112 | 1.6021 | 0.4909 | |
| PE (16:0/18:1) | q-value 1 | 0.7886 | 1 | 0.4757 |
| log2(FC) | 0.5419 | −0.1871 | −0.7290 | |
| SM (d18:1/23:0) | q-value | 1 | 1 | 1 |
| log2(FC) | −1.5919 | −0.6195 | 0.9724 | |
| SM (42:3) | q-value | 1 | 1 | 1 |
| log2(FC) | −0.5073 | 0.0852 | 0.5924 | |
| TG (54:1) | q-value | 1 | 1 | 1 |
| log2(FC) | −1.1873 | −2.9308 | −1.7435 |
| Adenoma vs. Control (AD vs. CTL) | ||
|---|---|---|
| FOB | q-value | 1 |
| log2(FC) | −0.3327 | |
| ChoE (16:0) | q-value | 1 |
| log2(FC) | 0.2828 | |
| ChoE (18:1) | q-value | 0.8655 |
| log2(FC) | 0.9083 | |
| ChoE (18:2) | q-value | 1 |
| log2(FC) | 0.5871 | |
| ChoE (18:3) | q-value | 0.6151 |
| log2(FC) | 0.7292 | |
| ChoE (20:2) | q-value | 0.7600 |
| log2(FC) | 1.0948 | |
| ChoE (20:4) | q-value | 1 |
| log2(FC) | 0.6010 | |
| ChoE (20:5) | q-value | 1 |
| log2(FC) | 0.4355 | |
| ChoE (22:4) | q-value | 1 |
| log2(FC) | 0.9314 | |
| ChoE (22:5) | q-value | 0.9695 |
| log2(FC) | 0.2337 | |
| ChoE (22:6) | q-value | 1 |
| log2(FC) | 0.3159 | |
| PE (16:0/18:1) | q-value | 0.7418 |
| log2(FC) | 0.0263 | |
| SM (d18:1/23:0) | q-value | 1 |
| log2(FC) | 0.0510 | |
| SM (42:3) | q-value | 0.4398 |
| log2(FC) | 0.3600 | |
| TG (54:1) | q-value | 1 |
| log2(FC) | 0.0009 |
| Experimental Group | Group Code | Batch | Number of Samples | Total Number of Samples |
|---|---|---|---|---|
| Control | CTL | 2 | 11 | 62 |
| 3 | 51 | |||
| Adenoma | AD | 2 | 11 | 56 |
| 3 | 45 | |||
| Colorectal cancer | CRC | 2 | 11 | 13 |
| 3 | 2 |
| AD vs. CTL | CRC vs. CTL | CRC vs. AD | ||
|---|---|---|---|---|
| FOB | q-value | 1 | 0.0186 | 0.0012 |
| log2(FC) | −0.6749 | 1.6339 | 2.3088 | |
| ChoE (16:0) | q-value | 1 | 1 | 1 |
| log2(FC) | 0.2446 | 0.1119 | −0.1327 | |
| ChoE (18:1) | q-value | 0.3847 | 0.8639 | 1 |
| log2(FC) | 0.7526 | −0.0471 | −0.7980 | |
| ChoE (18:2) | q-value | 1 | 0.0384 | 0.1852 |
| log2(FC) | 0.3965 | 0.4982 | 0.1017 | |
| ChoE (18:3) | q-value | 0.8146 | 0.3290 | 1 |
| log2(FC) | 0.5663 | 0.2705 | −0.2958 | |
| ChoE (20:2) | q-value | 0.6880 | 1 | 1 |
| log2(FC) | 1.0329 | 0.9844 | −0.0484 | |
| ChoE (20:4) | q-value | 0.2518 | 0.0384 | 0.1329 |
| log2(FC) | 0.4464 | 1.1898 | 0.7435 | |
| ChoE (20:5) | q-value | 1 | 0.2294 | 0.8909 |
| log2(FC) | 0.4231 | 2.0564 | 1.6333 | |
| ChoE (22:4) | q-value | 1 | 1 | 0.9124 |
| log2(FC) | 0.6970 | 0.1098 | −0.5872 | |
| ChoE (22:5) | q-value | 0.7798 | 1 | 1 |
| log2(FC) | 0.1128 | 1.0324 | 0.9195 | |
| ChoE (22:6) | q-value | 1 | 0.7371 | 1 |
| log2(FC) | 0.4306 | 0.7851 | 0.3545 | |
| PE (16:0/18:1) | q-value | 0.3456 | 1 | 1 |
| log2(FC) | 0.0966 | 0.5939 | 0.4973 | |
| SM (d18:1/23:0) | q-value | 1 | 0.4713 | 0.6079 |
| log2(FC) | −0.3906 | 0.3918 | 0.7824 | |
| SM (42:3) | q-value | 0.6272 | 0.0112 | 0.0589 |
| log2(FC) | 0.0754 | 1.1902 | 1.1148 | |
| TG (54:1) | q-value | 1 | 1 | 1 |
| log2(FC) | −0.2906 | −0.6918 | −0.4012 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Albóniga, O.E.; Cubiella, J.; Bujanda, L.; Blanco, M.E.; Lanza, B.; Alonso, C.; Nafría, B.; Falcón-Pérez, J.M. A Novel Approach on the Use of Samples from Faecal Occult Blood Screening Kits for Metabolomics Analysis: Application in Colorectal Cancer Population. Metabolites 2023, 13, 321. https://doi.org/10.3390/metabo13030321
Albóniga OE, Cubiella J, Bujanda L, Blanco ME, Lanza B, Alonso C, Nafría B, Falcón-Pérez JM. A Novel Approach on the Use of Samples from Faecal Occult Blood Screening Kits for Metabolomics Analysis: Application in Colorectal Cancer Population. Metabolites. 2023; 13(3):321. https://doi.org/10.3390/metabo13030321
Chicago/Turabian StyleAlbóniga, Oihane E., Joaquín Cubiella, Luis Bujanda, María Encarnación Blanco, Borja Lanza, Cristina Alonso, Beatriz Nafría, and Juan Manuel Falcón-Pérez. 2023. "A Novel Approach on the Use of Samples from Faecal Occult Blood Screening Kits for Metabolomics Analysis: Application in Colorectal Cancer Population" Metabolites 13, no. 3: 321. https://doi.org/10.3390/metabo13030321
APA StyleAlbóniga, O. E., Cubiella, J., Bujanda, L., Blanco, M. E., Lanza, B., Alonso, C., Nafría, B., & Falcón-Pérez, J. M. (2023). A Novel Approach on the Use of Samples from Faecal Occult Blood Screening Kits for Metabolomics Analysis: Application in Colorectal Cancer Population. Metabolites, 13(3), 321. https://doi.org/10.3390/metabo13030321









