Cupriavidus pinatubonensis JMP134 Alleviates Sulfane Sulfur Toxicity after the Loss of Sulfane Dehydrogenase through Oxidation by Persulfide Dioxygenase and Hydrogen Sulfide Release
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains, Primers and Media
2.2. Thiosulfate Oxidation by Whole-Cell and Product Detection
2.3. Real-Time Quantitative Reverse Transcription PCR (RT–qPCR)
2.4. Bioinformatics Analysis
2.5. Statistical Analysis
3. Results
3.1. Thiosulfate Oxidation by Wild-Type and Mutant Strains of C. pinatubonensis JMP134
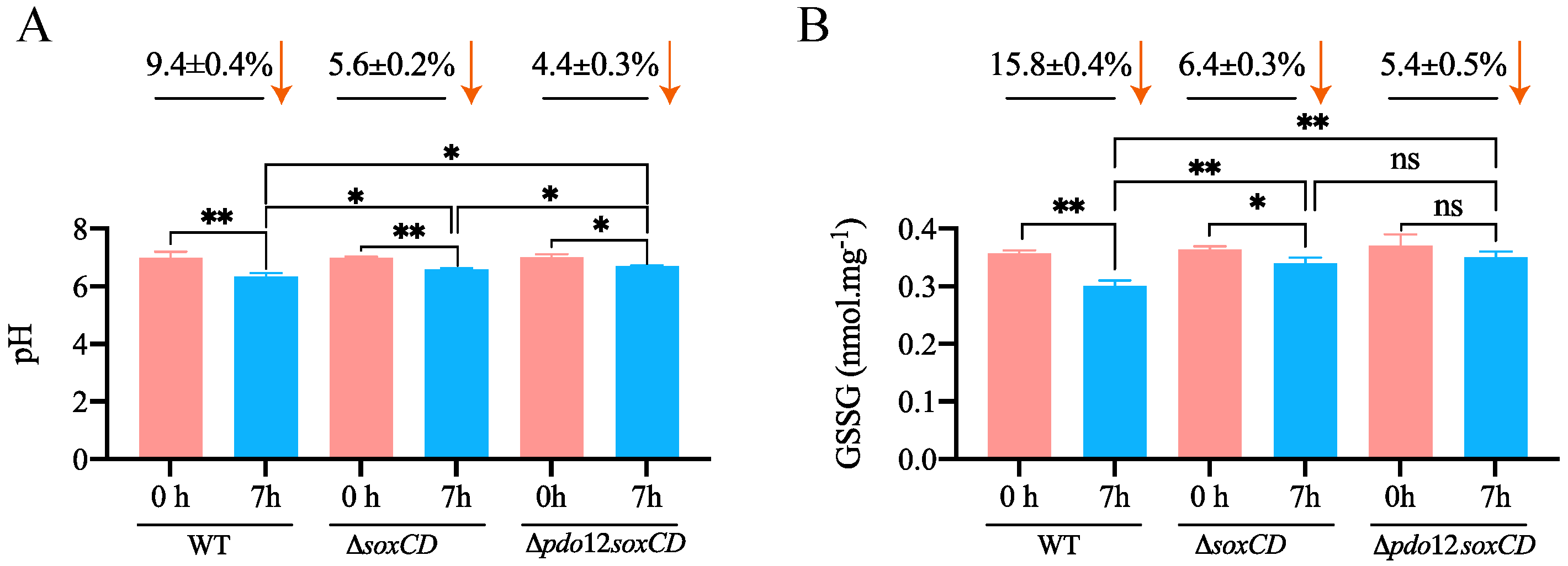
3.2. Changes in pH Values When Bacteria Oxidize Thiosulfate
3.3. GSSG Detection in the Cell Lysates
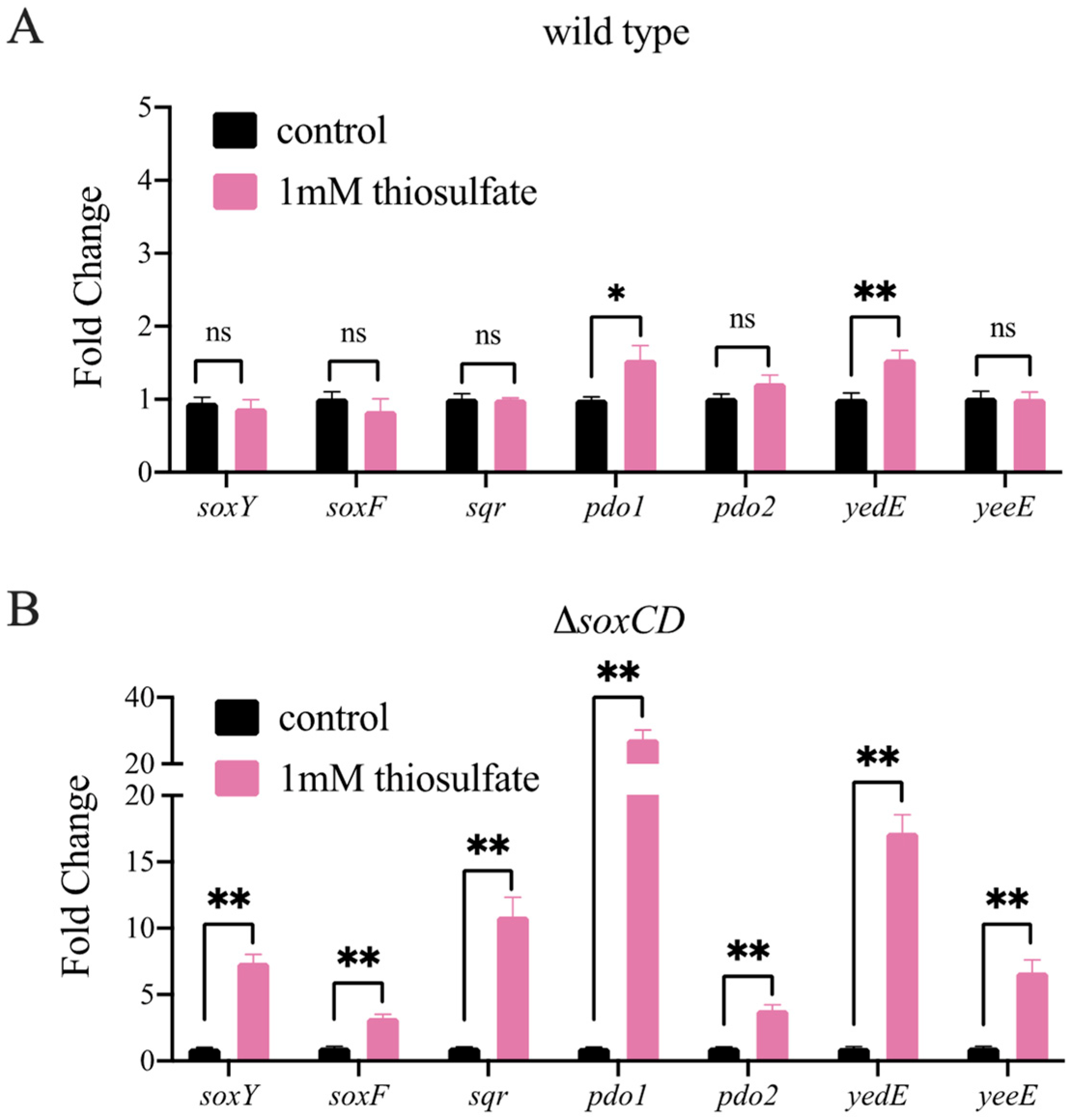
3.4. Gene Expression in the Wild-Type and Mutant Strains
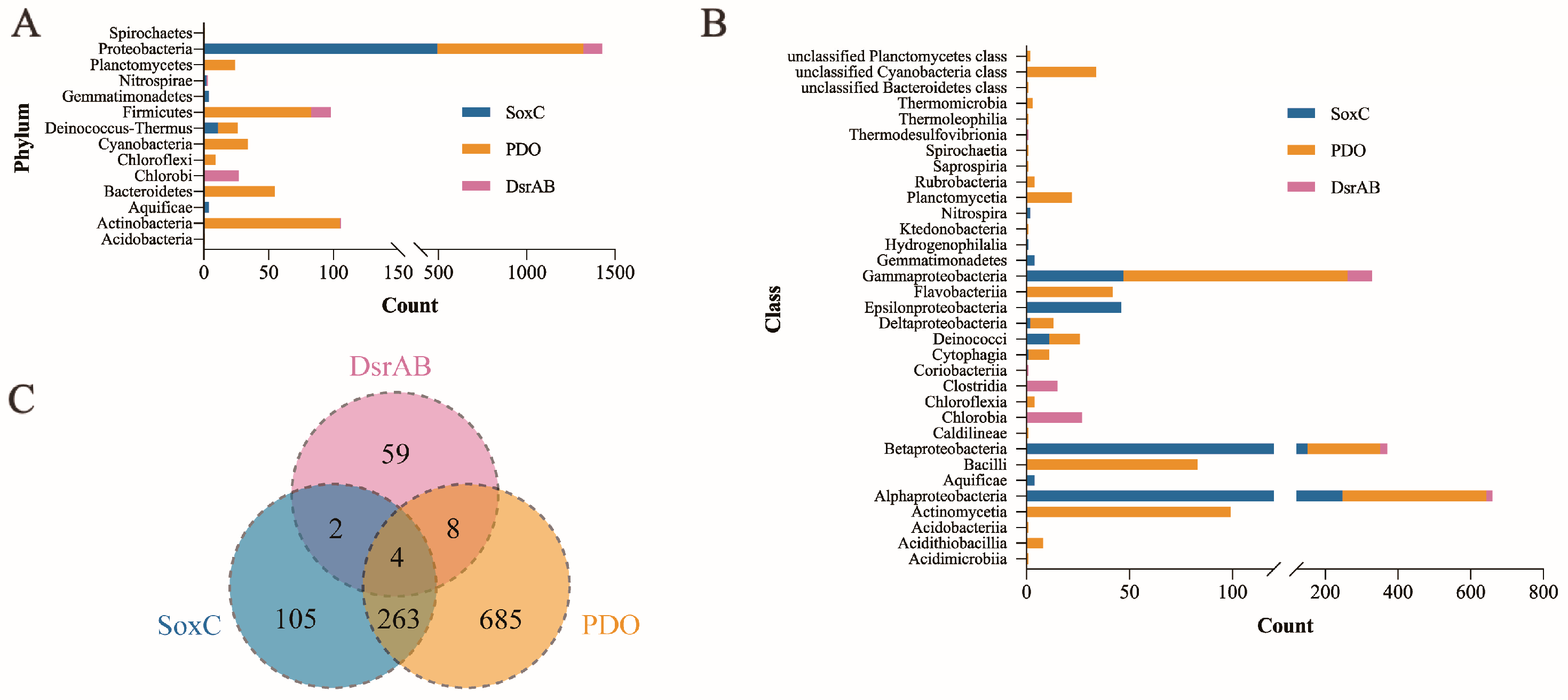
3.5. Distribution and Phylogenetic Analysis of SoxC, PDO and DsrAB
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lü, C.; Xia, Y.; Liu, D.; Zhao, R.; Gao, R.; Liu, H.; Xun, L. Cupriavidus necator H16 uses flavocytochrome c sulfide dehydrogenase to oxidize self-produced and added sulfide. Appl. Environ. Microbiol. 2017, 83, e01610-17. [Google Scholar] [CrossRef]
- Reinartz, M.; Tschape, J.; Bruser, T.; Truper, H.G.; Dahl, C. Sulfide oxidation in the phototrophic sulfur bacterium Chromatium vinosum. Arch. Microbiol. 1998, 170, 59–68. [Google Scholar] [CrossRef]
- Xin, Y.; Gao, R.; Cui, F.; Lü, C.; Liu, H.; Liu, H.; Xia, Y.; Xun, L. The heterotrophic bacterium Cupriavidus pinatubonensis JMP134 oxidizes sulfide to sulfate with thiosulfate as a key intermediate. Appl. Environ. Microbiol. 2020, 86, e01835-20. [Google Scholar] [CrossRef]
- Hensen, D.; Sperling, D.; Truper, H.G.; Brune, D.C.; Dahl, C. Thiosulphate oxidation in the phototrophic sulphur bacterium Allochromatium vinosum. Mol. Microbiol. 2006, 62, 794–810. [Google Scholar] [CrossRef] [PubMed]
- Rother, D.; Henrich, H.J.; Quentmeier, A.; Bardischewsky, F.; Friedrich, C.G. Novel genes of the sox gene cluster, mutagenesis of the flavoprotein SoxF, and evidence for a general sulfur-oxidizing system in Paracoccus pantotrophus GB17. J. Bacteriol. 2001, 183, 4499–4508. [Google Scholar] [CrossRef] [PubMed]
- Dahl, C.; Prange, A. Bacterial sulfur globules: Occurrence, structure and metabolism. In Inclusions in Prokaryotes; Shively, J.M., Ed.; Springer: Berlin/Heidelberg, Germany, 2006; pp. 21–51. [Google Scholar]
- Hou, N.; Yan, Z.; Fan, K.; Li, H.; Zhao, R.; Xia, Y.; Xun, L.; Liu, H. OxyR senses sulfane sulfur and activates the genes for its removal in Escherichia coli. Redox Biol. 2019, 26, 101293. [Google Scholar] [CrossRef] [PubMed]
- Schwedt, A.; Kreutzmann, A.-C.; Polerecky, L.; Schulz-Vogt, H.N. Sulfur respiration in a marine chemolithoautotrophic Beggiatoa strain. Front. Microbiol. 2012, 2, 276. [Google Scholar] [CrossRef] [PubMed]
- Williams, J.S.; Cooper, R.M. The oldest fungicide and newest phytoalexin—A reappraisal of the fungitoxicity of elemental sulphur. Plant Pathol. 2004, 53, 263–279. [Google Scholar] [CrossRef]
- Dahl, C.; Engels, S.; Pott-Sperling, A.S.; Schulte, A.; Sander, J.; Lubbe, Y.; Deuster, O.; Brune, D.C. Novel genes of the dsr gene cluster and evidence for close interaction of Dsr proteins during sulfur oxidation in the phototrophic sulfur bacterium Allochromatium vinosum. J. Bacteriol. 2005, 187, 1392–1404. [Google Scholar] [CrossRef] [PubMed]
- Müller, A.L.; Kjeldsen, K.U.; Rattei, T.; Pester, M.; Loy, A. Phylogenetic and environmental diversity of DsrAB-type dissimilatory (bi)sulfite reductases. ISME J. 2015, 9, 1152–1165. [Google Scholar] [CrossRef]
- Friedrich, C.G.; Rother, D.; Bardischewsky, F.; Quentmeier, A.; Fischer, J. Oxidation of reduced inorganic sulfur compounds by bacteria: Emergence of a common mechanism? Appl. Environ. Microbiol. 2001, 67, 2873–2882. [Google Scholar] [CrossRef] [PubMed]
- Friedrich, C.G.; Quentmeier, A.; Bardischewsky, F.; Rother, D.; Kraft, R.; Kostka, S.; Prinz, H. Novel genes coding for lithotrophic sulfur oxidation of Paracoccus pantotrophus GB17. J. Bacteriol. 2000, 182, 4677–4687. [Google Scholar] [CrossRef] [PubMed]
- Bamford, V.A.; Bruno, S.; Rasmussen, T.; Appia-Ayme, C.; Cheesman, M.R.; Berks, B.C.; Hemmings, A.M. Structural basis for the oxidation of thiosulfate by a sulfur cycle enzyme. EMBO J. 2002, 21, 5599–5610. [Google Scholar] [CrossRef]
- Sauvé, V.; Bruno, S.; Berks, B.C.; Hemmings, A.M. The SoxYZ complex carries sulfur cycle intermediates on a peptide swinging arm. J. Biol. Chem. 2007, 282, 23194–23204. [Google Scholar] [CrossRef]
- Sauvé, V.; Roversi, P.; Leath, K.J.; Garman, E.F.; Antrobus, R.; Lea, S.M.; Berks, B.C. Mechanism for the hydrolysis of a sulfur-sulfur bond based on the crystal structure of the thiosulfohydrolase SoxB. J. Biol. Chem. 2009, 284, 21707–21718. [Google Scholar] [CrossRef]
- Zander, U.; Faust, A.; Klink, B.U.; de Sanctis, D.; Panjikar, S.; Quentmeier, A.; Bardischewsky, F.; Friedrich, C.G.; Scheidig, A.J. Structural basis for the oxidation of protein-bound sulfur by the sulfur cycle molybdohemo-enzyme sulfane dehydrogenase SoxCD. J. Biol. Chem. 2011, 286, 8349–8360. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Xin, Y.; Xun, L. Distribution, diversity, and activities of sulfur dioxygenases in heterotrophic bacteria. Appl. Environ. Microbiol. 2014, 80, 1799–1806. [Google Scholar] [CrossRef]
- Hildebrandt, T.M.; Grieshaber, M.K. Three enzymatic activities catalyze the oxidation of sulfide to thiosulfate in mammalian and invertebrate mitochondria. FEBS J. 2008, 275, 3352–3361. [Google Scholar] [CrossRef]
- Holdorf, M.M.; Owen, H.A.; Lieber, S.R.; Yuan, L.; Adams, N.; Dabney-Smith, C.; Makaroff, C.A. Arabidopsis ETHE1 encodes a sulfur dioxygenase that is essential for embryo and endosperm development. Plant Physiol. 2012, 160, 226–236. [Google Scholar] [CrossRef]
- Gao, R.; Liu, H.; Xun, L. Cytoplasmic localization of sulfide:quinone oxidoreductase and persulfide dioxygenase of Cupriavidus pinatubonensis JMP134. Appl. Environ. Microbiol. 2017, 83, e01820-17. [Google Scholar] [CrossRef]
- Cherney, M.M.; Zhang, Y.; Solomonson, M.; Weiner, J.H.; James, M.N. Crystal structure of sulfide:quinone oxidoreductase from Acidithiobacillus ferrooxidans: Insights into sulfidotrophic respiration and detoxification. J. Mol. Biol. 2010, 398, 292–305. [Google Scholar] [CrossRef]
- Rohwerder, T.; Sand, W. The sulfane sulfur of persulfides is the actual substrate of the sulfur-oxidizing enzymes from Acidithiobacillus and Acidiphilium spp. Microbiology 2003, 149, 1699–1710. [Google Scholar] [CrossRef]
- Xia, Y.; Li, K.; Li, J.; Wang, T.; Gu, L.; Xun, L. T5 exonuclease-dependent assembly offers a low-cost method for efficient cloning and site-directed mutagenesis. Nucleic Acids Res. 2019, 47, e15. [Google Scholar] [CrossRef]
- Cline, J.D. Spectrophotometric determination of hydrogen sulfide in natural waters. Limnol. Oceanogr. 1969, 13, 454–458. [Google Scholar] [CrossRef]
- Xin, Y.; Liu, H.; Cui, F.; Liu, H.; Xun, L. Recombinant Escherichia coli with sulfide:quinone oxidoreductase and persulfide dioxygenase rapidly oxidises sulfide to sulfite and thiosulfate via a new pathway. Environ. Microbiol. 2016, 18, 5123–5136. [Google Scholar] [CrossRef]
- Kamyshny, A., Jr. Improved cyanolysis protocol for detection of zero-valent sulfur in natural aquatic systems. Limnol. Oceanogr. 2009, 7, 442–448. [Google Scholar] [CrossRef]
- Rahman, I.; Kode, A.; Biswas, S.K. Assay for quantitative determination of glutathione and glutathione disulfide levels using enzymatic recycling method. Nat. Protoc. 2006, 1, 3159–3165. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Xia, Y.; Lu, C.; Hou, N.; Xin, Y.; Liu, J.; Liu, H.; Xun, L. Sulfide production and oxidation by heterotrophic bacteria under aerobic conditions. ISME J. 2017, 11, 2754–2766. [Google Scholar] [CrossRef]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef] [PubMed]
- Shen, W.; Ren, H. TaxonKit: A practical and efficient NCBI taxonomy toolkit. J. Genet. Genom. 2021, 48, 844–850. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef]
- Andreetto, F.; Dela Pierre, F.; Gibert, L.; Natalicchio, M.; Ferrando, S. Potential fossilized sulfide-oxidizing bacteria in the upper miocene sulfur-bearing limestones from the Lorca Basin (SE Spain): Paleoenvironmental implications. Front. Microbiol. 2019, 10, 1031. [Google Scholar] [CrossRef] [PubMed]
- Schafer, F.Q.; Buettner, G.R. Redox environment of the cell as viewed through the redox state of the glutathione disulfide/glutathione couple. Free Radic. Biol. Med. 2001, 30, 1191–1212. [Google Scholar] [CrossRef] [PubMed]
- Bardischewsky, F.; Quentmeier, A.; Rother, D.; Hellwig, P.; Kostka, S.; Friedrich, C.G. Sulfur dehydrogenase of Paracoccus pantotrophus: The heme-2 domain of the molybdoprotein cytochrome c complex is dispensable for catalytic activity. Biochemistry 2005, 44, 7024–7034. [Google Scholar] [CrossRef]
- Kolluru, G.K.; Shen, X.; Bir, S.C.; Kevil, C.G. Hydrogen sulfide chemical biology: Pathophysiological roles and detection. Nitric Oxide 2013, 35, 5–20. [Google Scholar] [CrossRef]
- Masip, L.; Veeravalli, K.; Georgiou, G. The many faces of glutathione in bacteria. Antioxid. Redox Signal. 2006, 8, 753–762. [Google Scholar] [CrossRef]
- Kabil, O.; Banerjee, R. Enzymology of H2S biogenesis, decay and signaling. Antioxid. Redox Signal. 2014, 20, 770–782. [Google Scholar] [CrossRef]
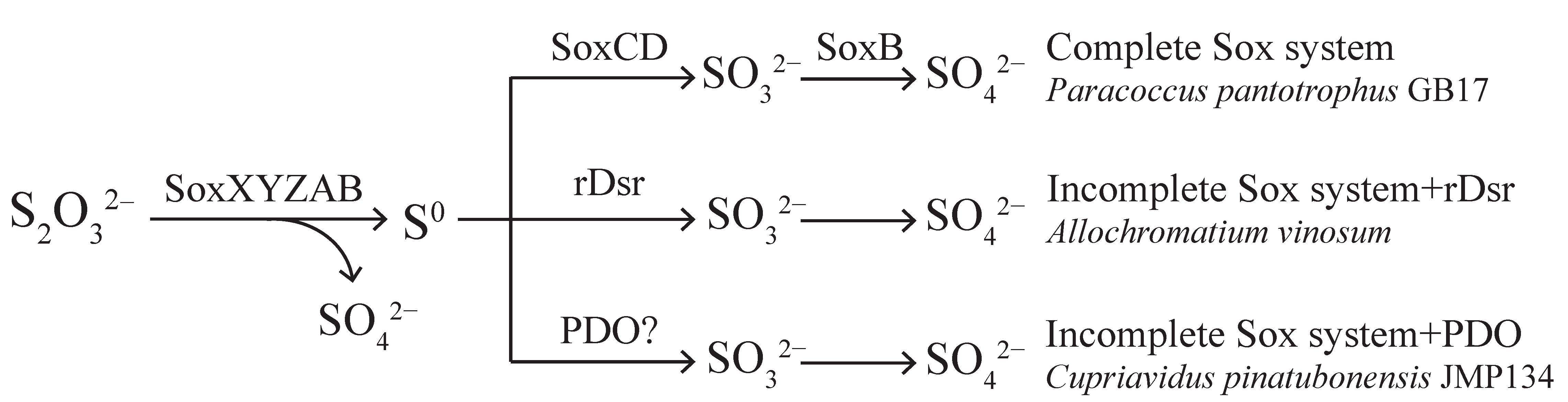


| Strains | Characteristics | Source |
|---|---|---|
| Wild-type strains | ||
| C. pinatubonensis JMP134 | Wild type | Our lab |
| E. coli S17-1 | recA pro thi hsdS, RP4 tra functions, supE44 | Invitrogen |
| Deletion strains | ||
| ΔsoxCD | soxC and soxD deleted in JMP134 | This study |
| Δpdo12soxCD | pdo1, pdo2, soxC and soxD deleted in JMP134 | This study |
| Δpdo12 | pdo1 and pdo2 deleted in JMP134 | [3] |
| Δpdo1soxCD | pdo1, soxC and soxD deleted in JMP134 | This study |
| Δpdo2soxCD | pdo2, soxC and soxD deleted in JMP134 | This study |
| Complementation strains | ||
| Δpdo12soxCD::pdo1 | Δpdo12soxCD with plasmid pBBR-pdo1 | This study |
| ΔsoxCD:soxCD | ΔsoxCD with plasmid pBBR-soxCD | This study |
| Plasmids | ||
| pBBR1MCS2 | Kanamycin resistance, mob+, pBBR1 replicon, cloning vector | Qi qingsheng |
| pK18mobsacB | Widely used gene knockout vector, kanamycin resistance | Our lab |
| pBBR-pdo1 | pBBR1MCS2 containing pdo1 | This study |
| pBBR-soxCD | pBBR1MCS2 containing soxCD | This study |
| Target Gene | Primers | Sequence (5′-3′) d |
|---|---|---|
| Deletion | ||
| soxC and soxD | a Up-f | CAGGAAACAGCTATGACATGATTACGAATTCACCGCCGGGTTTCTGTTG |
| Up-r | CCTACCATCGGTTCCTGCAATGCCGTCTCCT | |
| b Down-f | TTGCAGGAACCGATGGTAGGGTGGATTCTTGAG | |
| Down-r | TTCAGGATCCCCGGGTACCGAGCTCGAATTCTGCCATTGCTCTCTCCTGTTG | |
| c V-f | GGTGTTCGGCTACACCATGT | |
| V-r | AGCTTTGCTCTCCGGCTAC | |
| pdo1 | Up-f | CAGGAAACAGCTATGACATGATTACGAATTAGACGATTACCTGGTCTACACCTTC |
| Up-r | CAGCTGTTCGTACAGGCGCGTCAAATCCTTCTAT | |
| Down-f | CGCGCCTGTACGAACAGCTGATAGAAGGTTTGCAT | |
| Down-r | TTCAGGATCCCCGGGTACCGAGCTCGAATTGGCTGATGATGGAGAACGAAC | |
| V-f | TATTGGCTGCCATCTGCT | |
| V-r | GCTCTACAAGCTCAATGCG | |
| pdo2 | Up-f | CAGGAAACAGCTATGACATGATTACGAATTCGAGGTCGTAGCGGTAGTTG |
| Up-r | ACACACATGAGCTATCTGAAGATTCCCCTCAAC | |
| Down-f | TTCAGATAGCTCATGTGTGTCTATCCGTGGTTAGC | |
| Down-r | TTCAGGATCCCCGGGTACCGAGCTCGAATTCCATTTCATCGAGGAATAGCGT | |
| V-f | ATGGCGTCCCAATCCAGCTT | |
| V-r | TTGCCTGGAGAGTGGCTTTG | |
| Complementation | ||
| soxC and soxD | Forward | CACACAGGAAACAGCTATGCAGGAACGCACACC e |
| Reverse | TTCCATTCGCCATTCACTATTTTGCCTCAAGAATCCA e | |
| pdo1 | Forward | CACACAGGAAACAGCTATGACACCGACCATGCCAAG e |
| Reverse | TTCCATTCGCCATTCATCAGAGGGCGTTGAGGGG e | |
| Linearization | ||
| pBBR1MCS2 | Forward | TGAATGGCGAATGGAAATTGTAAG |
| Reverse | AGCTGTTTCCTGTGTGAAATTGTTATC | |
| RT‒PCR | ||
| soxY | Forward | GAGTGGAACAAGACCGCTTT |
| Reverse | ATCGCGATCTGCTCGGTATC | |
| pdo1 | Forward | CACGCTCTACCGTTCCATCA |
| Reverse | CACGTGGATGTTGTTCTCGC | |
| pdo2 | Forward | CATGCCCATGCCGACCACATC |
| Reverse | CGTGCCGAAGGTGAGCGTATC | |
| sqr | Forward | GCGTGGTGAAGTACGAACAA |
| Reverse | AGGTCGTAGCGGTAGTTGGA | |
| soxF | Forward | GCGTGAGTGGAGCGGACATG |
| Reverse | GATTGGACAGCGGACACGAGAC | |
| yedE | Forward | TTGCACGAAGACGGTCAGGAAAC |
| Reverse | CACGGCGTGTGCGGAATCTC | |
| yeeE | Forward | GGAAAGCCACCAGCCCGATG |
| Reverse | CCGCCAAGGTGCAGGGATTC |
| Phylum | Class | Complete Sox System | Incomplete Sox System | ||||
|---|---|---|---|---|---|---|---|
| Total | PDO | DsrAB | Total | PDO | DsrAB | ||
| Aquificae | Aquificae | 4 | 0 | 0 | 3 | 0 | 0 |
| Chlorobi | Chlorobia | 0 | 0 | 0 | 7 | 0 | 7 |
| Deinococcus-Thermus | Deinococci | 8 | 8 | 0 | 0 | 0 | 0 |
| Proteobacteria | - | 288 | 230 | 8 | 52 | 36 | 27 |
| Acidithiobacillia | 0 | 0 | 0 | 2 | 2 | 0 | |
| Alphaproteobacteria | 146 | 140 | 2 | 8 | 5 | 3 | |
| Betaproteobacteria | 75 | 63 | 1 | 20 | 16 | 5 | |
| Deltaproteobacteria | 2 | 0 | 0 | 0 | 0 | 0 | |
| Epsilonproteobacteria | 35 | 0 | 0 | 1 | 0 | 0 | |
| Gammaproteobacteria | 29 | 27 | 5 | 21 | 13 | 19 | |
| Hydrogenophilalia | 1 | 0 | 0 | 0 | 0 | 0 | |
| Total | 300 | 238 | 8 | 62 | 36 | 34 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xin, Y.; Wang, Y.; Zhang, H.; Wu, Y.; Xia, Y.; Li, H.; Qu, X. Cupriavidus pinatubonensis JMP134 Alleviates Sulfane Sulfur Toxicity after the Loss of Sulfane Dehydrogenase through Oxidation by Persulfide Dioxygenase and Hydrogen Sulfide Release. Metabolites 2023, 13, 218. https://doi.org/10.3390/metabo13020218
Xin Y, Wang Y, Zhang H, Wu Y, Xia Y, Li H, Qu X. Cupriavidus pinatubonensis JMP134 Alleviates Sulfane Sulfur Toxicity after the Loss of Sulfane Dehydrogenase through Oxidation by Persulfide Dioxygenase and Hydrogen Sulfide Release. Metabolites. 2023; 13(2):218. https://doi.org/10.3390/metabo13020218
Chicago/Turabian StyleXin, Yufeng, Yaxin Wang, Honglin Zhang, Yu Wu, Yongzhen Xia, Huanjie Li, and Xiaohua Qu. 2023. "Cupriavidus pinatubonensis JMP134 Alleviates Sulfane Sulfur Toxicity after the Loss of Sulfane Dehydrogenase through Oxidation by Persulfide Dioxygenase and Hydrogen Sulfide Release" Metabolites 13, no. 2: 218. https://doi.org/10.3390/metabo13020218
APA StyleXin, Y., Wang, Y., Zhang, H., Wu, Y., Xia, Y., Li, H., & Qu, X. (2023). Cupriavidus pinatubonensis JMP134 Alleviates Sulfane Sulfur Toxicity after the Loss of Sulfane Dehydrogenase through Oxidation by Persulfide Dioxygenase and Hydrogen Sulfide Release. Metabolites, 13(2), 218. https://doi.org/10.3390/metabo13020218







