Synthesis of Metabolites and Metabolite-like Compounds Using Biocatalytic Systems
Abstract
1. Introduction
2. Design and Engineering of Biocatalytic Systems
3. Synthesis of Naturally Occurring Metabolites
4. Synthesis of Isotope-Labelled Metabolites
5. Synthesis of Pharmaceutical Drug Metabolites
6. Synthesis of Metabolite-like Compounds
7. Discussion
8. Future Directions
| Metabolites | Metabolic Pathway | Reference |
|---|---|---|
| Acetic acid | Carbon metabolism | [58] |
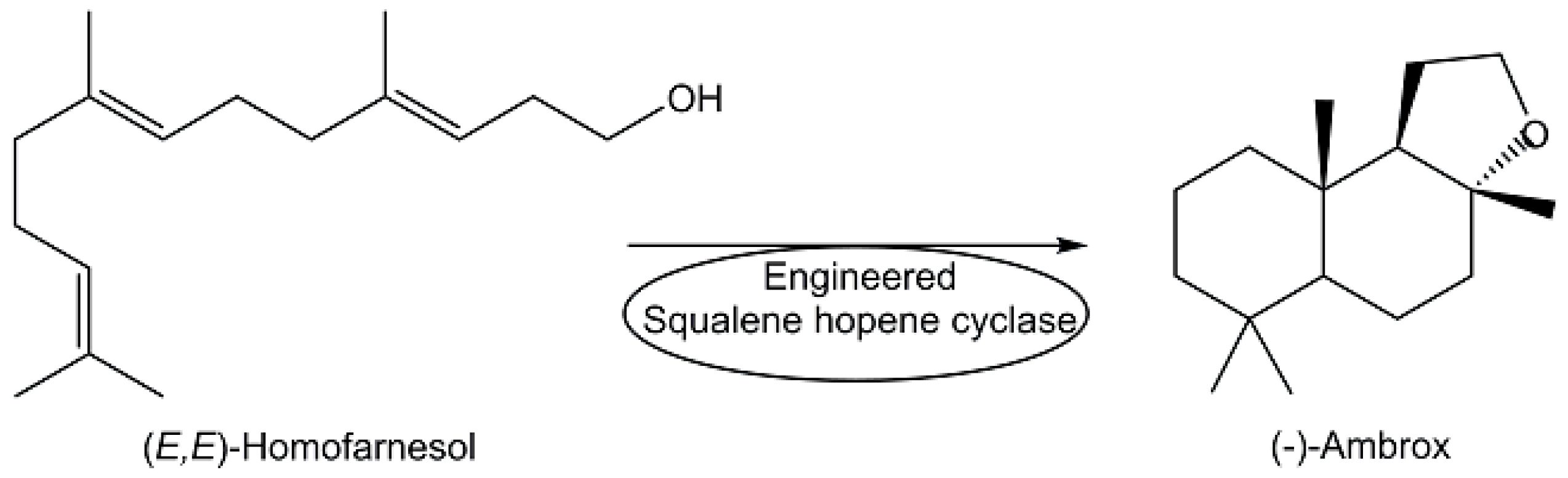
| (-)-Ambrox | Homofarnesol cyclization | [108] |
| 4-Androstene-3,17-dione | Steroid biosynthesis | [90] |
| L-Argininosuccinate | Urea cycle | [129,130] |
| Artemisinin | Artemisinin bioynthesis | [103] |
| Avermectin | Polyketide biosynthesis | [102] |
| Azadirachtin | Tetranortriterpenoid biosynthesis | [61] |
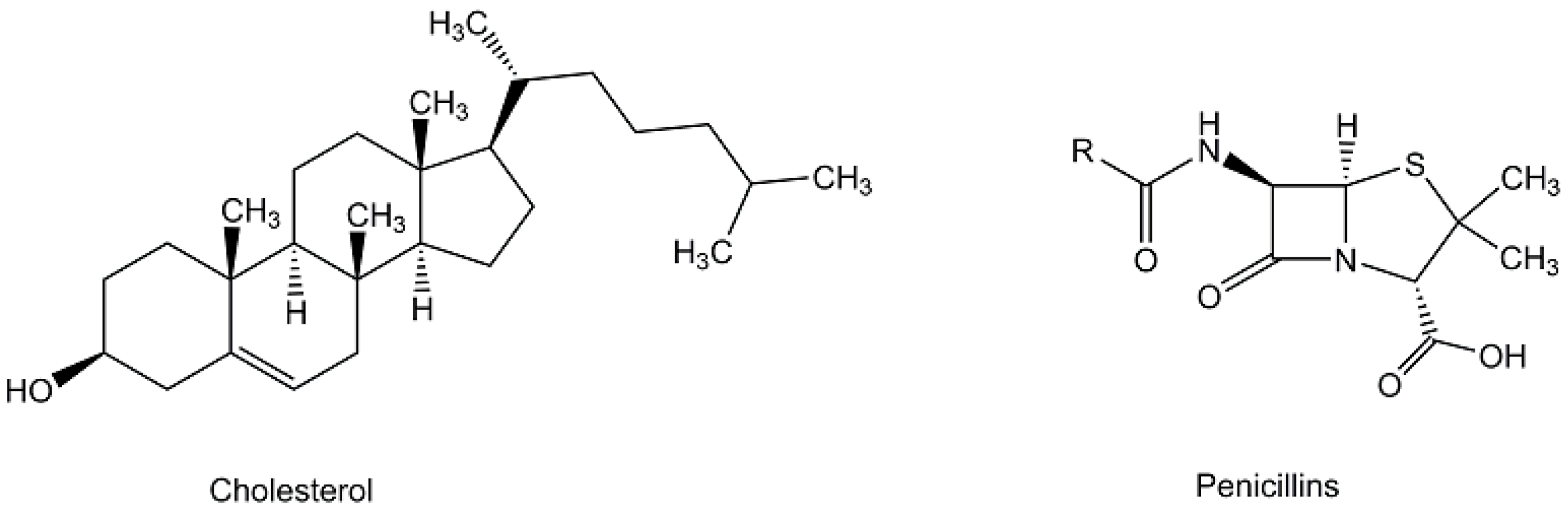
| Cholesterol | Steroid biosynthesis | [67,69] |
| Citric acid | Citrate cycle (TCA cycle) | [81] |
| 1α,25-Dihydroxyvitamin D2 | Steroid biosynthesis | [137] |
| 1α,25-Dihydroxyvitamin D3 | Steroid biosynthesis | [135] |
| 24R,25-Dihydroxyvitamin D2 | Steroid biosynthesis | [138] |
| 24R,25-Dihydroxyvitamin D3 | Steroid biosynthesis | [136] |
| 17β-Estradiol | Steroid biosynthesis | [89] |
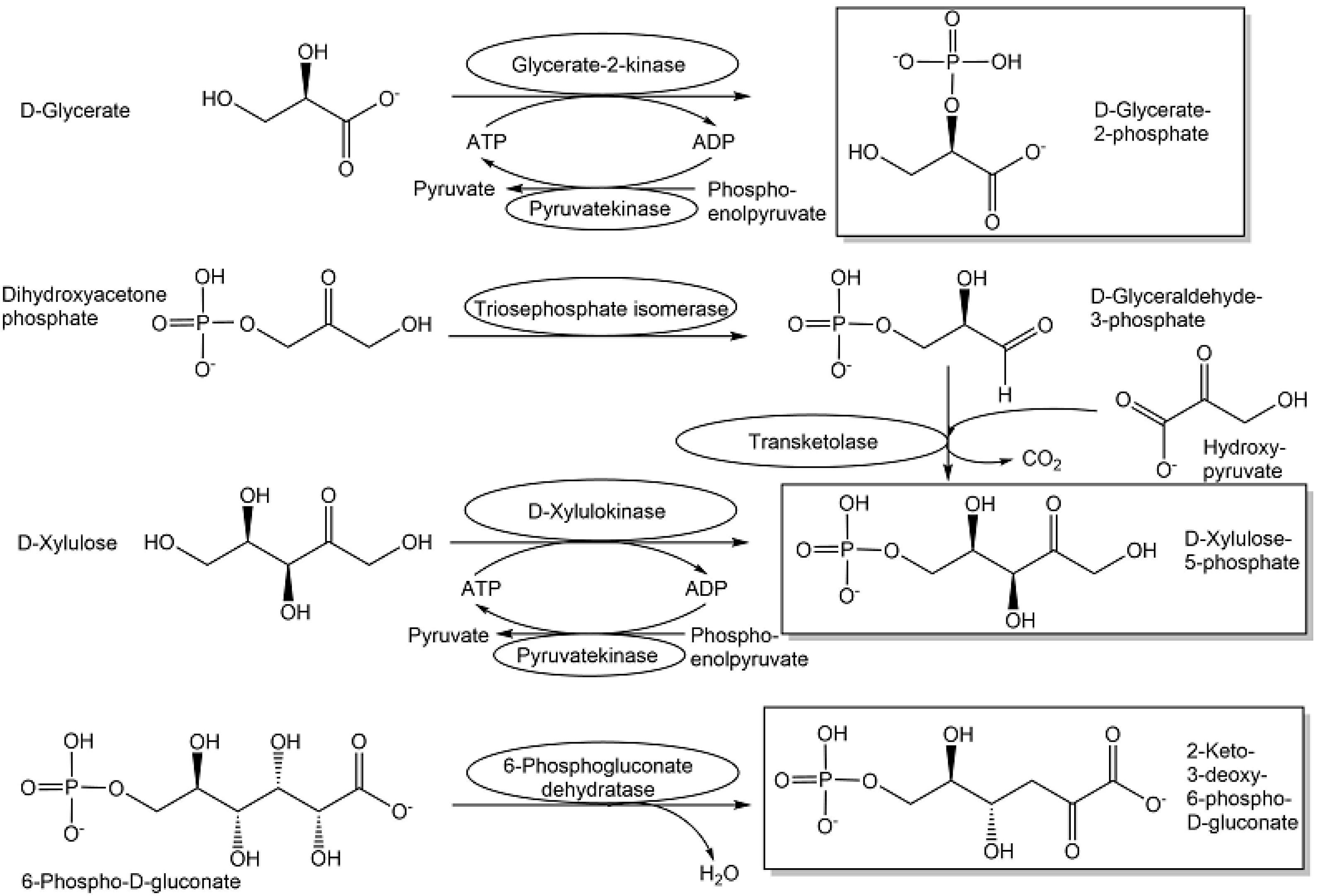
| L-Glyceraldehyde | Pentose and glucuronate interconversions | [115,116] |
| D-Glyceraldehyde-3-phosphate | Embden-Meyerhof-Parnas pathway Carbon fixation in photosynthesis Pentose phosphate pathway | [117] |
| L-Glyceraldehyde-3-phosphate | Isomerase bypass | [113,114] |
| D-Glycerate-2-phosphate | Embden-Meyerhof-Parnas pathway | [118] |
| 25-Hydroxyvitamin D2 | Steroid biosynthesis | [133] |
| 25-Hydroxyvitamin D3 | Steroid biosynthesis | [131,132,133,134] |
| 2-Keto-3-deoxy-D-galactonate | Galactose metabolism | [124] |
| 2-Keto-3-deoxy-D-gluconate | Non-phosphorylative Entner-Doudoroff pathway Pentose phosphate pathway | [125] |
| 2-Keto-3-deoxy-6-phosphogluconate | Entner-Doudoroff pathway Pentose phosphate pathway | [123] |
| 2-Keto-3-deoxy-D-xylonate | Pentose and glucuronate interconversions | [124] |
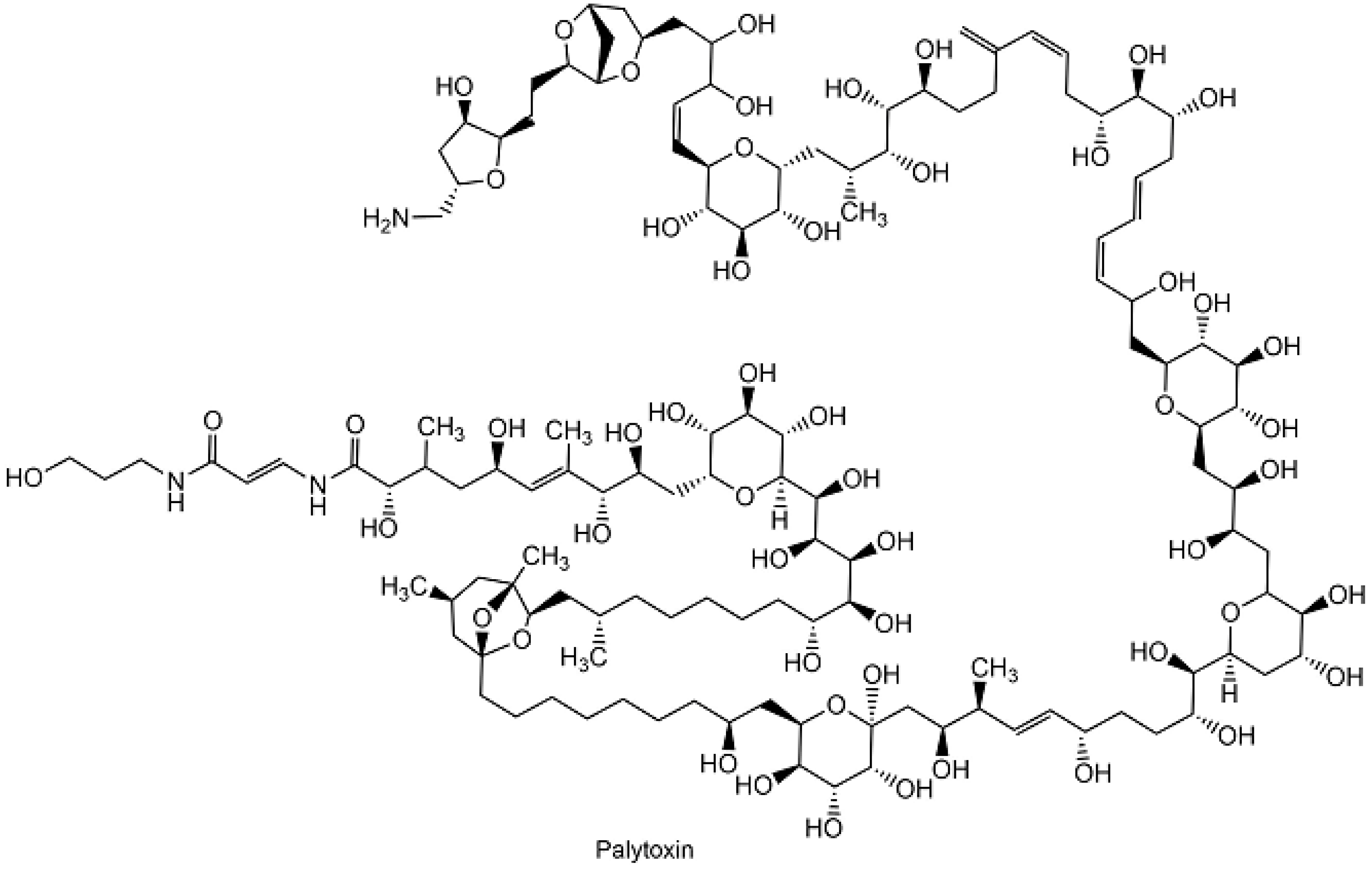
| Palytoxin | Palytoxin biosynthesis | [74] |
| Penicillin V | Penicillin biosynthesis | [59] |
| Nω-Phospho-L-arginine | Phosphagen pathway | [126] |
| Pyridoxamine-5’-phosphate | Vitamin B6 metabolism | [128] |
| Shikimic acid-3-phosphate | Shikimate pathway | [127] |
| D-Tagatose-1,6-diphosphate | Galactose metabolism D-Tagatose pathway | [122] |
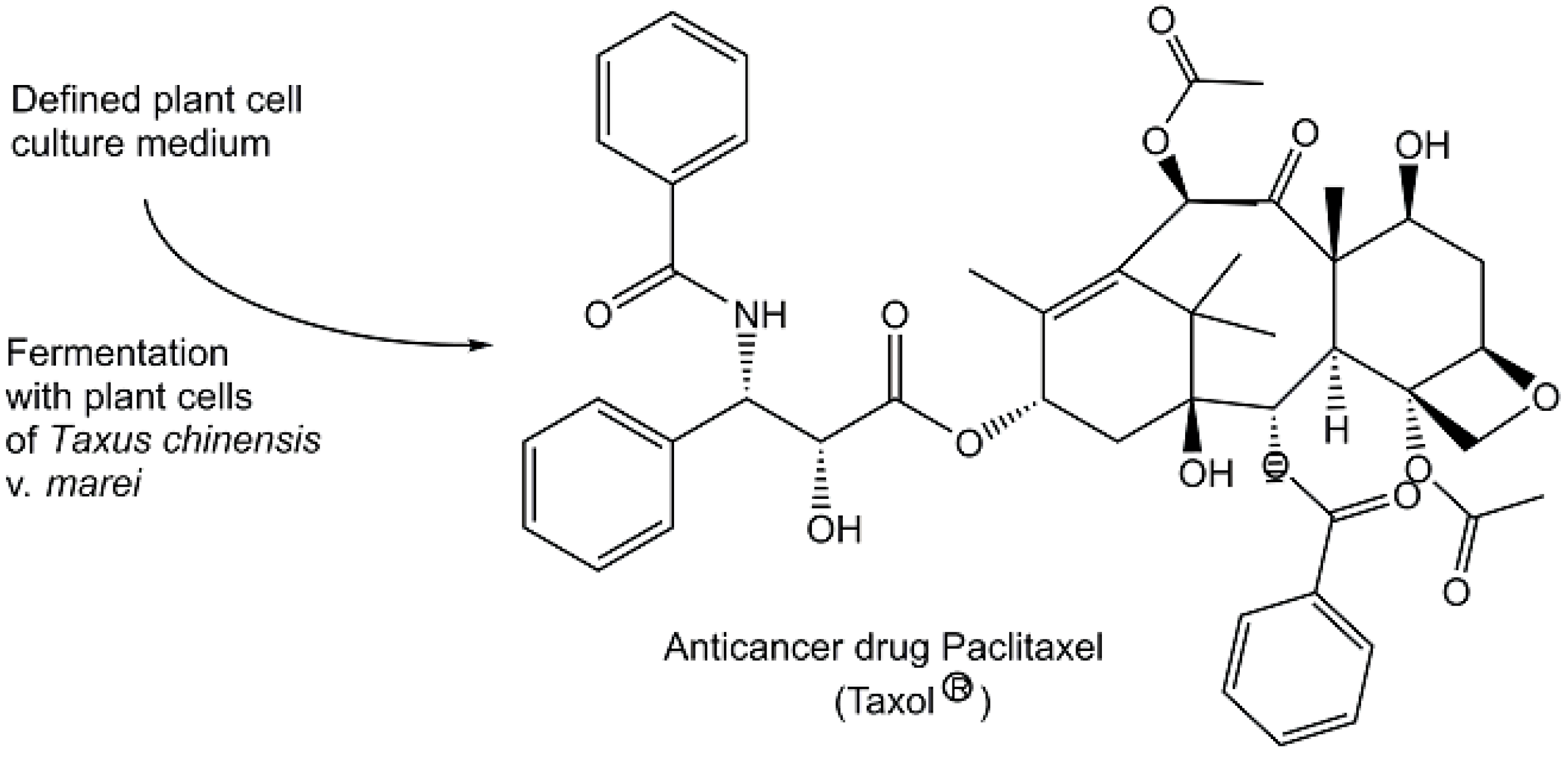
| Taxol | Taxol biosynthesis | [63,100,101] |
| Testosterone | Steroid biosynthesis | [91] |
| Urea | Urea cycle | [57,79,80] |
| Vinblastine | Indole alkaloid biosynthesis | [75,238] |
| Vitamin B12 | Cobalamine biosynthesis | [70,71,72,73,107] |
| D-Xylulose-5-phosphate | Pentose phosphate pathway | [119,120,121] |
| L-Xylulose-5-phosphate | Pentose and glucuronate interconversions | [121] |
Funding
Acknowledgments
Conflicts of Interest
References
- Schreiber, S.L. Small molecules: The missing link in the central dogma. Nat. Chem. Biol. 2005, 1, 64–66. [Google Scholar] [CrossRef]
- McKnight, S.L. Back to the future: Molecular biology meets metabolism. Cold Spring Harb. Symp. Quant. Biol. 2011, 76, 403–411. [Google Scholar] [CrossRef] [PubMed]
- Aharoni, A.; Goodacre, R.; Fernie, A.R. Plant and microbial sciences as key drivers in the development of metabolomics research. Proc. Natl. Acad. Sci. USA 2023, 120, e2217383120. [Google Scholar] [CrossRef] [PubMed]
- Kell, D.B.; Oliver, S. The metabolome 18 years on: A concept comes of age. Metabolomics 2016, 12, 148. [Google Scholar] [CrossRef]
- Fiehn, O. Metabolomics by Gas Chromatography-Mass Spectrometry: The combination of targeted and untargeted profiling. Curr. Protoc. Mol. Biol. 2017, 114, 30.4.1–30.4.32. [Google Scholar] [CrossRef] [PubMed]
- Plumb, R.S.; Gethings, L.A.; Rainville, P.D.; Isaac, G.; Trengove, R.; King, A.M.; Wilson, I.D. Advances in high throughput LC/MS based metabolomics: A review. Trends Anal. Chem. 2023, 160, 116954. [Google Scholar] [CrossRef]
- Emwas, A.H.; Roy, R.; McKay, R.T.; Tenori, L.; Saccenti, E.; Gowda, G.N.; Raftery, D.; Alahmari, F.; Jaremko, L.; Jaremko, M.; et al. NMR spectroscopy for metabolomics research. Metabolites 2019, 9, 123. [Google Scholar] [CrossRef]
- Guijas, C.; Montenegro-Burke, J.R.; Domingo-Almenara, X.; Palermo, A.; Warth, B.; Hermann, G.; Koellensperger, G.; Huan, T.; Uritboonthai, W.; Aisporna, A.E.; et al. METLIN: A Technology Platform for Identifying Knowns and Unknowns. Anal. Chem. 2018, 90, 3156–3164. [Google Scholar] [CrossRef]
- Wishart, D.S.; Cheng, L.L.; Copié, V.; Edison, A.S.; Eghbalnia, H.R.; Hoch, J.C.; Gouveia, G.J.; Pathmasiri, W.; Powers, R.; Schock, T.B.; et al. NMR and metabolomics—A roadmap for the future. Metabolites 2022, 12, 678. [Google Scholar] [CrossRef]
- Hoch, J.C.; Baskaran, K.; Burr, H.; Chin, J.; Eghbalnia, H.R.; Fujiwara, T.; Gryk, M.R.; Iwata, T.; Kojima, C.; Kurisu, G.; et al. Biological Magnetic Resonance Data Bank. Nucleic Acids Res. 2023, 51, D368–D376. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S.; Sato, Y.; Kawashima, M.; Furumichi, M.; Tanabe, M. Data, information, knowledge and principle: Back to metabolism in KEGG. Nucleic Acids Res. 2014, 42, D199–D205. [Google Scholar] [CrossRef]
- Hastings, J.; Owen, G.; Dekker, A.; Ennis, M.; Kale, N.; Muthukrishnan, V.; Turner, S.; Swainston, N.; Mendes, P.; Steinbeck, C. ChEBI in 2016: Improved services and an expanding collection of metabolites. Nucleic Acids Res. 2016, 44, D1214–D1219. [Google Scholar] [CrossRef] [PubMed]
- van Santen, J.A.; Jacob, G.; Singh, A.L.; Aniebok, V.; Balunas, M.J.; Bunsko, D.; Neto, F.C.; Castaño-Espriu, L.; Chang, C.; Clark, T.N.; et al. The Natural Products Atlas: An Open Access Knowledge Base for Microbial Natural Products Discovery. ACS Cent. Sci. 2019, 5, 1824–1833. [Google Scholar] [CrossRef] [PubMed]
- Wishart, D.S.; Guo, A.; Oler, E.; Wang, F.; Anjum, A.; Peters, H.; Dizon, R.; Sayeeda, Z.; Tian, S.; Lee, B.L.; et al. HMDB 5.0: The human metabolome database for 2022. Nucleic Acids Res. 2022, 50, D622–D631. [Google Scholar] [CrossRef] [PubMed]
- Wishart, D.S.; Oler, E.; Peters, H.; Guo, A.; Girod, S.; Han, S.; Saha, S.; Lui, V.W.; LeVatte, M.; Gautam, V.; et al. MiMeDB: The Human Microbial Metabolome Database. Nucleic Acids Res. 2023, 51, D611–D620. [Google Scholar] [CrossRef]
- Sajed, T.; Marcu, A.; Ramirez, M.; Pon, A.; Guo, A.; Knox, C.; Wilson, M.; Grant, J.; Djoumbou, Y.; Wishart, D. ECMDB 2.0: A richer resource for understanding the biochemistry of E. coli. Nucleic Acids Res. 2016, 44, D495–D501. [Google Scholar] [CrossRef]
- Huang, W.; Luke, K.; Brewer, L.K.; Jace, W.; Jones, J.W.; Angela, T.; Nguyen, A.T.; Ana Marcu, A.; David, S.; Wishart, D.S.; et al. PAMDB: A comprehensive Pseudomonas aeruginosa metabolome database. Nucleic Acids Res. 2018, 46, D575–D580. [Google Scholar] [CrossRef] [PubMed]
- Moumbock, A.F.A.; Gao, M.; Qaseem, A.; Li, J.; Kirchner, P.A.; Ndingkokhar, B.; Bekono, B.D.; Simoben, C.V.; Babiaka, S.M.; Malange, Y.I.; et al. StreptomeDB 3.0: An updated compendium of streptomycetes natural products. Nucleic Acids Res. 2021, 49, D600–D604. [Google Scholar] [CrossRef]
- Jones, M.R.; Pinto, E.; Torres, M.A.; Dörr, F.; Mazur-Marzec, H.; Szubert, K.; Tartaglione, L.; Dell’Aversano, C.; Miles, C.O.; Beach, D.G.; et al. CyanoMetDB, a comprehensive public database of secondary metabolites from cyanobacteria. Water Res. 2021, 196, 117017. [Google Scholar] [CrossRef]
- Wang, D.G.; Wang, C.Y.; Hu, J.Q.; Wang, J.J.; Liu, W.C.; Zhang, W.J.; Du, X.R.; Wang, H.; Zhu, L.L.; Sui, H.Y.; et al. Constructing a Myxobacterial Natural Product Database to Facilitate NMR-Based Metabolomics Bioprospecting of Myxobacteria. Anal. Chem. 2023, 95, 5256–5266. [Google Scholar] [CrossRef]
- Ramirez-Gaona, M.; Marcu, A.; Pon, A.; Guo, A.C.; Sajed, T.; Wishart, N.A.; Karu, N.; Djoumbou Feunang, Y.; Arndt, D.; Wishart, D.S. YMDB 2.0: A significantly expanded version of the yeast metabolome database. Nucleic Acids Res. 2017, 45, D440–D445. [Google Scholar] [CrossRef] [PubMed]
- Foroutan, A.; Fitzsimmons, C.; Mandal, R.; Piri-Moghadam, H.; Zheng, J.; Guo, A.; Li, C.; Guan, L.L.; Wishart, D.S. The Bovine Metabolome. Metabolites 2020, 10, 233. [Google Scholar] [CrossRef] [PubMed]
- Takeshi Ara, T.; Sakurai, N.; Takahashi, S.; Waki, N.; Suganuma, H.; Aizawa, K.; Matsumura, Y.; Kawada, T.; Shibata, D. TOMATOMET: A metabolome database consists of 7118 accurate mass values detected in mature fruits of 25 tomato cultivars. Plant Direct. 2021, 5, e00318. [Google Scholar] [CrossRef]
- Link, H.; Kochanowski, K.; Sauer, U. Systematic identification of allosteric protein-metabolite interactions that control enzyme activity in vivo. Nat. Biotechnol. 2013, 31, 357–361. [Google Scholar] [CrossRef]
- Piazza, I.; Kochanowski, K.; Cappelletti, V.; Fuhrer, T.; Noor, E.; Sauer, U.; Picotti, P. A map of protein-metabolite interactions reveals principles of chemical communication. Cell 2018, 172, 358–372. [Google Scholar] [CrossRef]
- Rinschen, M.R.; Ivanisevic, J.; Giera, M.; Siuzdak, G. Identification of bioactive metabolites using activity metabolomics. Nat. Rev. Mol. Cell Biol. 2019, 20, 353–367. [Google Scholar] [CrossRef]
- Buescher, J.M.; Antoniewicz, M.R.; Boros, L.G.; Burgess, S.C.; Brunengraber, H.; Clish, C.B.; DeBerardinis, R.J.; Feron, O.; Frezza, C.; Ghesquiere, B.; et al. A roadmap for interpreting 13C metabolite labeling patterns from cells. Curr. Opin. Biotechnol. 2015, 34, 189–201. [Google Scholar] [CrossRef] [PubMed]
- Yu, D.; Zhou, L.; Liu, X.; Xu, G. Stable isotope-resolved metabolomics based on mass spectrometry: Methods and their applications. Trends Anal. Chem. 2023, 160, 116985. [Google Scholar] [CrossRef]
- Han, S.; Van Treuren, W.; Fischer, C.R.; Merrill, B.D.; DeFelice, B.C.; Sanchez, J.M.; Higginbottom, S.K.; Guthrie, L.; Fall, L.A.; Dodd, D.; et al. A metabolomics pipeline for the mechanistic interrogation of the gut microbiome. Nature 2021, 595, 415–420. [Google Scholar] [CrossRef]
- Schultheisz, H.L.; Szymczyna, B.R.; Scott, L.G.; Williamson, J.R. Pathway Engineered Enzymatic de novo Purine Nucleotide Synthesis. ACS Chem Biol. 2008, 3, 499–511. [Google Scholar] [CrossRef]
- Rowbotham, J.S.; Ramirez, M.A.; Lenz, O.; Reeve, H.A.; Vincent, K.A. Bringing biocatalytic deuteration into the toolbox of asymmetric isotopic labelling techniques. Nat. Commun. 2020, 11, 1454. [Google Scholar] [CrossRef] [PubMed]
- Nicolaou, K.C.; Snyder, S.A. Chasing molecules that were never there: Misassigned natural products and the role of chemical synthesis in modern structure elucidation. Angew. Chem. Int. Ed. 2005, 44, 1012–1044. [Google Scholar] [CrossRef] [PubMed]
- Sunazuka, T.; Hirose, T.; Omura, S. Efficient Total Synthesis of Novel Bioactive Microbial Metabolites. Acc. Chem. Res. 2008, 41, 302–314. [Google Scholar] [CrossRef] [PubMed]
- Nicolaou, K.C. Organic synthesis: The art and science of replicating the molecules of living nature and creating others like them in the laboratory. Proc. R. Soc. A 2014, 470, 20130690. [Google Scholar] [CrossRef] [PubMed]
- Wohlgemuth, R. Route Selection and Reaction Engineering for Sustainable Metabolite Synthesis. React. Chem. Eng. 2023, 8, 2109–2118. [Google Scholar] [CrossRef]
- Arnold, F.H. Directed evolution: Bringing new chemistry to life. Angew. Chem. Int. Ed. 2018, 57, 4143–4148. [Google Scholar] [CrossRef]
- List, B. Introduction: Organocatalysis. Chem. Rev. 2007, 107, 5413–5415. [Google Scholar] [CrossRef]
- MacMillan, D. The advent and development of organocatalysis. Nature 2008, 455, 304–308. [Google Scholar] [CrossRef]
- Murray, J.; Hodgson, D.R.W.; O’Donoghue, A.C. Going Full Circle with Organocatalysis and Biocatalysis: The Latent Potential of Cofactor Mimics in Asymmetric Synthesis. J. Org. Chem. 2023, 88, 7619–7629. [Google Scholar] [CrossRef]
- Demain, A.L. From natural products discovery to commercialization: A success story. J. Ind. Microbiol. Biotechnol. 2006, 33, 486–495. [Google Scholar] [CrossRef]
- Hoff, B.; Plassmeier, J.; Blankschien, M.; Letzel, A.C.; Kourtz, L.; Schröder, H.; Koch, W.; Zelder, O. Unlocking Nature’s Biosynthetic Power—Metabolic Engineering for the Fermentative Production of Chemicals. Angew. Chem. Int. Ed. 2021, 60, 2258–2278. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.; Eun, H.; Prabowo, C.P.S.; Cho, S.; Lee, S.Y. Metabolic and cellular engineering for the production of natural products. Curr. Opin. Biotechnol. 2022, 77, 102760. [Google Scholar] [CrossRef] [PubMed]
- Wohlgemuth, R. Selective biocatalytic defunctionalization of raw materials. ChemSusChem 2022, 15, e202200402. [Google Scholar] [CrossRef] [PubMed]
- Kaiser, R. Scent of the Vanishing Flora; Wiley-VHCA AG: Zürich, Switzerland, 2010; ISBN 978-3-906390-64-2. [Google Scholar]
- Walsh, C.T.; Tang, Y. Natural Product Biosynthesis—Chemical Logic and Enzymatic Machinery; Royal Society of Chemistry: London, UK, 2017. [Google Scholar]
- Alcántara, A.R.; Dominguez de Maria, P.; Littlechild, J.A.; Schürmann, M.; Sheldon, R.A.; Wohlgemuth, R. Biocatalysis as key to sustainable industrial chemistry. ChemSusChem 2022, 15, e202102709. [Google Scholar] [CrossRef]
- Wohlgemuth, R. Tools and ingredients for the biocatalytic synthesis of metabolites. Biotechnol. J. 2009, 4, 1253–1265. [Google Scholar] [CrossRef]
- Wohlgemuth, R. Horizons of systems biocatalysis and renaissance of metabolite synthesis. Biotechnol. J. 2018, 13, 1700620. [Google Scholar] [CrossRef]
- Oberg, N.; Zallot, R.; Gerlt, J.A. EFI-EST, EFI-GNT, and EFI-CGFP: Enzyme Function Initiative (EFI) Web Resource for Genomic Enzymology Tools. J. Mol. Biol. 2023, 435, 168018. [Google Scholar] [CrossRef]
- Wittmann, C.; Liao, J.C. (Eds.) Industrial Biotechnology: Microorganism, 1st ed.; Wiley-VCH: Weinheim, Germany, 2017; ISBN 978-3-527-34179-5. [Google Scholar]
- Wittmann, C.; Liao, J.C. (Eds.) Industrial Biotechnology: Products and Processes, 1st ed.; Wiley-VCH: Weinheim, Germany, 2017; ISBN 978-3-527-34181-8. [Google Scholar]
- Lee, S.Y.; Nielsen, J.; Stephanopoulos, G. (Eds.) Metabolic Engineering—Concepts and Applications, 1st ed.; Wiley-VCH: Weinheim, Germany, 2021; ISBN 978-3-527-34662-2. [Google Scholar]
- Flickinger, M.C. (Ed.) Encyclopedia of Industrial Biotechnology: Bioprocess, Bioseparation, and Cell Technology; John Wliey & Sons: Hoboken, NJ, USA, 2010; Volumes 1–7, ISBN 978-0-471-79930-6. [Google Scholar]
- Wohlgemuth, R.; Littlechild, J. Complexity reduction and opportunities in the design, integration and intensification of biocata-lytic processes for metabolite synthesis. Front. Bioeng. Biotechnol. 2022, 10, 958606. [Google Scholar] [CrossRef]
- Wohlgemuth, R. Biocatalysis–Key enabling tools from biocatalytic one-step and multi-step reactions to biocatalytic total synthesis. New Biotechnol. 2021, 60, 113–123. [Google Scholar] [CrossRef]
- Wentrup, C. Origins of Organic Chemistry and Organic Synthesis. Eur. J. Org. Chem. 2022, 2022, e202101492. [Google Scholar] [CrossRef]
- Wöhler, F. Ueber künstliche Bildung des Harnstoffs. Ann. Phys. 1828, 88, 253–256. [Google Scholar] [CrossRef]
- Kolbe, H. Beiträge zur Kenntnis der gepaarten Verbindungen. Ann. Chem. Pharm. 1845, 54, 145–188. [Google Scholar] [CrossRef]
- Sheehan, J.C.; Henery-Logan, K.R. The total synthesis of penicillin V. J. Am. Chem. Soc. 1957, 79, 1262–1263. [Google Scholar] [CrossRef]
- Nicolaou, K.C.; Vourloumis, D.; Winssinger, N.; Baran, P.S. The art and science of total synthesis at the dawn of the twenty-first century. Angew. Chem. Int. Ed. 2000, 39, 44–122. [Google Scholar] [CrossRef]
- Veitch, G.E.; Boyer, A.; Ley, S.V. The Azadirachtin Story. Angew. Chem. Int. Ed. 2008, 47, 9402–9429. [Google Scholar] [CrossRef]
- Nicolaou, K.C.; Rigol, S. Perspectives from nearly five decades of total synthesis of natural products and their analogues for biology and medicine. Nat. Prod. Rep. 2020, 37, 1404–1435. [Google Scholar] [CrossRef] [PubMed]
- Min, L.; Han, J.C.; Zhang, W.; Gu, C.C.; Zou, Y.P.; Li, C.C. Strategies and Lessons Learned from Total Synthesis of Taxol. Chem. Rev. 2023, 123, 4934–4971. [Google Scholar] [CrossRef]
- Nicolaou, K.C.; Rigol, S.; Yu, R. Total synthesis endeavors and their contributions to science and society: A personal account. CCS Chemistry 2019, 1, 3–37. [Google Scholar] [CrossRef]
- Peters, D.S.; Pitts, C.R.; McClymont, K.S.; Stratton, T.P.; Bi, C.; Baran, P.S. Ideality in Context: Motivations for Total Synthesis. Acc. Chem. Res. 2021, 54, 605–617. [Google Scholar] [CrossRef]
- Cardwell, H.M.E.; Cornforth, J.W.; Duff, S.R.; Holtermann, H.; Robinson, R. Total synthesis of androgenic hormones. Chem. Ind. 1951, 20, 389–390. [Google Scholar]
- Woodward, R.B.; Sondheimer, F.; Taub, D. The total synthesis of cholesterol. J. Am. Chem. Soc. 1951, 73, 3548. [Google Scholar] [CrossRef]
- Woodward, R.B.; Sondheimer, F.; Taub, D.; Heusler, K.; McLamore, W.M. The Total Synthesis of Steroids. J. Am. Chem. Soc. 1952, 74, 4223–4251. [Google Scholar] [CrossRef]
- Munt, M.; Spieß, O.; Indolese, A.; Roux, L.; Giraud, M.; Schinzer, D. Short and Scalable Synthesis of Plant-Based Cholesterol in GMP Grade. Adv. Synth. Catal. 2023, 365, 2406–2409. [Google Scholar] [CrossRef]
- Eschenmoser, A.; Wintner, C.E. Natural Product Synthesis and Vitamin B12: Total synthesis of vitamin B12 provided a framework for exploration in several areas of organic chemistry. Science 1977, 196, 1410–1420. [Google Scholar] [CrossRef]
- Woodward, R.B. The total synthesis of vitamin B12. Pure Appl. Chem. 1973, 33, 145–178. [Google Scholar] [CrossRef] [PubMed]
- Scott, A.I. Discovering Nature’s Diverse Pathways to Vitamin B12: A 35-Year Odyssey. J. Org. Chem. 2003, 68, 2529–2539. [Google Scholar] [CrossRef]
- Eschenmoser, A. Vitamin B12: Experiments concerning the origin of its molecular structure. Angew. Chem. Int. Ed. 1988, 27, 5–39. [Google Scholar] [CrossRef]
- Kishi, Y. Palytoxin: An inexhaustible source of inspiration—Personal perspective. Tetrahedron 2002, 58, 6239–6258. [Google Scholar] [CrossRef]
- Sears, J.E.; Boger, D.L. Total Synthesis of Vinblastine, Related Natural Products, and Key Analogues and Development of Inspired Methodology Suitable for the Systematic Study of Their Structure−Function Properties. ACC Chem. Res. 2015, 48, 653–662. [Google Scholar] [CrossRef]
- Baran, P.S.; Maimone, T.J.; Richter, J.M. Total synthesis of marine natural products without using protecting groups. Nature 2007, 446, 404–408. [Google Scholar] [CrossRef]
- Pasteur, L. Mémoire sur la fermentation appelée lactique. Comptes Rendus Chim. 1857, 45, 913–916. [Google Scholar]
- Buchner, E. Alkoholische Gährung ohne Hefezellen. Berichte Der Dtsch. Chem. Ges. 1897, 30, 117–124. [Google Scholar] [CrossRef]
- Krebs, H.A.; Henseleit, K. Untersuchungen über die Harnstoffbildung im Tierkörper. Klin. Wochenschr. 1932, 11, 757–759. [Google Scholar] [CrossRef]
- Krebs, H.A.; Henseleit, K. Untersuchungen über die Harnstoffbildung im Tierkörper. II. Klin. Wochenschr. 1932, 11, 1137–1139. [Google Scholar] [CrossRef]
- Amato, A.; Becci, A.; Beolchini, F. Citric acid bioproduction: The technological innovation change. Crit. Rev. Biotechnol. 2020, 40, 199–212. [Google Scholar] [CrossRef]
- Feng, J.; Wu, Q.; Zhu, D.; Ma, Y. Biotransformation Enables Innovations Toward Green Synthesis of Steroidal Pharmaceuticals. ChemSusChem 2022, 15, e2021023. [Google Scholar] [CrossRef]
- Elander, R.P. Industrial production of β-lactam antibiotics. Appl. Microbiol. Biotechnol. 2003, 61, 385–392. [Google Scholar] [CrossRef]
- Meyer, H.P.; Robins, K.T. Large scale bioprocess for the production of optically pure L-carnitine. Monatsh. Chem. 2005, 136, 1269–1277. [Google Scholar] [CrossRef]
- Sanchez, S.; Rodríguez-Sanoja, R.; Ramos, A.; Demain, A.L. Our microbes not only produce antibiotics, they also overproduce amino acids. J. Antibiot. 2018, 71, 26–36. [Google Scholar] [CrossRef]
- Vandamme, E.J.; Revuelta, J.L. Industrial Biotechnology of Vitamins, Biopigments, and Antioxidants; Wiley-VCH: Weinheim, Germany, 2016. [Google Scholar] [CrossRef]
- Ramírez-Rendon, D.; Passari, A.K.; Ruiz-Villafán, B.; Rodríguez-Sanoja, R.; Sánchez, S.; Demain, A.L. Impact of novel microbial secondary metabolites on the pharma industry. Appl. Microbiol. Biotechnol. 2022, 106, 1855–1878. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Cabezón, L.; Galán, B.; García, J.L. New Insights on Steroid Biotechnology. Front. Microbiol. 2018, 9, 958. [Google Scholar] [CrossRef]
- Cheng, C.; Tsai, H.R. Yeast-mediated stereo-selective reduction of estrone by continuous cell culture with dual stirred tanks for product yield improvement. J. Chem. Technol. Biotechnol. 2011, 86, 601–607. [Google Scholar] [CrossRef]
- Chang, H.; Zhang, H.; Zhu, L.; Zhang, W.; You, S.; Qi, W.; Qian, J.; Su, R.; He, Z. A combined strategy of metabolic pathway regulation and two-step bioprocess for improved 4-androstene-3, 17-dione production with an engineered Mycobacterium neoaurum. Biochem. Eng. J. 2020, 164, 107789. [Google Scholar] [CrossRef]
- Su, B.M.; Zhao, H.R.; Xu, L.; Xu, X.Q.; Wang, L.C.; Lin, J.; Lin, W. Construction of an Efficient Non-natural Enzyme System for Preparation of Testosterone in High Space-Time Yield. ACS Sustain. Chem. Eng. 2022, 10, 3373–3382. [Google Scholar] [CrossRef]
- Kille, S.; Zilly, F.E.; Acevedo, J.P.; Reetz, M.T. Regio- and stereoselectivity of P450-catalysed hydroxylation of steroids controlled by laboratory evolution. Nat. Chem. 2011, 3, 738–743. [Google Scholar] [CrossRef]
- Pan, H.; Chang, S.; Qu, Y.; Liu, M.; Tian, W.; Chang, Z. Hydrocortisone production using whole-cell biocatalysts in recombinant Escherichia coli. Biochem. Eng. J. 2023, 198, 109023. [Google Scholar] [CrossRef]
- Gu, Y.; Jiao, X.; Ye, L.; Yu, H. Metabolic engineering strategies for de novo biosynthesis of sterols and steroids in yeast. Bioresour. Bioprocess. 2021, 8, 110. [Google Scholar] [CrossRef]
- Thoma, R.; Schulz-Gasch, T.; D’Arcy, B.; Benz, J.; Aebi, J.; Dehmlow, H.; Hennig, M.; Stihle, M.; Ruf, A. Insight into steroid scaffold formation from the structure of human oxidosqualene cyclase. Nature 2004, 432, 118–122. [Google Scholar] [CrossRef] [PubMed]
- Muthulakshmi, M.V.; Srinivasan, A.; Srivastava, S. Antioxidant Green Factories: Toward Sustainable Production of Vitamin E in Plant In Vitro Cultures. ACS Omega 2023, 8, 3586–3605. [Google Scholar] [CrossRef]
- Ye, Z.; Shi, B.; Huang, Y.; Ma, T.; Xiang, Z.; Hu, B.; Kuang, Z.; Huang, M.; Lin, X.; Tian, Z.; et al. Revolution of vitamin E production by starting from microbial fermented farnesene to isophytol. Innovation 2022, 3, 100228. [Google Scholar] [CrossRef]
- Zhu, K.; Jiang, M.; Ye, B.; Zhang, G.T.; Li, W.; Tang, P.; Huang, Z.; Chen, F. A unified strategy to prostaglandins: Chemoenzymatic total synthesis of cloprostenol, bimatoprost, PGF2a, fluprostenol, and travoprost guided by biocatalytic retrosynthesis. Chem. Sci. 2021, 12, 10362–10370. [Google Scholar] [CrossRef]
- Newman, D.J.; Cragg, G.M. Natural Products as Sources of New Drugs over the Nearly Four Decades from 01/1981 to 09/2019. J. Nat. Prod. 2020, 83, 770–803. [Google Scholar] [CrossRef]
- Bringi, V.; Kadkade, P.G.; Prince, C.L.; Roach, B.L. Enhanced Production of Taxol and Taxanes by Cell Cultures of Taxus Species. U.S. Patent 2013/0017582 A1, 17 January 2013. [Google Scholar]
- Sharma, A.; Bhatia, S.K.; Banyal, A.; Chanana, I.; Kumar, A.; Chand, D.; Kulshrestha, S.; Kumar, P. An Overview on Taxol Production Technology and Its Applications as Anticancer Agent. Biotechnol. Bioprocess Engin. 2022, 27, 706–728. [Google Scholar] [CrossRef]
- Omura, S. A Splendid Gift from the Earth: The Origins and Impact of the Avermectins (Nobel Lecture). Angew. Chem. Int. Ed. 2016, 55, 10190–10209. [Google Scholar] [CrossRef] [PubMed]
- Tu, Y. Artemisinin—A Gift from Traditional Chinese Medicine to the World (Nobel Lecture). Angew. Chem. Int. Ed. 2016, 55, 10210–10226. [Google Scholar] [CrossRef] [PubMed]
- Fischbach, M.A.; Walsh, C.T. Antibiotics for Emerging Pathogens. Science 2009, 325, 1089–1093. [Google Scholar] [CrossRef]
- Lewis, K. The Science of Antibiotic Discovery. Cell 2020, 181, 29–45. [Google Scholar] [CrossRef] [PubMed]
- Miethke, M.; Pieroni, M.; Weber, T.; Brönstrup, M.; Hammann, P.; Halby, L.; Arimondo, P.B.; Glaser, P.; Aigle, B.; Bode, H.B.; et al. Towards the sustainable discovery and development of new antibiotics. Nat. Rev. Chem. 2021, 5, 726–749. [Google Scholar] [CrossRef]
- Calvillo, A.; Pellicer, T.; Carnicer, M.; Planas, A. Bioprocess Strategies for Vitamin B12 Production by Microbial Fermentation and Market Applications. Bioengineering 2022, 9, 365. [Google Scholar] [CrossRef]
- Eichhorn, E.; Locher, E.; Guillemer, S.; Wahler, D.; Fourage, L.; Schilling, B. Biocatalytic process for (−)-Ambrox production using squalene hopene cyclase. Adv. Synth. Catal. 2018, 360, 2339–2351. [Google Scholar] [CrossRef]
- Meyer, H.P.; Eichhorn, E.; Hanlon, S.; Lütz, S.; Schürmann, M.; Wohlgemuth, R.; Coppolecchia, R. The use of enzymes in organic synthesis and the life sciences: Perspectives from the Swiss Industrial Biocatalysis Consortium (SIBC). Catal. Sci. Technol. 2013, 3, 29–40. [Google Scholar] [CrossRef]
- Bühlmann, P.; Pretsch, E.; Bakker, E. Carrier-Based Ion-Selective Electrodes and Bulk Optodes. 2. Ionophores for Potentiometric and Optical Sensors. Chem. Rev. 1998, 98, 1593–1687. [Google Scholar] [CrossRef] [PubMed]
- Jani, P.; Emmert, J.; Wohlgemuth, R. Process analysis of macrotetrolide biosynthesis during fermentation by means of direct infusion LC-MS. Biotechnol. J. 2008, 3, 202–208. [Google Scholar] [CrossRef] [PubMed]
- Cuartero, M.; Colozza, N.; Fernández-Pérez, B.M.; Crespo, G.A. Why ammonium detection is particularly challenging but insightful with ionophore-based potentiometric sensors—An overview of the progress in the last 20 years. Analyst 2020, 145, 3188–3210. [Google Scholar] [CrossRef] [PubMed]
- Gauss, D.; Schoenenberger, B.; Wohlgemuth, R. Chemical and enzymatic methodologies for the synthesis of enantiomerically pure glyceraldehyde 3-phosphates. Carbohydr. Res. 2014, 389, 18–24. [Google Scholar] [CrossRef] [PubMed]
- Molla, G.S.; Kinfu, B.M.; Chow, J.; Streit, W.; Wohlgemuth, R.; Liese, A. Bioreaction engineering leading to efficient synthesis of L-glyceraldehyd-3-phosphate. Biotechnol. J. 2017, 12, 1600625. [Google Scholar] [CrossRef] [PubMed]
- Richter, N.; Neumann, M.; Liese, A.; Wohlgemuth, R.; Eggert THummel, W. Characterisation of a Recombinant NADP-Dependent Glycerol Dehydrogenase from Gluconobacter oxydans and its Application in the Production of L-Glyceraldehyde. ChemBioChem 2009, 10, 1888–1896. [Google Scholar] [CrossRef]
- Richter, N.; Neumann, M.; Liese, A.; Wohlgemuth, R.; Weckbecker, A.; Eggert, T.; Hummel, W. Characterization of a whole-cell catalyst co-expressing glycerol dehydrogenase and glucose dehydrogenase and its application in the synthesis of L-glyceraldehyde. Biotechnol. Bioeng. 2010, 106, 541–552. [Google Scholar] [CrossRef]
- Gauss, D.; Sánchez-Moreno, I.; Oroz-Guinea, I.; García-Junceda, E.; Wohlgemuth, R. Phosphorylation catalyzed by dihydroxyacetone kinase. Eur. J. Org. Chem. 2018, 23, 2892–2895. [Google Scholar] [CrossRef]
- Hardt, N.; Kinfu, B.M.; Chow, J.; Schoenenberger, B.; Streit, W.R.; Obkircher, M.; Wohlgemuth, R. Biocatalytic Asymmetric Phosphorylation Catalyzed by Recombinant Glycerate-2-Kinase. ChemBioChem 2017, 18, 1518–1522. [Google Scholar] [CrossRef]
- Shaeri, J.; Wohlgemuth, R.; Woodley, J.M. Semiquantitative process screening for the biocatalytic synthesis of D-xylulose-5-phosphate. Org. Proc. Res. Dev. 2006, 10, 605–610. [Google Scholar] [CrossRef]
- Shaeri, J.; Wright, I.; Rathbone, E.B.; Wohlgemuth, R.; Woodley, J.M. Characterization of enzymatic D-xylulose 5-phosphate synthesis. Biotechnol. Bioeng. 2008, 101, 761–767. [Google Scholar] [CrossRef] [PubMed]
- Hardt, N.; Kind, S.; Schoenenberger, B.; Eggert, T.; Obkircher, M.; Wohlgemuth, R. Facile synthesis of D-xylulose-5-phosphate and L-xylulose-5-phosphate by xylulokinase-catalyzed phosphorylation. Biocatal. Biotransform. 2020, 38, 35–45. [Google Scholar] [CrossRef]
- Schoenenberger, B.; Kind, S.; Meier, R.; Eggert, T.; Obkircher, M.; Wohlgemuth, R. Efficient biocatalytic synthesis of D-tagatose 1, 6-diphosphate by LacC-catalysed phosphorylation of D-tagatose 6-phosphate. Biocatal. Biotransform. 2020, 38, 53–63. [Google Scholar] [CrossRef]
- Krevet, S.; Shen, L.; Bohnen, T.; Schoenenberger, B.; Meier, R.; Obkircher, M.; Bangert, K.; Koehling, R.; Allenspach, E.; Wohlgemuth, R.; et al. Enzymatic synthesis of 2-keto-3-deoxy-6-phosphogluconate by the 6-phosphogluconate-dehydratase from Caulobacter crescentus. Front. Bioeng. Biotechnol. 2020, 8, 185. [Google Scholar] [CrossRef] [PubMed]
- Shen, L.; Kohlhaas, M.; Enoki, J.; Meier, R.; Schönenberger, B.; Wohlgemuth, R.; Kourist, R.; Niemeyer, F.; van Niekerk, D.; Bräsen, C.; et al. A combined experimental and modelling approach for the Weimberg pathway optimisation. Nat. Commun. 2020, 11, 1098. [Google Scholar] [CrossRef]
- Matsubara, K.; Köhling, R.; Schönenberger, B.; Kouril, T.; Esser, D.; Bräsen, C.; Siebers, B.; Wohlgemuth, R. One-step synthesis of 2-keto-3-deoxy-D-gluconate by biocatalytic dehydration of D-gluconate. J. Biotechnol. 2014, 191, 69–77. [Google Scholar] [CrossRef]
- Schoenenberger, B.; Wszolek, A.; Milesi, T.; Brundiek, H.; Obkircher, M.; Wohlgemuth, R. Synthesis of Nω-Phospho-L-arginine by Biocatalytic Phosphorylation of L-Arginine. ChemCatChem 2017, 9, 121–126. [Google Scholar] [CrossRef]
- Schoenenberger, B.; Wszolek, A.; Meier, R.; Brundiek, H.; Obkircher, M.; Wohlgemuth, R. Recombinant AroL-Catalyzed Phosphorylation for the Efficient Synthesis of Shikimic Acid 3-Phosphate. Biotechnol. J. 2018, 13, 1700529. [Google Scholar] [CrossRef]
- Schell, U.; Wohlgemuth, R.; Ward, J.M. Synthesis of pyridoxamine 5′-phosphate using an MBA: Pyruvate transaminase as biocatalyst. J. Mol. Catal. B Enzym. 2009, 59, 279–285. [Google Scholar] [CrossRef]
- Schoenenberger, B.; Wszolek, A.; Meier, R.; Brundiek, H.; Obkircher, M.; Wohlgemuth, R. Biocatalytic asymmetric Michael addition reaction of l-arginine to fumarate for the green synthesis of N-(([(4S)-4-amino-4-carboxy-butyl] amino) iminomethyl)-L-aspartic acid lithium salt (L-argininosuccinic acid lithium salt). RSC Adv. 2017, 7, 48952–48957. [Google Scholar] [CrossRef]
- Schoenenberger, B.; Wszolek, A.; Meier, R.; Brundiek, H.; Obkircher, M.; Wohlgemuth, R. Biocatalytic Asymmetric Aza-Michael Addition Reactions and Synthesis of L-Argininosuccinate by Argininosuccinate Lyase ARG4-Catalysed Aza-Michael Addition of L-Arginine to Fumarate. In Applied Biocatalysis; Whittall, J., Sutton, P.W., Eds.; John Wiley & Sons: Hoboken, NJ, USA, 2021; pp. 204–210. [Google Scholar]
- Yasutake, Y.; Nishioka, T.; Imoto, N.; Tamura, T. A Single Mutation at the Ferredoxin Binding Site of P450 Vdh Enables Efficient Biocatalytic Production of 25-Hydroxyvitamin D3. ChemBioChem 2013, 14, 2284–2291. [Google Scholar] [CrossRef] [PubMed]
- Tang, D.; Liu, W.; Huang, L.; Cheng, L.; Xu, Z. Efficient biotransformation of vitamin D3 to 25-hydroxyvitamin D3 by a newly isolated Bacillus cereus strain. Appl. Microbiol. Biotechnol. 2020, 104, 765–774. [Google Scholar] [CrossRef] [PubMed]
- Babot, E.D.; del Río, J.C.; Kalum, L.; Martínez, A.T.; Gutiérrez, A. Regioselective Hydroxylation in the Production of 25-Hydroxyvitamin D by Coprinopsis cinerea Peroxygenase. ChemCatChem 2015, 7, 283–290. [Google Scholar] [CrossRef]
- Warnke, M.; Jung, T.; Dermer, J.; Hipp, K.; Jehmlich, N.; von Bergen, M.; Ferlaino, S.; Fries, A.; Müller, M.; Boll, M. 25-Hydroxyvitamin D3 Synthesis by Enzymatic Steroid Side-Chain Hydroxylation with Water. Angew. Chem. Int. Ed. 2016, 55, 1881–1884. [Google Scholar] [CrossRef]
- Kang, D.J.; Im, J.H.; Kang, J.H.; Kim, K.H. Whole cell bioconversion of vitamin D3 to calcitriol using Pseudonocardia sp. KCTC 1029BP. Bioprocess Biosyst. Eng. 2015, 38, 1281–1290. [Google Scholar] [CrossRef]
- Taniguchi, T.; Eto, T.A.; Shiotsuki, H.; Sueta, H.; Higashi, S.; Iwamura, T.; Okuda, K.I.; Setoguchi, T. Newly established assay method for 25-hydroxyvitamin D3 24-hydroxylase revealed much lower Km for 25-hydroxyvitamin D3 than for 1alpha,25-dihydroxyvitamin D3. J. Bone Miner. Res. 2001, 16, 57–62. [Google Scholar] [CrossRef]
- Yasuda, K.; Yogo, Y.; Sugimoto, H.; Mano, H.; Takita, T.; Ohta, M.; Kamakura, M.; Ikushiro, S.; Yasukawa, K.; Shiro, Y.; et al. Production of an active form of vitamin D2 by genetically engineered CYP105A1. Biochem. Biophys. Res. Commun. 2017, 486, 336–341. [Google Scholar] [CrossRef]
- Putkaradze, N.; König, L.; Kattner, L.; Hutter, M.C.; Bernhardt, R. Highly regio- and stereoselective hydroxylation of vitamin D2 by CYP109E1. Biochem. Biophys. Res. Commun. 2020, 524, 295–300. [Google Scholar] [CrossRef]
- Coene, K.L.; Kluijtmans, L.A.; van der Heeft, E.; Engelke, U.F.; de Boer, S.; Hoegen, B.; Kwast, H.J.; van de Vorst, M.; Huigen, M.C.; Keularts, I.M.; et al. Next-generation metabolic screening: Targeted and untargeted metabolomics for the diagnosis of inborn errors of metabolism in individual patients. J. Inherit. Metab. Dis. 2018, 41, 337–353. [Google Scholar] [CrossRef]
- Pavlova, N.N.; Thompson, C.B. The emerging hallmarks of cancer metabolism. Cell Metab. 2016, 23, 27–47. [Google Scholar] [CrossRef] [PubMed]
- Nemet, I.; Saha, P.P.; Gupta, N.; Zhu, W.; Romano, K.A.; Skye, S.M.; Cajka, T.; Mohan, M.L.; Li, L.; Wu, Y.; et al. A cardiovascular disease-linked gut microbial metabolite acts via adrenergic receptors. Cell 2020, 180, 862–877. [Google Scholar] [CrossRef] [PubMed]
- Calvin, M. The Path of Carbon in Photosynthesis: The carbon cycle is a tool for exploring chemical biodynamics and the mechanism of quantum conversion. Science 1962, 135, 879–889. [Google Scholar] [CrossRef] [PubMed]
- Rising, K.A.; Schramm, V.L. Enzymatic Synthesis of NAD+ with the Specific Incorporation of Atomic Labels. J. Am. Chem. Soc. 1994, 116, 6531–6536. [Google Scholar] [CrossRef]
- Tran, A.; Yokose, R.; Cen, Y. Chemo-enzymatic synthesis of isotopically labelled nicotinamide riboside. Org. Biomol. Chem. 2018, 16, 3662–3671. [Google Scholar] [CrossRef]
- Khoroshilov, A.V. Production of stable isotopes of light elements: Past, present and future. J. Phys. Conf. Ser. 2018, 1099, 012002. [Google Scholar] [CrossRef]
- Letertre, M.P.M.; Dervilly, G.; Giraudeau, P. Combined Nuclear Magnetic Resonance Spectroscopy and Mass Spectrometry Approaches for Metabolomics. Anal. Chem. 2021, 93, 500–518. [Google Scholar] [CrossRef]
- Sauer, U.; Lasko, D.R.; Fiaux, J.; Hochuli, M.; Glaser, R.; Szyperski, T.; Wüthrich, K.; Bailey, J.E. Metabolic flux ratio analysis of genetic and environmental modulations of Escherichia coli central carbon metabolism. J. Bacteriol. 1999, 181, 6679–6688. [Google Scholar] [CrossRef]
- Faubert, B.; Tasdogan, A.; Morrison, S.J.; Mathews, T.P.; DeBerardinis, R.J. Stable isotope tracing to assess tumor metabolism in vivo. Nat. Protoc. 2021, 16, 5123–5145. [Google Scholar] [CrossRef]
- Steinhauser, M.L.; Bailey, A.P.; Senyo, S.E.; Guillermier, C.; Perlstein, T.S.; Gould, A.P.; Lee, R.T.; Lechene, C.P. Multi-isotope imaging mass spectrometry quantifies stem cell division and metabolism. Nature 2012, 481, 516–519. [Google Scholar] [CrossRef]
- Grey, A.C.; Tang, M.; Zahraei, A.; Guo, G.; Demarais, N.J. Applications of stable isotopes in MALDI imaging: Current approaches and an eye on the future. Anal. Bioanal. Chem. 2021, 413, 2637–2653. [Google Scholar] [CrossRef]
- Fan, T.W.M.; Lane, A.N. Applications of NMR spectroscopy to systems biochemistry. Prog. Nucl. Magn. Reson. Spectrosc. 2016, 92–93, 18–53. [Google Scholar] [CrossRef]
- Giraudeau, P. Quantitative NMR spectroscopy of complex mixtures. Chem. Commun. 2023, 59, 6627–6642. [Google Scholar] [CrossRef]
- Wohlgemuth, R.; Waespe-Sarcevic, N.; Seelig, J. Bilayers of Phosphatidylglycerol. A Deuterium and Phosphorus NuclearMagnetic Resonance Study of the Head-Group Region. Biochemistry 1980, 19, 3315–3321. [Google Scholar] [CrossRef] [PubMed]
- Chun, S.W.; Narayan, A.R.H. Biocatalytic, Stereoselective Deuteration of α-Amino Acids and Methyl Esters. ACS Catal. 2020, 10, 7413–7418. [Google Scholar] [CrossRef] [PubMed]
- Doyon, T.J.; Buller, A.R. Site-Selective Deuteration of Amino Acids through Dual-Protein Catalysis. J. Am. Chem. Soc. 2022, 144, 7327–7336. [Google Scholar] [CrossRef] [PubMed]
- Rowbotham, J.S.; Hardy, A.P.; Reeve, H.A.; Vincent, K.A. Synthesis of [4S-2H] NADH, [4R-2H] NADH, [4-2H2] NADH and [4-2H] NAD+ cofactors through heterogeneous biocatalysis in heavy water. J. Label. Compd. Radiopharm. 2021, 64, 181–186. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Lou, Y.; Wang, L.; Wang, Z.; Xu, W.; Ma, W.; Chen, Z.; Chen, X.; Wu, Q. Rational Design of Biocatalytic Deuteration Platform of Aldehydes. ACS Catal. 2021, 11, 13348–13354. [Google Scholar] [CrossRef]
- Tolbert, T.J.; Williamson, J.R. Preparation of Specifically Deuterated RNA for NMR Studies Using a Combination of Chemical and Enzymatic Synthesis. J. Am. Chem. Soc. 1996, 118, 7929–7940. [Google Scholar] [CrossRef]
- Bennett, B.D.; Yuan, J.; Kimball, E.H.; Rabinowitz, J.D. Absolute quantitation of intracellular metabolite concentrations by an isotope ratio-based approach. Nat. Protoc. 2008, 3, 1299–1311. [Google Scholar] [CrossRef]
- Sauer, U. Metabolic networks in motion: 13C-based flux analysis. Mol. Syst. Biol. 2006, 2, 62. [Google Scholar] [CrossRef] [PubMed]
- Arrivault, S.; Guenther, M.; Fry, S.C.; Fuenfgeld, M.F.F.F.; Veyel, D.; Mettler-Altmann, T.; Stitt, M.; Lunn, J.E. Synthesis and Use of Stable-Isotope-Labelled Internal Standards for Quantification of Phosphorylated Metabolites by LC–MS/MS. Anal. Chem. 2015, 87, 6896–6904. [Google Scholar] [CrossRef]
- Eisenreich, W.; Schwarz, M.; Cartayrade, A.; Arigoni, D.; Zenk, M.H.; Bacher, A. The deoxyxylulose phosphate pathway of terpenoid biosynthesis in plants and microorganisms. Chem. Biol. 1998, 5, R221–R233. Available online: http://biomednet.com/elecref/10745521005R0221 (accessed on 8 August 2023). [CrossRef] [PubMed]
- Zhang, W.; Zhao, S.; Serianni, A.S. Labeling monosaccharides with stable isotopes. Methods Enzymol. 2015, 565, 423–458. [Google Scholar] [CrossRef] [PubMed]
- Goux, W.J.; Rench, L.; Weber, D.S. Stereoselective synthesis of stable isotope labeled L-α-amino acids: The enzymatic preparation of 13C-labeled L-glutamic acids. J. Label. Compd. Radiopharm. 1993, 33, 181–193. [Google Scholar] [CrossRef]
- Maeda, H.; Takata, K.; Toyoda, A.; Niitsu, T.; Iwakura, M.; Shibata, K. Production of L-[3-13C] serine from [13C] formaldehyde and glycine using an enzyme system combined with tetrahydrofolate regeneration. J. Ferment. Bioeng. 1997, 83, 113–115. [Google Scholar] [CrossRef]
- Jemielity, J.; Kańska, M.; Kański, R. Enzymatic Synthesis of [1-13C]-and [1-14C]-L-Phenyl-Alanine. Isot. Environ. Health Stud. 1998, 34, 335–339. [Google Scholar] [CrossRef]
- Akita, H.; Suzuki, H.; Doi, K.; Ohshima, T. Efficient synthesis of D-branched-chain amino acids and their labeled compounds with stable isotopes using D-amino acid dehydrogenase. Appl. Microbiol. Biotechnol. 2014, 98, 1135–1143. [Google Scholar] [CrossRef]
- Van Raad, D.; Huber, T.; Otting, G. Improved spectral resolution of [13C,1H]-HSQC spectra of aromatic amino acid residues in proteins produced by cell-free synthesis from inexpensive 13C-labelled precursors. J. Biomol. NMR 2023, 77, 183–190. [Google Scholar] [CrossRef]
- Guo, X.; Liu, Y.; Wang, Q.; Wang, X.; Li, Q.; Liu, W.; Zhao, Z.K. Non-natural Cofactor and Formate-Driven Reductive Carboxylation of Pyruvate. Angew. Chem. Int. Ed. 2020, 59, 3143–3146. [Google Scholar] [CrossRef]
- Morgan, K.D. The use of nitrogen-15 in microbial natural product discovery and biosynthetic characterization. Front. Microbiol. 2023, 14, 1174591. [Google Scholar] [CrossRef] [PubMed]
- Chiriaca, M.; Lupan, I.; Popa, F.; Palibroda, N.; Popescu, O. Enzymatic synthesis of some 15N-labelled L-amino acids. Isot. Environ. Health Stud. 2010, 46, 249–254. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Asam, S.; Chen, J.; Ehrmann, M.; Rychlik, M. Production of Four 15N-Labelled Cobalamins via Biosynthesis Using Propionibacterium freudenreichii. Front. Microbiol. 2021, 12, 713321. [Google Scholar] [CrossRef] [PubMed]
- Wishart, D.S.; Feunang, Y.D.; Guo, A.C.; Lo, E.J.; Marcu, A.; Grant, J.R.; Sajed, T.; Johnson, D.; Li, C.; Sayeeda, Z.; et al. DrugBank 5.0: A major update to the DrugBank database for 2018. Nucleic Acids Res. 2018, 46, D1074–D1082. [Google Scholar] [CrossRef]
- Clayton, T.A.; Baker, D.; Lindon, J.C.; Everett, J.R.; Nicholson, J.K. Pharmacometabonomic identification of a significant host-microbiome metabolic interaction affecting human drug metabolism. Proc. Natl. Acad. Sci. USA 2009, 106, 14728–14733. [Google Scholar] [CrossRef]
- Zimmermann, M.; Zimmermann-Kogadeeva, M.; Wegmann, R.; Goodman, A.L. Mapping human microbiome drug metabolism by gut bacteria and their genes. Nature 2019, 570, 462–467. [Google Scholar] [CrossRef]
- Javdan, B.; Lopez, J.G.; Chankhamjon, P.; Lee, Y.C.J.; Hull, R.; Wu, Q.; Wang, X.; Chatterjee, S.; Donia, M.S. Personalized mapping of drug metabolism by the human gut microbiome. Cell 2020, 181, 1661–1679.e22. [Google Scholar] [CrossRef]
- Heinken, A.; Hertel, J.; Acharya, G.; Ravcheev, D.A.; Nyga, M.; Okpala, O.E.; Hogan, M.; Magnúsdóttir, S.; Martinelli, F.; Nap, B.; et al. Genome-scale metabolic reconstruction of 7,302 human microorganisms for personalized medicine. Nat. Biotechnol. 2023, 41, 1320–1331. [Google Scholar] [CrossRef]
- Fura, A.; Shu, Y.Z.; Zhu, M.; Hanson, R.L.; Roongta, V.; Humphreys, W.G. Discovering Drugs through Biological Transformation: Role of Pharmacologically Active Metabolites in Drug Discovery. J. Med. Chem. 2004, 47, 4339–4351. [Google Scholar] [CrossRef]
- Rautio, J.; Meanwell, N.A.; Di, L.; Hageman, M.J. The expanding role of prodrugs in contemporary drug design and development. Nat. Rev. Drug Discov. 2018, 17, 559–587. [Google Scholar] [CrossRef]
- Schadt, S.; Bister, B.; Chowdhury, S.K.; Funk, C.; Hop, C.E.C.A.; Humphreys, W.G.; Igarashi, F.; James, A.D.; Kagan, M.; Khojasteh, S.C.; et al. A Decade in the MIST: Learnings from Investigations of Drug Metabolites in Drug Development under the “Metabolites in Safety Testing” Regulatory Guidance. Drug Metab. Dispos. 2018, 46, 865–878. [Google Scholar] [CrossRef] [PubMed]
- US Food and Drug Administration (FDA). Safety Testing of Drug Metabolites. 2020. Available online: https://www.fda.gov/media/72279/download (accessed on 8 August 2023).
- Luffer-Atlas, D.; Obach, R.S.; Smith, D.A. A MIST conception: What has been learned from twenty years of human metabolite safety assessment? Med. Chem. Res. 2023, 32, 1933–1949. [Google Scholar] [CrossRef]
- Chhatrapati Bisen, A.; Nashik Sanap, S.; Agrawal, S.; Biswas, A.; Sankar Bhatta, R. Chemical metabolite synthesis and profiling: Mimicking in vivo biotransformation reactions. Bioorg. Chem. 2023, 139, 106722. [Google Scholar] [CrossRef]
- Winkler, M.; Geier, M.; Hanlon, S.P.; Nidetzky, B.; Glieder, A. Human enzymes for organic synthesis. Angew. Chem. Int. Ed. 2018, 57, 13406–13423. [Google Scholar] [CrossRef] [PubMed]
- Naumann, J.M.; Zöllner, A.; Drăgan, C.A.; Messinger, J.; Adam, J.; Bureik, M. Biotechnological Production of 20-alpha-Dihydrodydrogesterone at Pilot Scale. Appl. Biochem. Biotechnol. 2011, 165, 190–203. [Google Scholar] [CrossRef] [PubMed]
- Brinkmann, V.; Billich, A.; Baumruker, T.; Heining, P.; Schmouder, R.; Francis, G.; Aradhye, S.; Burtin, P. Fingolimod (FTY720): Discovery and development of an oral drug to treat multiple sclerosis. Nat. Rev. Drug Discov. 2010, 9, 883–897. [Google Scholar] [CrossRef]
- Kittelmann, M.; Rheinegger, U.; Espigat, A.; Oberer, L.; Aichholz, R.; Francotte, E.; Ghisalba, O. Preparative Enzymatic Synthesis of the Acylglucuronide of Mycophenolic Acid. Adv. Synth. Catal. 2003, 345, 825–829. [Google Scholar] [CrossRef]
- Rautio, J.; Kumpulainen, H.; Heimbach, T.; Oliyai, R.; Oh, D.; Järvinen, T.; Savolainen, J. Prodrugs: Design and clinical applications. Nat. Rev. Drug Discov. 2008, 7, 255–270. [Google Scholar] [CrossRef]
- Wolff, N.A.; Burckhardt, B.C.; Burckhardt, G.; Oellerich, M.; Armstrong, V.W. Mycophenolic acid (MPA) and its glucuronide metabolites interact with transport systems responsible for excretion of organic anions in the basolateral membrane of the human kidney. Nephrol. Dial. Transplant. 2007, 22, 2497–2503. [Google Scholar] [CrossRef]
- Park, B.K.; Boobis, A.; Clarke, S.; Goldring, C.E.P.; Jones, D.; Kenna, J.G.; Lambert, C.; Laverty, H.G.; Naisbitt, D.J.; Nelson, S.; et al. Managing the challenge of chemically reactive metabolites in drug development. Nat. Rev. Drug Discov. 2011, 10, 292–306. [Google Scholar] [CrossRef]
- Tateishi, Y.; Ohe, T.; Ogawa, M.; Takahashi, K.; Nakamura, S.; Mashino, T. Development of Novel Diclofenac Analogs Designed to Avoid Metabolic Activation and Hepatocyte Toxicity. ACS Omega 2020, 5, 32608–32616. [Google Scholar] [CrossRef] [PubMed]
- Guengerich, F.P. A history of the roles of cytochrome P450 enzymes in the toxicity of drugs. Toxicol. Res. 2021, 37, 1–23. [Google Scholar] [CrossRef]
- Dahlin, D.C.; Miwa, G.T.; Lu, A.Y.; Nelson, S.D. N-acetyl-p-benzoquinone imine: A cytochrome P-450-mediated oxidation pro-duct of acetaminophen. Proc. Natl. Acad. Sci. USA 1984, 81, 1327–1331. [Google Scholar] [CrossRef] [PubMed]
- Bender, R.P.; Lindsey, R.H., Jr.; Burden, D.A.; Osheroff, N. N-Acetyl-p-benzoquinone Imine, the Toxic Metabolite of Acetaminophen, Is a Topoisomerase II Poison. Biochemistry 2004, 43, 3731–3739. [Google Scholar] [CrossRef] [PubMed]
- Ertl, P.; Roggo, S.; Schuffenhauer, A. Natural Product-likeness Score and Its Application for Prioritization of Compound Libraries. J. Chem. Inf. Model. 2008, 48, 68–74. [Google Scholar] [CrossRef] [PubMed]
- Tay, D.W.P.; Yeo, N.Z.X.; Adaikkappan, K.; Lim, Y.H.; Ang, S.J. 67 million natural product-like compound database generated via molecular language processing. Sci. Data 2023, 10, 296. [Google Scholar] [CrossRef] [PubMed]
- Dobson, P.D.; Patel, Y.; Kell, D.B. Metabolite-likeness’ as a criterion in the design and selection of pharmaceutical drug libraries. Drug Discov. Today 2009, 14, 31–40. [Google Scholar] [CrossRef]
- O’Hagan, S.; Kell, D.B. Understanding the foundations of the structural similarities between marketed drugs and endogenous human metabolites. Front. Pharmacol. 2015, 6, 105. [Google Scholar] [CrossRef][Green Version]
- O′Hagan, S.; Swainston, N.; Handl, J.; Kell, D.B. A ‘rule of 0.5’ for the metabolite-likeness of approved pharmaceutical drugs. Metabolomics 2015, 11, 323–339. [Google Scholar] [CrossRef]
- Ertl, P. Substituents of life: The most common substituent patterns present in natural products. Bioorg. Med. Chem. 2022, 54, 116562. [Google Scholar] [CrossRef]
- Shultz, M.D. Two Decades under the Influence of the Rule of Five and the Changing Properties of Approved Oral Drugs. J. Med. Chem. 2019, 62, 1701–1714. [Google Scholar] [CrossRef] [PubMed]
- Brown, D.G.; Wobst, J.J. A Decade of FDA-Approved Drugs (2010−2019): Trends and Future Directions. J. Med. Chem. 2021, 64, 2312–2338. [Google Scholar] [CrossRef] [PubMed]
- Young, R.J.; Flitsch, S.L.; Grigalunas, M.; Leeson, P.D.; Quinn, R.J.; Turner, N.J.; Waldmann, H. The Time and Place for Nature in Drug Discovery. JACS Au 2022, 2, 2400–2416. [Google Scholar] [CrossRef] [PubMed]
- Saha, S.; Rajpal, D.K.; Brown, J.R. Human microbial metabolites as a source of new drugs. Drug Discov. Today 2016, 21, 692–698. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Ranhotra, H.S.; Mani, S.; Dvořák, Z.; Sokol, H.; Müller, R. Human microbial metabolite mimicry as a strategy to expand the chemical space of potential drugs. Drug Discov. Today 2020, 25, 1575–1579. [Google Scholar] [CrossRef]
- Dvorák, Z.; Kopp, F.; Costello, C.M.; Kemp, J.S.; Li, H.; Vrzalová, A.; Stepánková, M.; Bartonková, I.; Jiskrová, E.; Poulíková, K.; et al. Targeting the pregnane X receptor using microbial metabolite mimicry. EMBO Mol. Med. 2020, 12, e11621. [Google Scholar] [CrossRef]
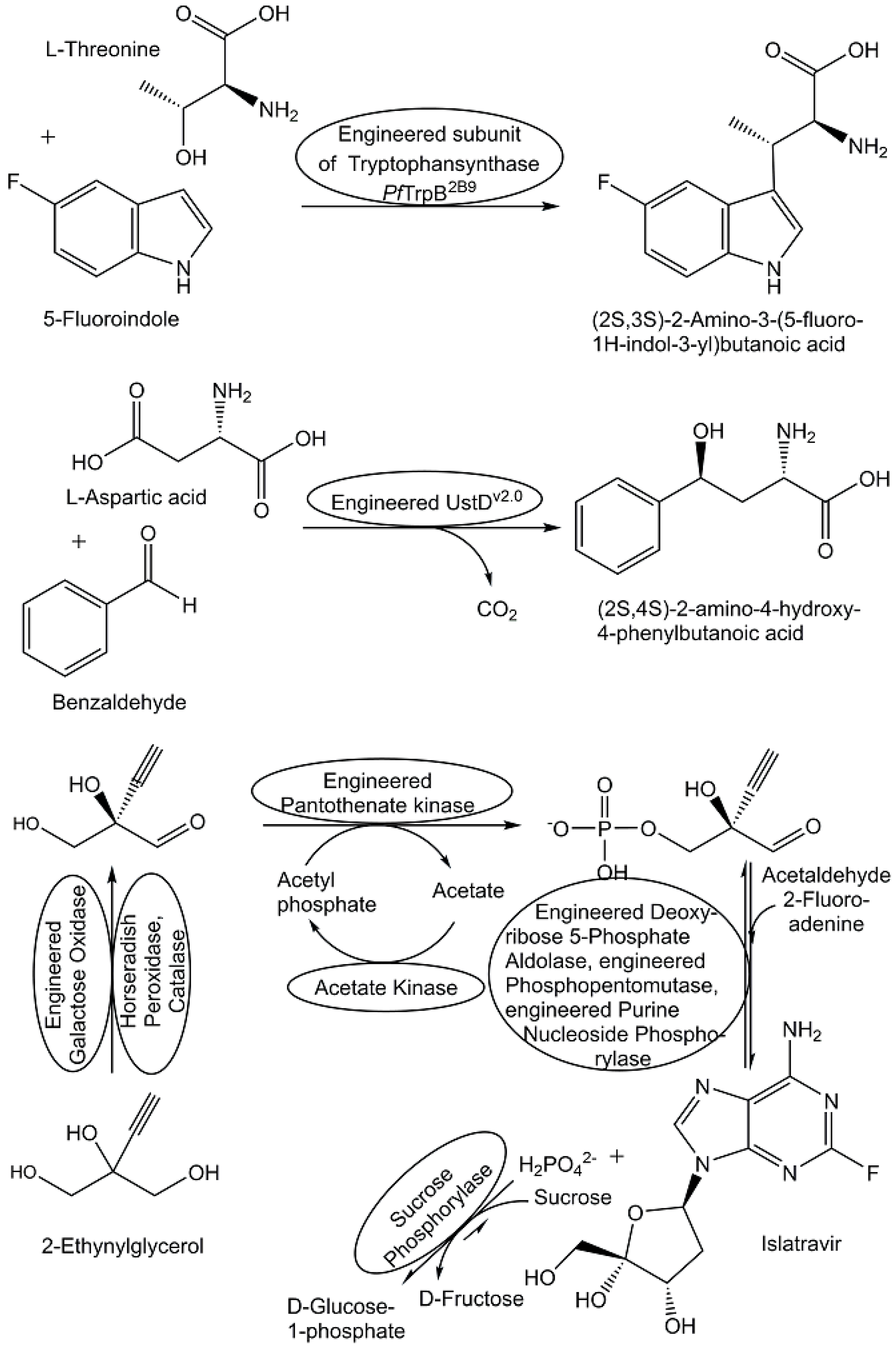
- Xue, Y.P.; Cao, C.H.; Zheng, Y.G. Enzymatic asymmetric synthesis of chiral amino acids. Chem. Soc. Rev. 2018, 47, 1516–1561. [Google Scholar] [CrossRef]
- Herger, M.; van Roye, P.; Romney, D.K.; Brinkmann-Chen, S.; Buller, A.R.; Arnold, F.H. Synthesis of β-branched tryptophan analogues using an engineered subunit of tryptophan synthase. J. Am. Chem. Soc. 2016, 138, 8388–8391. [Google Scholar] [CrossRef]
- Alfonzo, E.; Das, A.; Arnold, F.H. New Additions to the Arsenal of Biocatalysts for Noncanonical Amino Acid Synthesis. Curr. Opin. Green Sustain. Chem. 2022, 38, 100701. [Google Scholar] [CrossRef]
- Ellis, J.M.; Campbell, M.E.; Kumar, P.; Geunes, E.P.; Bingman, C.A.; Buller, A.R. Biocatalytic synthesis of non-standard amino acids by a decarboxylative aldol reaction. Nat. Catal. 2022, 5, 136–143. [Google Scholar] [CrossRef]
- Huffman, M.A.; Fryszkowska, A.; Alvizo, O.; Borra-Garske, M.; Campos, K.R.; Canada, K.A.; Devine, P.N.; Duan, D.; Forstater, J.H.; Grosser, S.T.; et al. Design of an in vitro biocatalytic cascade for the manufacture of islatravir. Science 2019, 366, 1255–1259. [Google Scholar] [CrossRef]
- Zetzsche, L.E.; Narayan, A.R.H. Broadening the scope of biocatalytic C–C bond formation. Nat. Rev. Chem. 2020, 4, 334–346. [Google Scholar] [CrossRef] [PubMed]
- Schneider, P.; Henßen, B.; Paschold, B.; Chapple, B.P.; Schatton, M.; Seebeck, F.P.; Classen, T.; Pietruszka, J. Biocatalytic C3-Indole Methylation—A Useful Tool for the Natural-Product-Inspired Stereoselective Synthesis of Pyrroloindoles. Angew. Chem. Int. Ed. 2021, 60, 23412–23418. [Google Scholar] [CrossRef]
- Fansher, D.J.; Palmer, D.R. A Type 1 Aldolase, NahE, Catalyzes a Stereoselective Nitro-Michael Reaction: Synthesis of β-Aryl-γ-nitrobutyric Acids. Angew. Chem. Int. Ed. 2023, 62, e202214539. [Google Scholar] [CrossRef]
- Walsh, C.T.; Tang, Y. Recent Advances in Enzymatic Complexity Generation: Cyclization Reactions. Biochemistry 2018, 57, 3087–3104. [Google Scholar] [CrossRef] [PubMed]
- Gao, L.; Su, C.; Du, X.; Wang, R.; Chen, S.; Zhou, Y.; Liu, C.; Liu, X.; Tian, R.; Zhang, L.; et al. FAD-dependent enzyme-catalysed intermolecular [4+2] cycloaddition in natural product biosynthesis. Nat. Chem. 2020, 12, 620–628. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Yang, J.; Gao, L.; Zhang, L.; Lei, X. Chemoenzymatic Total Syntheses of Artonin I with an Intermolecular Diels-Alderase. Biotechnol. J. 2020, 15, 2000119. [Google Scholar] [CrossRef]
- Basler, S.; Studer, S.; Zou, Y.; Mori, T.; Ota, Y.; Camus, A.; Bunzel, H.A.; Helgeson, R.C.; Houk, K.N.; Jiménez-Osés, G.; et al. Efficient Lewis acid catalysis of an abiological reaction in a de novo protein scaffold. Nat. Chem. 2021, 13, 231–235. [Google Scholar] [CrossRef]
- Gao, L.; Zou, Y.; Liu, X.; Yang, J.; Du, X.; Wang, J.; Yu, X.; Fan, J.; Jiang, M.; Li, Y.; et al. Enzymatic control of endo- and exo-stereoselective Diels–Alder reactions with broad substrate scope. Nat. Catal. 2021, 4, 1059–1069. [Google Scholar] [CrossRef]
- Löwe, J.; Dietz, K.J.; Gröger, H. From a Biosynthetic Pathway toward a Biocatalytic Process and Chemocatalytic Modifications: Three-Step Enzymatic Cascade to the Plant Metabolite cis-(+)-12-OPDA and Metathesis-Derived Products. Adv. Sci. 2020, 7, 1902973. [Google Scholar] [CrossRef]
- Westarp, S.; Kaspar, F.; Neubauer, P.; Kurreck, A. Industrial potential of the enzymatic synthesis of nucleoside analogs: Existing challenges and perspectives. Curr. Opin. Biotechnol. 2022, 78, 102829. [Google Scholar] [CrossRef]
- Cosgrove, S.C.; Miller, G.J. Advances in biocatalytic and chemoenzymatic synthesis of nucleoside analogues. Expert Opin. Drug Discov. 2022, 17, 355–364. [Google Scholar] [CrossRef]
- McIntosh, J.A.; Benkovics, T.; Silverman, S.M.; Huffman, M.A.; Kong, J.; Maligres, P.E.; Itoh, T.; Yang, H.; Verma, D.; Pan, W.; et al. Engineered ribosyl-1-kinase enables concise synthesis of molnupiravir, an antiviral for COVID-19. ACS Cent. Sci. 2021, 7, 1980–1985. [Google Scholar] [CrossRef] [PubMed]
- McIntosh, J.A.; Liu, Z.; Andresen, B.M.; Marzijarani, N.S.; Moore, J.C.; Marshall, N.M.; Borra-Garske, M.; Obligacion, J.V.; Fier, P.S.; Peng, F.; et al. A kinase-cGAS cascade to synthesize a therapeutic STING activator. Nature 2022, 603, 439–444. [Google Scholar] [CrossRef] [PubMed]
- Trung, M.N.; Kieninger, S.; Fandi, Z.; Qiu, D.; Liu, G.; Mehendale, N.K.; Saiardi, A.; Jessen, H.; Keller, B.; Fiedler, D. Stable Isotopomers of myo-Inositol Uncover a Complex MINPP1-Dependent Inositol Phosphate Network. ACS Cent. Sci. 2022, 8, 1683–1694. [Google Scholar] [CrossRef] [PubMed]
- Shen, B. A New Golden Age of Natural Products Drug Discovery. Cell 2015, 163, 1297–1300. [Google Scholar] [CrossRef]
- Atanasov, A.G.; Zotchev, S.B.; Dirsch, V.M.; Supuran, C.T. Natural products in drug discovery: Advances and opportunities. Nat. Rev. Drug Discov. 2021, 20, 200–216. [Google Scholar] [CrossRef]
- Reetz, M.T.; Sun, Z.; Qu, G. Enzyme Engineering: Selective Catalysts for Applications in Biotechnology, Organic Chemistry, and Life Science; Wiley-VCH: Weinhem, Germany, 2023; ISBN 978-3-527-35033-9. [Google Scholar]
- Chen, K.; Arnold, F.H. Engineering new catalytic activities in enzymes. Nat. Catal. 2020, 3, 203–213. [Google Scholar] [CrossRef]
- Zetzsche, L.E.; Chakrabarty, S.; Narayan, A.R. The transformative power of biocatalysis in convergent synthesis. J. Am. Chem. Soc. 2022, 144, 5214–5225. [Google Scholar] [CrossRef]
- Stout, C.N.; Wasfy, N.M.; Chen, F.; Renata, H. Charting the Evolution of Chemoenzymatic Strategies in the Syntheses of Complex Natural Products. J. Am. Chem. Soc. 2023, 145, 18161–18181. [Google Scholar] [CrossRef]
- Li, F.; Deng, H.; Renata, H. Chemoenzymatic approaches for exploring structure–activity relationship studies of bioactive natural products. Nat. Synth. 2023, 2, 708–718. [Google Scholar] [CrossRef]
- Lovelock, S.L.; Crawshaw, R.; Basler, S.; Levy, C.; Baker, D.; Hilvert, D.; Green, A.P. The road to fully programmable protein catalysis. Nature 2022, 606, 49–58. [Google Scholar] [CrossRef] [PubMed]
- Hermann, J.C.; Marti-Arbona, R.; Fedorov, A.A.; Fedorov, E.; Almo, S.C.; Shoichet, B.K.; Raushel, F.M. Structure-based activity prediction for an enzyme of unknown function. Nature 2007, 448, 775–779. [Google Scholar] [CrossRef] [PubMed]
- Zallot, R.; Oberg, N.; Gerlt, J.A. The EFI web resource for genomic enzymology tools: Leveraging protein, genome, and metagenome databases to discover novel enzymes and metabolic pathways. Biochemistry 2019, 58, 4169–4182. [Google Scholar] [CrossRef]
- Price, M.N.; Wetmore, K.M.; Waters, R.J.; Callaghan, M.; Ray, J.; Liu, H.; Kuehl, J.V.; Melnyk, R.A.; Lamson, J.S.; Suh, Y.; et al. Mutant phenotypes for thousands of bacterial genes of unknown function. Nature 2018, 557, 503–509. [Google Scholar] [CrossRef]
- Robinson, S.L.; Piel, J.; Sunagawa, S. A roadmap for metagenomic enzyme discovery. Nat. Prod. Rep. 2021, 38, 1994–2023. [Google Scholar] [CrossRef]
- Caputi, L.; Franke, J.; Farrow, S.C.; Chung, K.; Payne, R.M.; Nguyen, T.D.; Dang, T.T.T.; Soares Teto Carqueijeiro, I.; Koudounas, K.; Dugé de Bernonville, T.; et al. Missing enzymes in the biosynthesis of the anticancer drug vinblastine in Madagascar periwinkle. Science 2018, 360, 1235–1239. [Google Scholar] [CrossRef]
- Blin, K.; Shaw, S.; Augustijn, H.E.; Reitz, Z.L.; Biermann, F.; Alanjary, M.; Fetter, A.; Terlouw, B.R.; Metcalf, W.W.; Helfrich, E.J.N.; et al. antiSMASH 7.0: New and improved predictions for detection, regulation, chemical structures and visualisation. Nucleic Acids Res. 2023, 51, W46–W50. [Google Scholar] [CrossRef]
- Caesar, L.K.; Montaser, R.; Keller, N.P.; Kelleher, N.L. Metabolomics and genomics in natural products research: Complementary tools for targeting new chemical entities. Nat. Prod. Rep. 2021, 38, 2041–2065. [Google Scholar] [CrossRef]
- Scherlach, K.; Hertweck, C. Mining and unearthing hidden biosynthetic potential. Nat. Commun. 2021, 12, 3864. [Google Scholar] [CrossRef]
- Klapper, M.; Hübner, A.; Ibrahim, A.; Wasmuth, I.; Borry, M.; Haensch, V.G.; Zhang, S.; Al-Jammal, W.K.; Suma, H.; Fellows Yates, J.A.; et al. Natural products from reconstructed bacterial genomes of the Middle and Upper Paleolithic. Science 2023, 380, 619–624. [Google Scholar] [CrossRef] [PubMed]
- Caesar, L.K.; Butun, F.A.; Robey, M.T.; Ayon, N.J.; Gupta, R.; Dainko, D.; Bok, J.W.; Nickles, G.; Stankey, R.J.; Johnson, D.; et al. Correlative metabologenomics of 110 fungi reveals metabolite–gene cluster pairs. Nat. Chem. Biol. 2023, 19, 846–854. [Google Scholar] [CrossRef] [PubMed]
- Smanski, M.J.; Zhou, H.; Claesen, J.; Shen, B.; Fischbach, M.; Voigt, C.A. Synthetic biology to access and expand nature’s chemical diversity. Nat. Rev. Microbiol. 2016, 14, 135–149. [Google Scholar] [CrossRef] [PubMed]
- Erb, T.J.; Jones, P.R.; Bar-Even, A. Synthetic metabolism: Metabolic engineering meets enzyme design. Curr. Opin. Chem. Biol. 2017, 37, 56–62. [Google Scholar] [CrossRef]
- Yi, J.; Li, Z. Artificial multi-enzyme cascades for natural product synthesis. Curr. Opin. Biotechnol. 2022, 78, 102831. [Google Scholar] [CrossRef]






| Biological Species | Name of Species- Specific Metabolite Database | Abbreviation of Metabolite Database Name | Website of Metabolite Database | Reference | Accessed Date for the URL |
|---|---|---|---|---|---|
| Human | Human Metabolome Database | HMDB | https://hmdb.ca/ | [14] | accessed on 16 July 2023 |
| Human Microbiome | Human Microbial Metabolome Database i | MiMeDB | https://mimedb.org/ | [15] | accessed on 16 July 2023 |
| Escherichia coli | Escherichia coli Metabolome Database | ECMDB | http://www.ecmdb.ca/ | [16] | accessed on 16 July 2023 |
| Pseudomonas aeruginosa | Pseudomonas aeruginosa Metabolome Database | PAMDB | http://pseudomonas.umaryland.edu/ | [17] | accessed on 16 July 2023 |
| Streptomyces sp. | Streptomyces Natural Products Database | StreptomeDB | http://www.pharmbioinf.uni-freiburg.de/streptomedb/ | [18] | accessed on 30 July 2023 |
| Cyanobacteria | Comprehensive database of secondary metabolites from cyanobacteria | CyanoMetDB | https://zenodo.org/record/7922070/ | [19] | accessed on 30 July 2023 |
| Myxobacteria | Myxobacterial Natural Product Database | MyxoDB | https://www.myxonpdb.sdu.edu.cn/ | [20] | accessed on 30 July 2023 |
| Yeast | Yeast Metabolome Database | YMDB | http://www.ymdb.ca/ | [21] | accessed on 16 July 2023 |
| Bovine | Bovine Metabolome Database | BMDB | https://bovinedb.ca/ | [22] | accessed on 16 July 2023 |
| Tomato | Tomato Metabolome Database | TOMATOMET | http://metabolites.in/tomato-fruits/ | [23] | accessed on 24 July 2023 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wohlgemuth, R. Synthesis of Metabolites and Metabolite-like Compounds Using Biocatalytic Systems. Metabolites 2023, 13, 1097. https://doi.org/10.3390/metabo13101097
Wohlgemuth R. Synthesis of Metabolites and Metabolite-like Compounds Using Biocatalytic Systems. Metabolites. 2023; 13(10):1097. https://doi.org/10.3390/metabo13101097
Chicago/Turabian StyleWohlgemuth, Roland. 2023. "Synthesis of Metabolites and Metabolite-like Compounds Using Biocatalytic Systems" Metabolites 13, no. 10: 1097. https://doi.org/10.3390/metabo13101097
APA StyleWohlgemuth, R. (2023). Synthesis of Metabolites and Metabolite-like Compounds Using Biocatalytic Systems. Metabolites, 13(10), 1097. https://doi.org/10.3390/metabo13101097







