The Sterol Transporter Npc2c Controls Intestinal Stem Cell Mitosis and Host–Microbiome Interactions in Drosophila
Abstract
:1. Introduction
2. Materials and Methods
2.1. Drosophila Stocks and Rearing
2.2. MARCM Clones
2.3. Oral Administration of Bacteria
2.4. Oral Administration of Chemicals
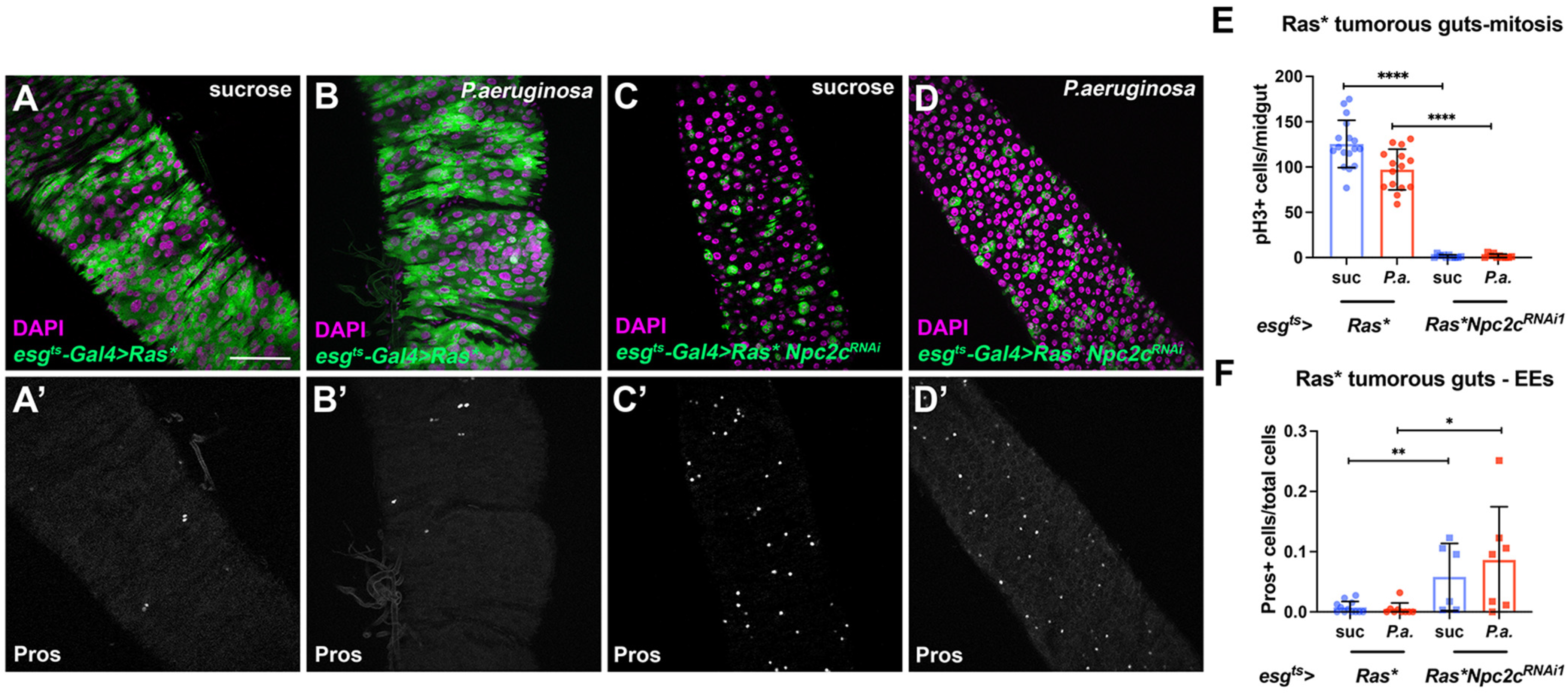
2.5. Midgut Tumorigenesis
2.6. Dissections and Immunohistochemistry
2.7. Npc2c Polyclonal Antibody
2.8. Filipin Staining
2.9. Survival Assay
2.10. Smurf Assay/Gut Permeability
2.11. RNA Isolation and RT-qPCR
2.12. Bacterial Load
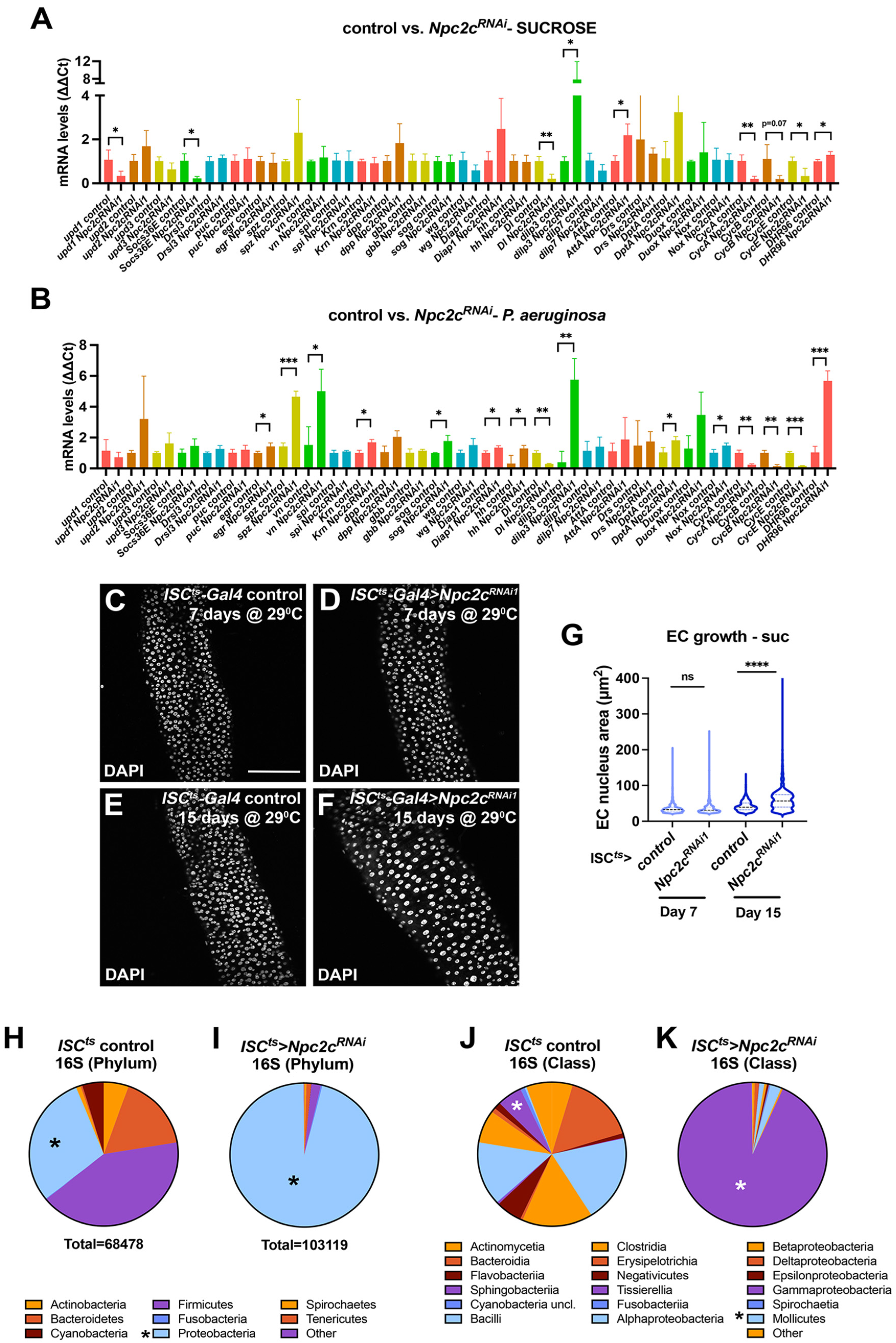
2.13. Microbiota Analysis
2.14. Image Acquisition and Analysis
2.15. Statistical Analysis
3. Results
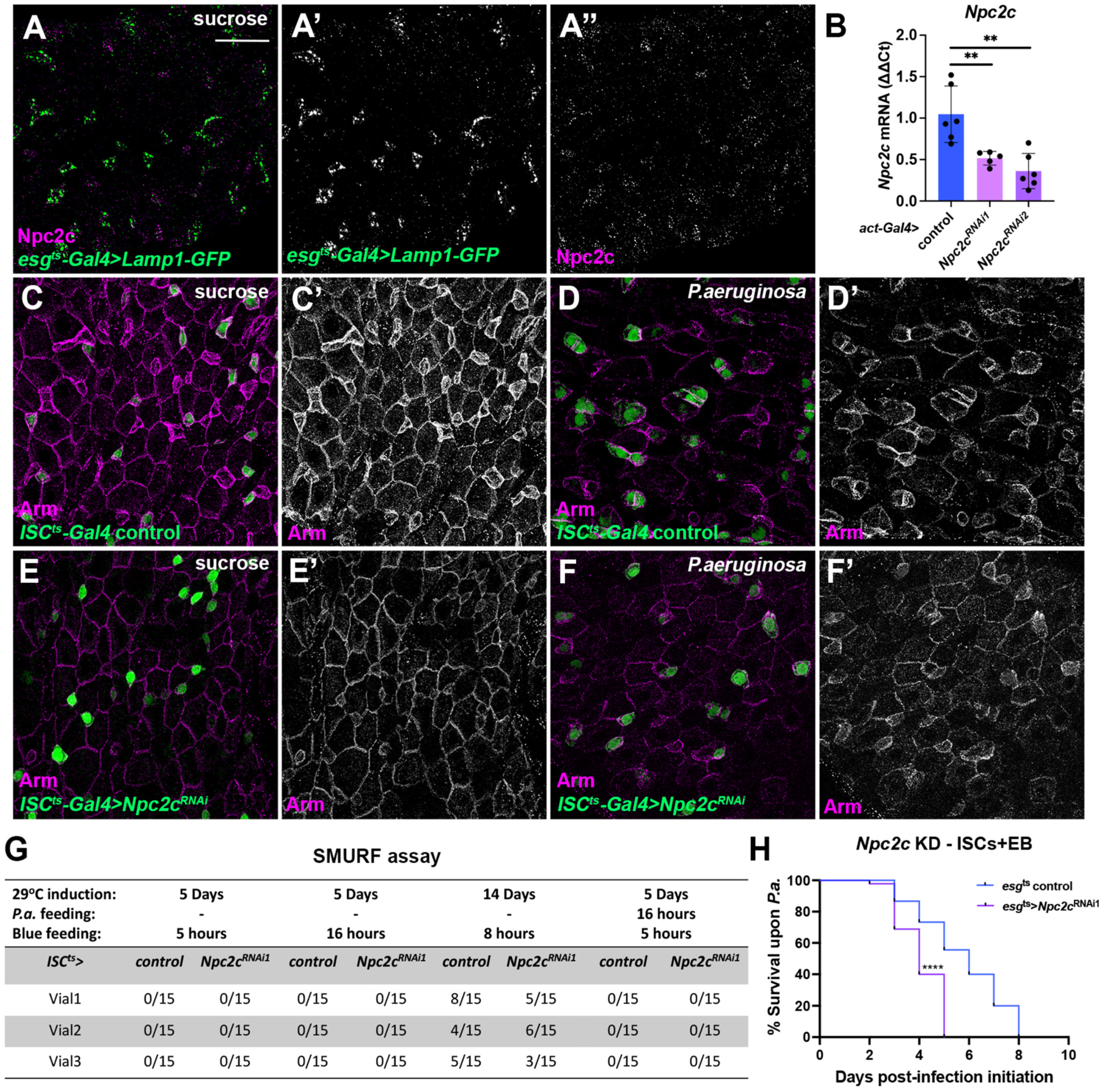
3.1. Npc2c Is Expressed in the Midgut Epithelium and Affects Physiology
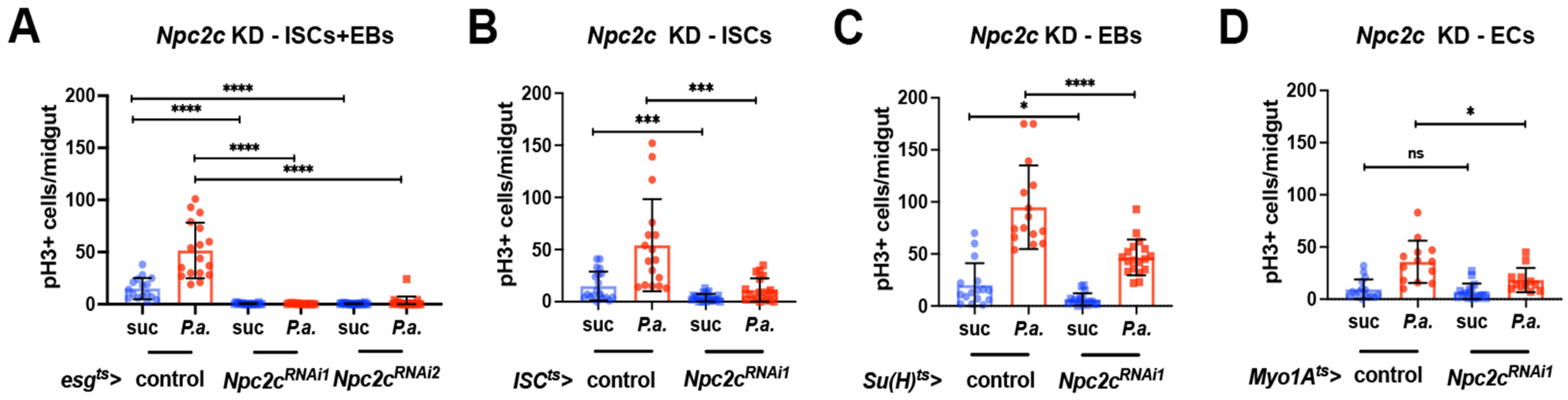
3.2. Npc2c Autonomously Controls ISC Mitosis
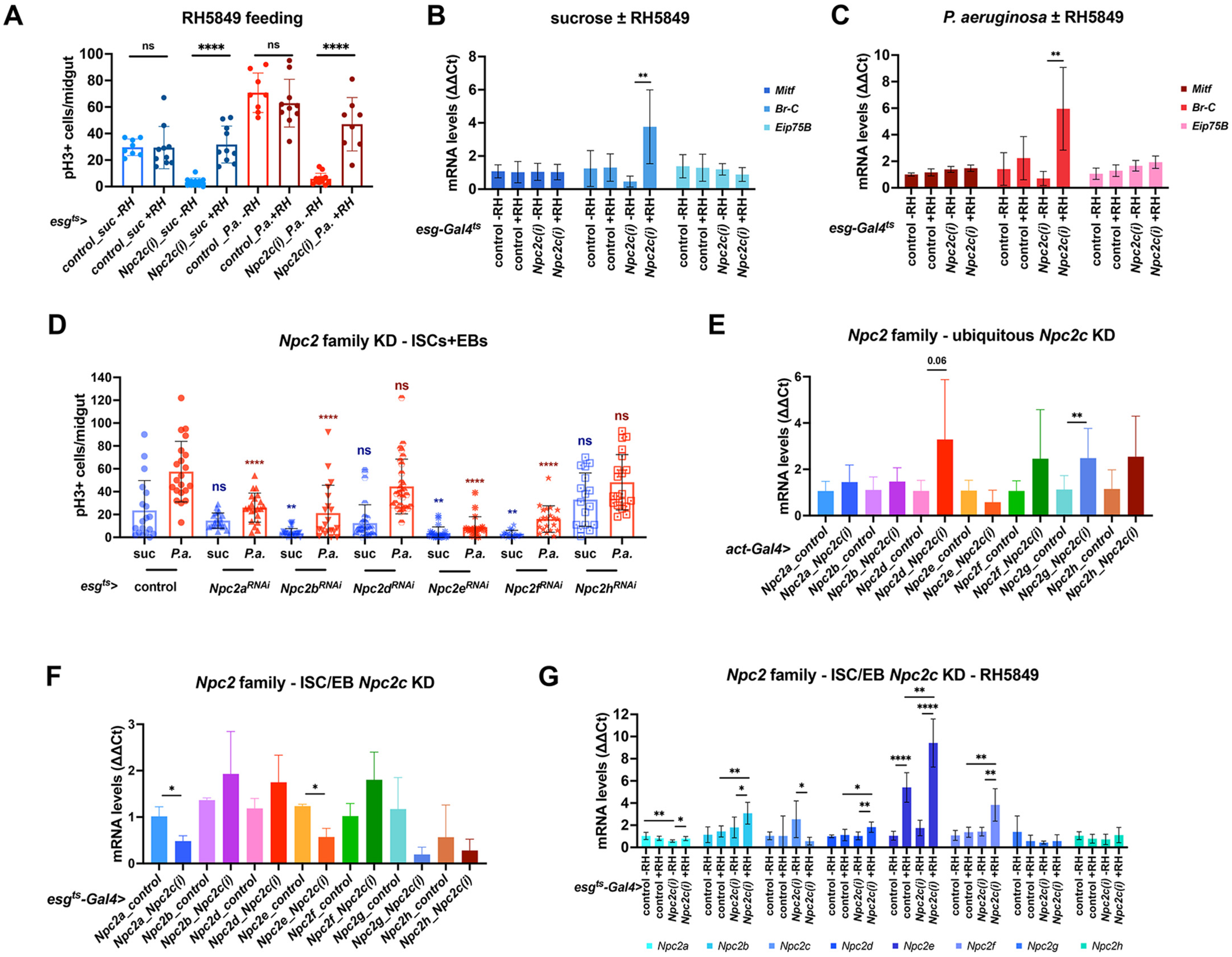
3.3. Npc2c Silencing Affects Expression of Mitotic Regulators and Leads to Intestinal Dysbiosis
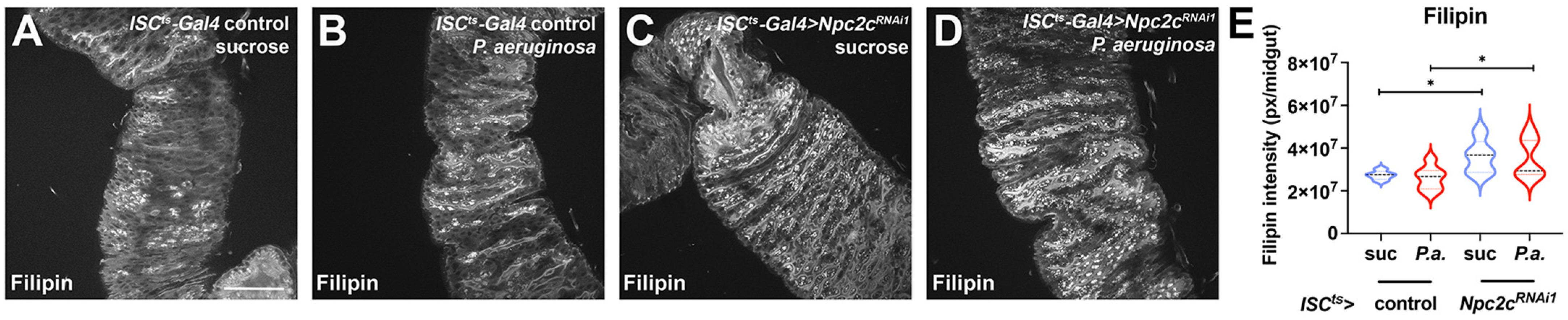
3.4. ISC-Specific Npc2c Silencing Impairs Cholesterol Trafficking and Controls Ecdysone Signaling
3.5. Additional Npc2 Family Genes Control Intestinal Mitosis
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Heiden, M.G.V.; Lunt, S.Y.; Dayton, T.L.; Fiske, B.P.; Israelsen, W.J.; Mattaini, K.R.; Vokes, N.I.; Stephanopoulos, G.; Cantley, L.C.; Metallo, C.M.; et al. Metabolic pathway alterations that support cell proliferation. Cold Spring Harb. Symp. Quant. Biol. 2011, 76, 325–334. [Google Scholar] [CrossRef] [PubMed]
- Miller, W.L.; Auchus, R.J. The molecular biology, biochemistry, and physiology of human steroidogenesis and its disorders. Endocr. Rev. 2011, 32, 81–151. [Google Scholar] [CrossRef]
- Yang, S.T.; Kreutzberger, A.J.B.; Lee, J.; Kiessling, V.; Tamm, L.K. The role of cholesterol in membrane fusion. Chem. Phys. Lipids 2016, 199, 136–143. [Google Scholar] [CrossRef] [PubMed]
- Jefcoate, C.R.; Lee, J. Cholesterol signaling in single cells: Lessons from STAR and sm-FISH. J. Mol. Endocrinol. 2018, 60, R213–R235. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Patel, S.B. Recent insights into the Smith-Lemli-Opitz syndrome. Clin. Genet. 2005, 68, 383–391. [Google Scholar] [CrossRef]
- Jung, E.; Kong, S.Y.; Ro, Y.S.; Ryu, H.H.; Shin, S.D. Serum Cholesterol Levels and Risk of Cardiovascular Death: A Systematic Review and a Dose-Response Meta-Analysis of Prospective Cohort Studies. Int. J. Environ. Res. Public Health 2022, 19, 8272. [Google Scholar] [CrossRef]
- Klop, B.; Elte, J.W.; Cabezas, M.C. Dyslipidemia in obesity: Mechanisms and potential targets. Nutrients 2013, 5, 1218–1240. [Google Scholar] [CrossRef]
- Holtzman, E.J.; Yaari, S.; Goldbourt, U. Serum cholesterol and the risk of colorectal cancer. N. Engl. J. Med. 1987, 317, 114. [Google Scholar] [CrossRef]
- Baek, A.E.; Nelson, E.R. The Contribution of Cholesterol and Its Metabolites to the Pathophysiology of Breast Cancer. Horm. Cancer 2016, 7, 219–228. [Google Scholar] [CrossRef]
- Ma, Y.; Yang, Y.; Wang, F.; Zhang, P.; Shi, C.; Zou, Y.; Qin, H. Obesity and risk of colorectal cancer: A systematic review of prospective studies. PLoS ONE 2013, 8, e53916. [Google Scholar] [CrossRef]
- Neophytou, C.; Pitsouli, C. How Gut Microbes Nurture Intestinal Stem Cells: A Drosophila Perspective. Metabolites 2022, 12, 169. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Rong, X.; Palladino, E.N.D.; Wang, J.; Fogelman, A.M.; Martín, M.G.; Alrefai, W.A.; Ford, D.A.; Tontonoz, P. Phospholipid Remodeling and Cholesterol Availability Regulate Intestinal Stemness and Tumorigenesis. Cell Stem Cell 2018, 22, 206–220.e4. [Google Scholar] [CrossRef] [PubMed]
- Obniski, R.; Sieber, M.; Spradling, A.C. Dietary Lipids Modulate Notch Signaling and Influence Adult Intestinal Development and Metabolism in Drosophila. Dev. Cell 2018, 47, 98–111.e5. [Google Scholar] [CrossRef] [PubMed]
- Brown, M.S.; Goldstein, J.L. A Receptor-Mediated Pathway for Cholesterol Homeostasis (Nobel Lecture). Angew. Chem. Int. Ed. Engl. 1986, 25, 583–602. [Google Scholar] [CrossRef]
- Clark, A.J.; Bloch, K. The Absence of Sterol Synthesis in Insects. J. Biol. Chem. 1959, 234, 2578–2582. [Google Scholar] [CrossRef]
- Van Hoof, D.; Rodenburg, K.W.; Van der Horst, D.J. Receptor-mediated endocytosis and intracellular trafficking of lipoproteins and transferrin in insect cells. Insect Biochem. Mol. Biol. 2005, 35, 117–128. [Google Scholar] [CrossRef]
- Horner, M.A.; Pardee, K.; Liu, S.; King-Jones, K.; Lajoie, G.; Edwards, A.; Krause, H.M.; Thummel, C.S. The Drosophila DHR96 nuclear receptor binds cholesterol and regulates cholesterol homeostasis. Genes. Dev. 2009, 23, 2711–2716. [Google Scholar] [CrossRef]
- Huang, X.; Warren, J.T.; Buchanan, J.; Gilbert, L.I.; Scott, M.P. Drosophila Niemann-Pick Type C-2 genes control sterol homeostasis and steroid biosynthesis: A model of human neurodegenerative disease. Development 2007, 134, 3733–3742. [Google Scholar] [CrossRef]
- Niwa, R.; Niwa, Y.S. The Fruit Fly Drosophila melanogaster as a Model System to Study Cholesterol Metabolism and Homeostasis. Cholesterol 2011, 2011, 176802. [Google Scholar] [CrossRef]
- Bujold, M.; Gopalakrishnan, A.; Nally, E.; King-Jones, K. Nuclear receptor DHR96 acts as a sentinel for low cholesterol concentrations in Drosophila melanogaster. Mol. Cell Biol. 2010, 30, 793–805. [Google Scholar] [CrossRef]
- Huang, X.; Suyama, K.; Buchanan, J.; Zhu, A.J.; Scott, M.P. A Drosophila model of the Niemann-Pick type C lysosome storage disease: dnpc1a is required for molting and sterol homeostasis. Development 2005, 132, 5115–5124. [Google Scholar] [CrossRef]
- Brown, M.S.; Goldstein, J.L. The SREBP pathway: Regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell 1997, 89, 331–340. [Google Scholar] [CrossRef]
- Kalaany, N.Y.; Mangelsdorf, D.J. LXRS and FXR: The yin and yang of cholesterol and fat metabolism. Annu. Rev. Physiol. 2006, 68, 159–191. [Google Scholar] [CrossRef] [PubMed]
- Seegmiller, A.C.; Dobrosotskaya, I.; Goldstein, J.L.; Ho, Y.K.; Brown, M.S.; Rawson, R.B. The SREBP Pathway in Drosophila: Regulation by Palmitate, Not Sterols. Dev. Cell 2002, 2, 229–238. [Google Scholar] [CrossRef] [PubMed]
- Bonneton, F.; Laudet, V. 6—Evolution of Nuclear Receptors in Insects. In Insect Endocrinology; Gilbert, L.I., Ed.; Academic Press: San Diego, CA, USA, 2012; pp. 219–252. [Google Scholar]
- Riddiford, L.M.; Cherbas, P.; Truman, J.W. Ecdysone receptors and their biological actions. Vitam. Horm. 2000, 60, 1–73. [Google Scholar] [CrossRef]
- Petryk, A.; Warren, J.T.; Marqués, G.; Jarcho, M.P.; Gilbert, L.I.; Kahler, J.; Parvy, J.P.; Li, Y.; Dauphin-Villemant, C.; O’Connor, M.B. Shade is the Drosophila P450 enzyme that mediates the hydroxylation of ecdysone to the steroid insect molting hormone 20-hydroxyecdysone. Proc. Natl. Acad. Sci. USA 2003, 100, 13773–13778. [Google Scholar] [CrossRef] [PubMed]
- Simon, A.F.; Shih, C.; Mack, A.; Benzer, S. Steroid control of longevity in Drosophila melanogaster. Science 2003, 299, 1407–1410. [Google Scholar] [CrossRef]
- Tricoire, H.; Battisti, V.; Trannoy, S.; Lasbleiz, C.; Pret, A.M.; Monnier, V. The steroid hormone receptor EcR finely modulates Drosophila lifespan during adulthood in a sex-specific manner. Mech. Ageing Dev. 2009, 130, 547–552. [Google Scholar] [CrossRef]
- Ahmed, S.M.H.; Maldera, J.A.; Krunic, D.; Paiva-Silva, G.O.; Pénalva, C.; Teleman, A.A.; Edgar, B.A. Fitness trade-offs incurred by ovary-to-gut steroid signalling in Drosophila. Nature 2020, 584, 415–419. [Google Scholar] [CrossRef]
- Zipper, L.; Jassmann, D.; Burgmer, S.; Görlich, B.; Reiff, T. Ecdysone steroid hormone remote controls intestinal stem cell fate decisions via the PPARγ-homolog Eip75B in Drosophila. eLife 2020, 9, e55795. [Google Scholar] [CrossRef]
- Miguel-Aliaga, I.; Jasper, H.; Lemaitre, B. Anatomy and Physiology of the Digestive Tract of Drosophila melanogaster. Genetics 2018, 210, 357–396. [Google Scholar] [CrossRef] [PubMed]
- Reiff, T.; Jacobson, J.; Cognigni, P.; Antonello, Z.; Ballesta, E.; Tan, K.J.; Yew, J.Y.; Dominguez, M.; Miguel-Aliaga, I. Endocrine remodelling of the adult intestine sustains reproduction in Drosophila. eLife 2015, 4, e06930. [Google Scholar] [CrossRef] [PubMed]
- Marvin, K.A.; Reinking, J.L.; Lee, A.J.; Pardee, K.; Krause, H.M.; Burstyn, J.N. Nuclear receptors homo sapiens Rev-erbbeta and Drosophila melanogaster E75 are thiolate-ligated heme proteins which undergo redox-mediated ligand switching and bind CO and NO. Biochemistry 2009, 48, 7056–7071. [Google Scholar] [CrossRef] [PubMed]
- King-Jones, K.; Thummel, C.S. Nuclear receptors—A perspective from Drosophila. Nat. Rev. Genet. 2005, 6, 311–323. [Google Scholar] [CrossRef] [PubMed]
- Carstea, E.D.; Morris, J.A.; Coleman, K.G.; Loftus, S.K.; Zhang, D.; Cummings, C.; Gu, J.; Rosenfeld, M.A.; Pavan, W.J.; Krizman, D.B.; et al. Niemann-Pick C1 Disease Gene: Homology to Mediators of Cholesterol Homeostasis. Science 1997, 277, 228–231. [Google Scholar] [CrossRef] [PubMed]
- Vanier, M.T. Niemann-Pick disease type C. Orphanet J. Rare Dis. 2010, 5, 16. [Google Scholar] [CrossRef]
- Hammond, N.; Munkacsi, A.B.; Sturley, S.L. The complexity of a monogenic neurodegenerative disease: More than two decades of therapeutic driven research into Niemann-Pick type C disease. Biochim. Biophys. Acta (BBA)—Mol. Cell Biol. Lipids 2019, 1864, 1109–1123. [Google Scholar] [CrossRef]
- Nganso, B.T.; Mani, K.; Eliash, N.; Rafaeli, A.; Soroker, V. Towards disrupting Varroa–honey bee chemosensing: A focus on a Niemann-Pick type C2 transcript. Insect Mol. Biol. 2021, 30, 519–531. [Google Scholar] [CrossRef]
- Mani, K.; Nganso, B.T.; Rodin, P.; Otmy, A.; Rafaeli, A.; Soroker, V. Effects of Niemann-Pick type C2 (NPC2) gene transcripts silencing on behavior of Varroa destructor and molecular changes in the putative olfactory gene networks. Insect Biochem. Mol. Biol. 2022, 148, 103817. [Google Scholar] [CrossRef]
- Hovakimyan, M.; Meyer, A.; Lukas, J.; Luo, J.; Gudziol, V.; Hummel, T.; Rolfs, A.; Wree, A.; Witt, M. Olfactory deficits in Niemann-Pick type C1 (NPC1) disease. PLOS ONE 2013, 8, e82216. [Google Scholar] [CrossRef]
- Neufeld, E.B.; Wastney, M.; Patel, S.; Suresh, S.; Cooney, A.M.; Dwyer, N.K.; Roff, C.F.; Ohno, K.; Morris, J.A.; Carstea, E.D.; et al. The Niemann-Pick C1 Protein Resides in a Vesicular Compartment Linked to Retrograde Transport of Multiple Lysosomal Cargo. J. Biol. Chem. 1999, 274, 9627–9635. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Farver, W.; Kodukula, S.; Storch, J. Regulation of sterol transport between membranes and NPC2. Biochemistry 2008, 47, 11134–11143. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Warren, J.T.; Gilbert, L.I. New players in the regulation of ecdysone biosynthesis. J. Genet. Genom. 2008, 35, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Fluegel, M.L.; Parker, T.J.; Pallanck, L.J. Mutations of a Drosophila NPC1 gene confer sterol and ecdysone metabolic defects. Genetics 2006, 172, 185–196. [Google Scholar] [CrossRef] [PubMed]
- Voght, S.P.; Fluegel, M.L.; Andrews, L.A.; Pallanck, L.J. Drosophila NPC1b Promotes an Early Step in Sterol Absorption from the Midgut Epithelium. Cell Metab. 2007, 5, 195–205. [Google Scholar] [CrossRef]
- Sieber, M.H.; Thummel, C.S. The DHR96 nuclear receptor controls triacylglycerol homeostasis in Drosophila. Cell Metab. 2009, 10, 481–490. [Google Scholar] [CrossRef]
- Shi, X.Z.; Zhong, X.; Yu, X.Q. Drosophila melanogaster NPC2 proteins bind bacterial cell wall components and may function in immune signal pathways. Insect Biochem. Mol. Biol. 2012, 42, 545–556. [Google Scholar] [CrossRef]
- Micchelli, C.A.; Perrimon, N. Evidence that stem cells reside in the adult Drosophila midgut epithelium. Nature 2006, 439, 475–479. [Google Scholar] [CrossRef]
- Zeng, X.; Hou, S.X. Enteroendocrine cells are generated from stem cells through a distinct progenitor in the adult Drosophila posterior midgut. Development 2015, 142, 644–653. [Google Scholar] [CrossRef]
- Tamamouna, V.; Panagi, M.; Theophanous, A.; Demosthenous, M.; Michail, M.; Papadopoulou, M.; Teloni, S.; Pitsouli, C.; Apidianakis, Y. Evidence of two types of balance between stem cell mitosis and enterocyte nucleus growth in the Drosophila midgut. Development 2020, 147, dev189472. [Google Scholar] [CrossRef]
- Apidianakis, Y.; Pitsouli, C.; Perrimon, N.; Rahme, L. Synergy between bacterial infection and genetic predisposition in intestinal dysplasia. Proc. Natl. Acad. Sci. USA 2009, 106, 20883–20888. [Google Scholar] [CrossRef] [PubMed]
- Neophytou, C.; Pitsouli, C. Biotin controls intestinal stem cell mitosis and host-microbiome interactions. Cell Rep. 2022, 38, 110505. [Google Scholar] [CrossRef] [PubMed]
- Pitsouli, C.; Delidakis, C. The interplay between DSL proteins and ubiquitin ligases in Notch signaling. Development 2005, 132, 4041–4050. [Google Scholar] [CrossRef]
- Brand, A.H.; Perrimon, N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 1993, 118, 401–415. [Google Scholar] [CrossRef]
- McGuire, S.E.; Mao, Z.; Davis, R.L. Spatiotemporal gene expression targeting with the TARGET and gene-switch systems in Drosophila. Sci. STKE 2004, 2004, pl6. [Google Scholar] [CrossRef] [PubMed]
- Lee, T.; Luo, L. Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron 1999, 22, 451–461. [Google Scholar] [CrossRef]
- Apidianakis, Y.; Rahme, L.G. Drosophila melanogaster as a model host for studying Pseudomonas aeruginosa infection. Nat. Protoc. 2009, 4, 1285–1294. [Google Scholar] [CrossRef]
- Rera, M.; Clark, R.I.; Walker, D.W. Intestinal barrier dysfunction links metabolic and inflammatory markers of aging to death in Drosophila. Proc. Natl. Acad. Sci. USA 2012, 109, 21528–21533. [Google Scholar] [CrossRef]
- Leader, D.P.; Krause, S.A.; Pandit, A.; Davies, S.A.; Dow, J.A.T. FlyAtlas 2: A new version of the Drosophila melanogaster expression atlas with RNA-Seq, miRNA-Seq and sex-specific data. Nucleic Acids Res. 2017, 46, D809–D815. [Google Scholar] [CrossRef]
- Buchon, N.; Osman, D.; David, F.P.; Fang, H.Y.; Boquete, J.P.; Deplancke, B.; Lemaitre, B. Morphological and molecular characterization of adult midgut compartmentalization in Drosophila. Cell Rep. 2013, 3, 1725–1738. [Google Scholar] [CrossRef]
- Pulipparacharuvil, S.; Akbar, M.A.; Ray, S.; Sevrioukov, E.A.; Haberman, A.S.; Rohrer, J.; Krämer, H. Drosophila Vps16A is required for trafficking to lysosomes and biogenesis of pigment granules. J. Cell Sci. 2005, 118, 3663–3673. [Google Scholar] [CrossRef] [PubMed]
- Peifer, M.; Wieschaus, E. The segment polarity gene armadillo encodes a functionally modular protein that is the Drosophila homolog of human plakoglobin. Cell 1990, 63, 1167–1176. [Google Scholar] [CrossRef] [PubMed]
- de Navascués, J.; Perdigoto, C.N.; Bian, Y.; Schneider, M.H.; Bardin, A.J.; Martínez-Arias, A.; Simons, B.D. Drosophila midgut homeostasis involves neutral competition between symmetrically dividing intestinal stem cells. EMBO J. 2012, 31, 2473–2485. [Google Scholar] [CrossRef] [PubMed]
- Jin, Y.; Patel, P.H.; Kohlmaier, A.; Pavlovic, B.; Zhang, C.; Edgar, B.A. Intestinal Stem Cell Pool Regulation in Drosophila. Stem Cell Rep. 2017, 8, 1479–1487. [Google Scholar] [CrossRef] [PubMed]
- Ohlstein, B.; Spradling, A. The adult Drosophila posterior midgut is maintained by pluripotent stem cells. Nature 2006, 439, 470–474. [Google Scholar] [CrossRef] [PubMed]
- Tomasetti, C.; Li, L.; Vogelstein, B. Stem cell divisions, somatic mutations, cancer etiology, and cancer prevention. Science 2017, 355, 1330–1334. [Google Scholar] [CrossRef]
- Ohlstein, B.; Spradling, A. Multipotent Drosophila intestinal stem cells specify daughter cell fates by differential notch signaling. Science 2007, 315, 988–992. [Google Scholar] [CrossRef]
- Osman, D.; Buchon, N.; Chakrabarti, S.; Huang, Y.T.; Su, W.C.; Poidevin, M.; Tsai, Y.C.; Lemaitre, B. Autocrine and paracrine unpaired signaling regulate intestinal stem cell maintenance and division. J. Cell Sci. 2012, 125, 5944–5949. [Google Scholar] [CrossRef]
- Buonocore, F.; Fausto, A.M.; Della Pelle, G.; Roncevic, T.; Gerdol, M.; Picchietti, S. Attacins: A Promising Class of Insect Antimicrobial Peptides. Antibiotics 2021, 10, 212. [Google Scholar] [CrossRef]
- Marianes, A.; Spradling, A.C. Physiological and stem cell compartmentalization within the Drosophila midgut. ELife 2013, 2, e00886. [Google Scholar] [CrossRef]
- Louwette, S.; Régal, L.; Wittevrongel, C.; Thys, C.; Vandeweeghde, G.; Decuyper, E.; Leemans, P.; De Vos, R.; Van Geet, C.; Jaeken, J.; et al. NPC1 defect results in abnormal platelet formation and function: Studies in Niemann–Pick disease type C1 patients and zebrafish. Hum. Mol. Genet. 2012, 22, 61–73. [Google Scholar] [CrossRef] [PubMed]
- Warren, J.T.; Petryk, A.; Marques, G.; Jarcho, M.; Parvy, J.P.; Dauphin-Villemant, C.; O’Connor, M.B.; Gilbert, L.I. Molecular and biochemical characterization of two P450 enzymes in the ecdysteroidogenic pathway of Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 2002, 99, 11043–11048. [Google Scholar] [CrossRef]
- Wing, K.D. RH 5849, a nonsteroidal ecdysone agonist: Effects on a Drosophila cell line. Science 1988, 241, 467–469. [Google Scholar] [CrossRef]
- Robinson, P.D.; Morgan, E.D.; Wilson, I.D.; Lafont, R. The metabolism of ingested and injected [3H]ecdysone by final instar larvae of Heliothis armigera. Physiol. Entomol. 1987, 12, 321–330. [Google Scholar] [CrossRef]
- Uyehara, C.M.; Leatham-Jensen, M.; McKay, D.J. Opportunistic binding of EcR to open chromatin drives tissue-specific developmental responses. Proc. Natl. Acad. Sci. USA 2022, 119, e2208935119. [Google Scholar] [CrossRef] [PubMed]
- Chandra, S.; Roy, A.; Jana, M.; Pahan, K. Cinnamic acid activates PPARα to stimulate Lysosomal biogenesis and lower Amyloid plaque pathology in an Alzheimer’s disease mouse model. Neurobiol. Dis. 2019, 124, 379–395. [Google Scholar] [CrossRef]
- Bouché, V.; Espinosa, A.P.; Leone, L.; Sardiello, M.; Ballabio, A.; Botas, J. Drosophila Mitf regulates the V-ATPase and the lysosomal-autophagic pathway. Autophagy 2016, 12, 484–498. [Google Scholar] [CrossRef]
- Meng, Y.; Heybrock, S.; Neculai, D.; Saftig, P. Cholesterol Handling in Lysosomes and Beyond. Trends Cell Biol. 2020, 30, 452–466. [Google Scholar] [CrossRef]
- Chikh, K.; Vey, S.; Simonot, C.; Vanier, M.T.; Millat, G. Niemann–Pick type C disease: Importance of N-glycosylation sites for function and cellular location of the NPC2 protein. Mol. Genet. Metab. 2004, 83, 220–230. [Google Scholar] [CrossRef]
- Liou, H.-L.; Dixit, S.S.; Xu, S.; Tint, G.S.; Stock, A.M.; Lobel, P. NPC2, the Protein Deficient in Niemann-Pick C2 Disease, Consists of Multiple Glycoforms That Bind a Variety of Sterols. J. Biol. Chem. 2006, 281, 36710–36723. [Google Scholar] [CrossRef]
- Uyehara, C.M.; McKay, D.J. Direct and widespread role for the nuclear receptor EcR in mediating the response to ecdysone in Drosophila. Proc. Natl. Acad. Sci. USA 2019, 116, 9893–9902. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.; Wang, M.; Shi, H. Cholesterol Promotes Colorectal Cancer Growth by Activating the PI3K/AKT Pathway. J. Oncol. 2022, 2022, 1515416. [Google Scholar] [CrossRef] [PubMed]
- Ouahoud, S.; Jacobs, R.J.; Peppelenbosch, M.P.; Fühler, G.M.; Heijmans, J.; Diks, S.; Wildenberg, M.E.; Hawinkels, L.; Kodach, L.L.; Voorneveld, P.W.; et al. Kinome-wide analysis of the effect of statins in colorectal cancer. Br. J. Cancer 2021, 124, 1978–1987. [Google Scholar] [CrossRef]
- Voorneveld, P.W.; Reimers, M.S.; Bastiaannet, E.; Jacobs, R.J.; van Eijk, R.; Zanders, M.M.J.; Herings, R.M.C.; van Herk-Sukel, M.P.P.; Kodach, L.L.; van Wezel, T.; et al. Statin Use After Diagnosis of Colon Cancer and Patient Survival. Gastroenterology 2017, 153, 470–479.e4. [Google Scholar] [CrossRef] [PubMed]
- Dobrzycka, M.; Spychalski, P.; Łachiński, A.J.; Kobiela, P.; Jędrusik, P.; Kobiela, J. Statins and Colorectal Cancer—A Systematic Review. Exp. Clin. Endocrinol. Diabetes 2020, 128, 255–262. [Google Scholar] [CrossRef] [PubMed]
- Lim, S.H.; Kim, T.W.; Hong, Y.S.; Han, S.W.; Lee, K.H.; Kang, H.J.; Hwang, I.G.; Lee, J.Y.; Kim, H.S.; Kim, S.T.; et al. A randomised, double-blind, placebo-controlled multi-centre phase III trial of XELIRI/FOLFIRI plus simvastatin for patients with metastatic colorectal cancer. Br. J. Cancer 2015, 113, 1421–1426. [Google Scholar] [CrossRef]
- Xiao, X.; Liao, X.; Zhou, Y.; Weng, L.; Guo, L.; Zhou, L.; Wang, X.; Liu, X.; Liu, H.; Bi, X.; et al. Variants in the Niemann-pick type C genes are not associated with Alzheimer’s disease: A large case-control study in the Chinese population. Neurobiol. Aging 2022, 116, 49–54. [Google Scholar] [CrossRef]
- Rudge, J.D. The Lipid Invasion Model: Growing Evidence for This New Explanation of Alzheimer’s Disease. J. Alzheimers Dis. 2023, 94, 457–470. [Google Scholar] [CrossRef]
- Rudge, J.D. A New Hypothesis for Alzheimer’s Disease: The Lipid Invasion Model. J. Alzheimers Dis. Rep. 2022, 6, 129–161. [Google Scholar] [CrossRef]
- Kiani, L. 27-Hydroxycholesterol propagates α-synuclein pathology in Parkinson disease. Nat. Rev. Neurol. 2023, 19, 573. [Google Scholar] [CrossRef]
- Cougnoux, A.; Movassaghi, M.; Picache, J.A.; Iben, J.R.; Navid, F.; Salman, A.; Martin, K.; Farhat, N.Y.; Cluzeau, C.; Tseng, W.C.; et al. Gastrointestinal Tract Pathology in a BALB/c Niemann-Pick Disease Type C1 Null Mouse Model. Dig. Dis. Sci. 2018, 63, 870–880. [Google Scholar] [CrossRef] [PubMed]
- Rizzatti, G.; Lopetuso, L.R.; Gibiino, G.; Binda, C.; Gasbarrini, A. Proteobacteria: A Common Factor in Human Diseases. Biomed. Res. Int. 2017, 2017, 9351507. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Neophytou, C.; Soteriou, E.; Pitsouli, C. The Sterol Transporter Npc2c Controls Intestinal Stem Cell Mitosis and Host–Microbiome Interactions in Drosophila. Metabolites 2023, 13, 1084. https://doi.org/10.3390/metabo13101084
Neophytou C, Soteriou E, Pitsouli C. The Sterol Transporter Npc2c Controls Intestinal Stem Cell Mitosis and Host–Microbiome Interactions in Drosophila. Metabolites. 2023; 13(10):1084. https://doi.org/10.3390/metabo13101084
Chicago/Turabian StyleNeophytou, Constantina, Euripides Soteriou, and Chrysoula Pitsouli. 2023. "The Sterol Transporter Npc2c Controls Intestinal Stem Cell Mitosis and Host–Microbiome Interactions in Drosophila" Metabolites 13, no. 10: 1084. https://doi.org/10.3390/metabo13101084
APA StyleNeophytou, C., Soteriou, E., & Pitsouli, C. (2023). The Sterol Transporter Npc2c Controls Intestinal Stem Cell Mitosis and Host–Microbiome Interactions in Drosophila. Metabolites, 13(10), 1084. https://doi.org/10.3390/metabo13101084





