Opening the Random Forest Black Box of 1H NMR Metabolomics Data by the Exploitation of Surrogate Variables
Abstract
1. Introduction
2. Materials and Methods
2.1. Samples and Data Acquisition
2.2. Identification of Truffle Metabolites
2.3. Software and Data Analysis
3. Results and Discussion
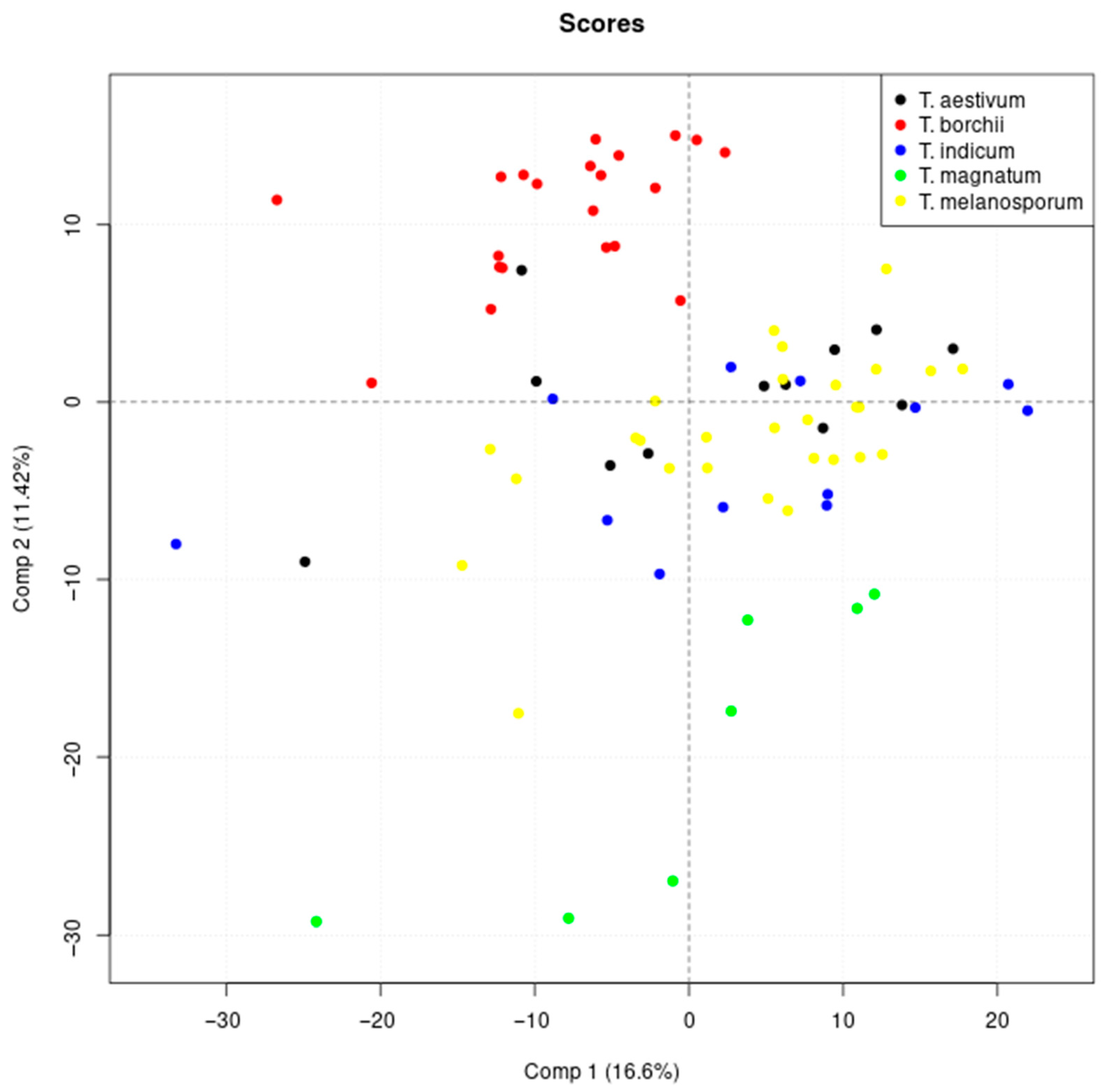
3.1. Classification of Truffle Samples
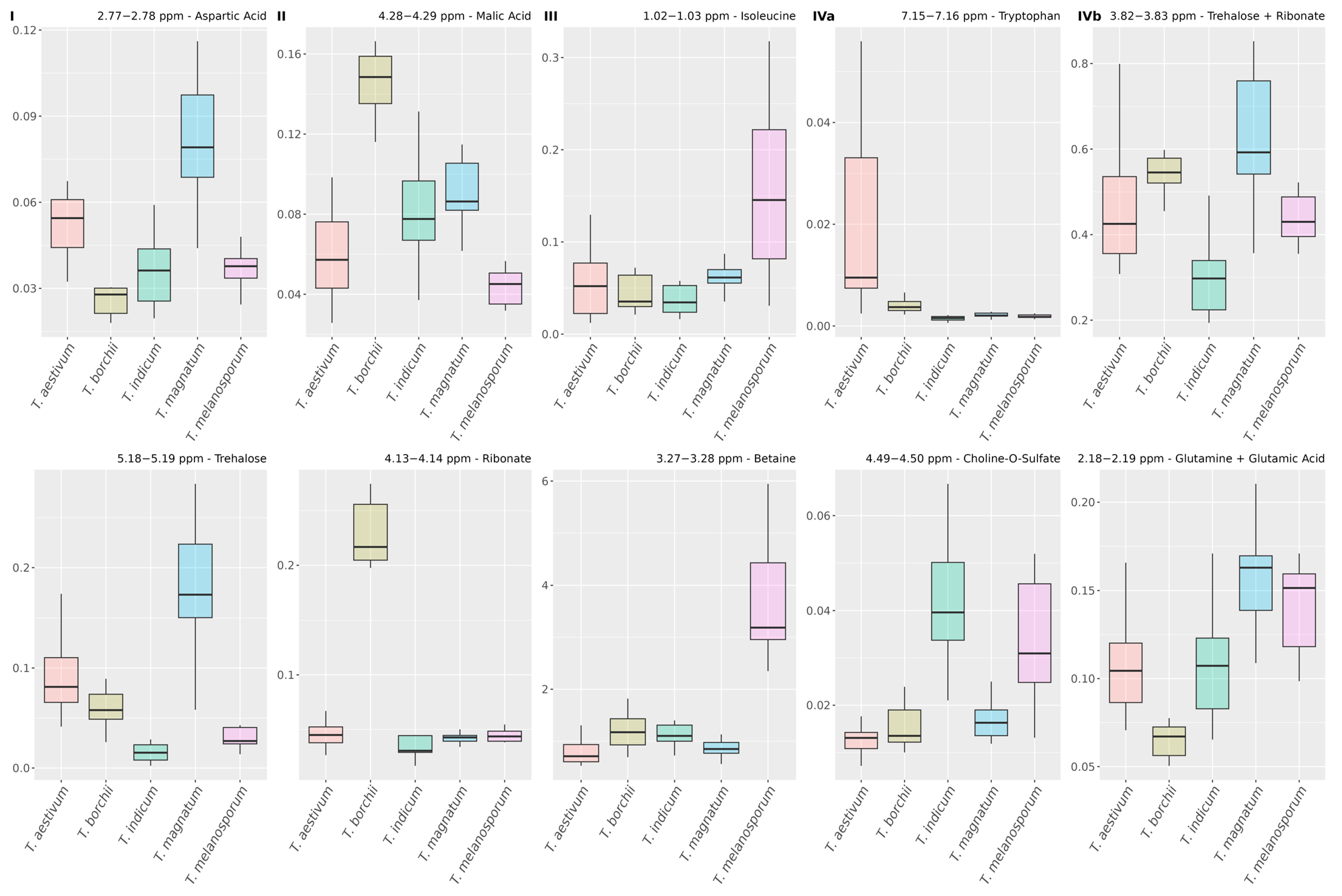
3.2. Bucket Assignment for Truffle Metabolites
3.3. Variable Selection
3.4. Analysis of Variable Relations
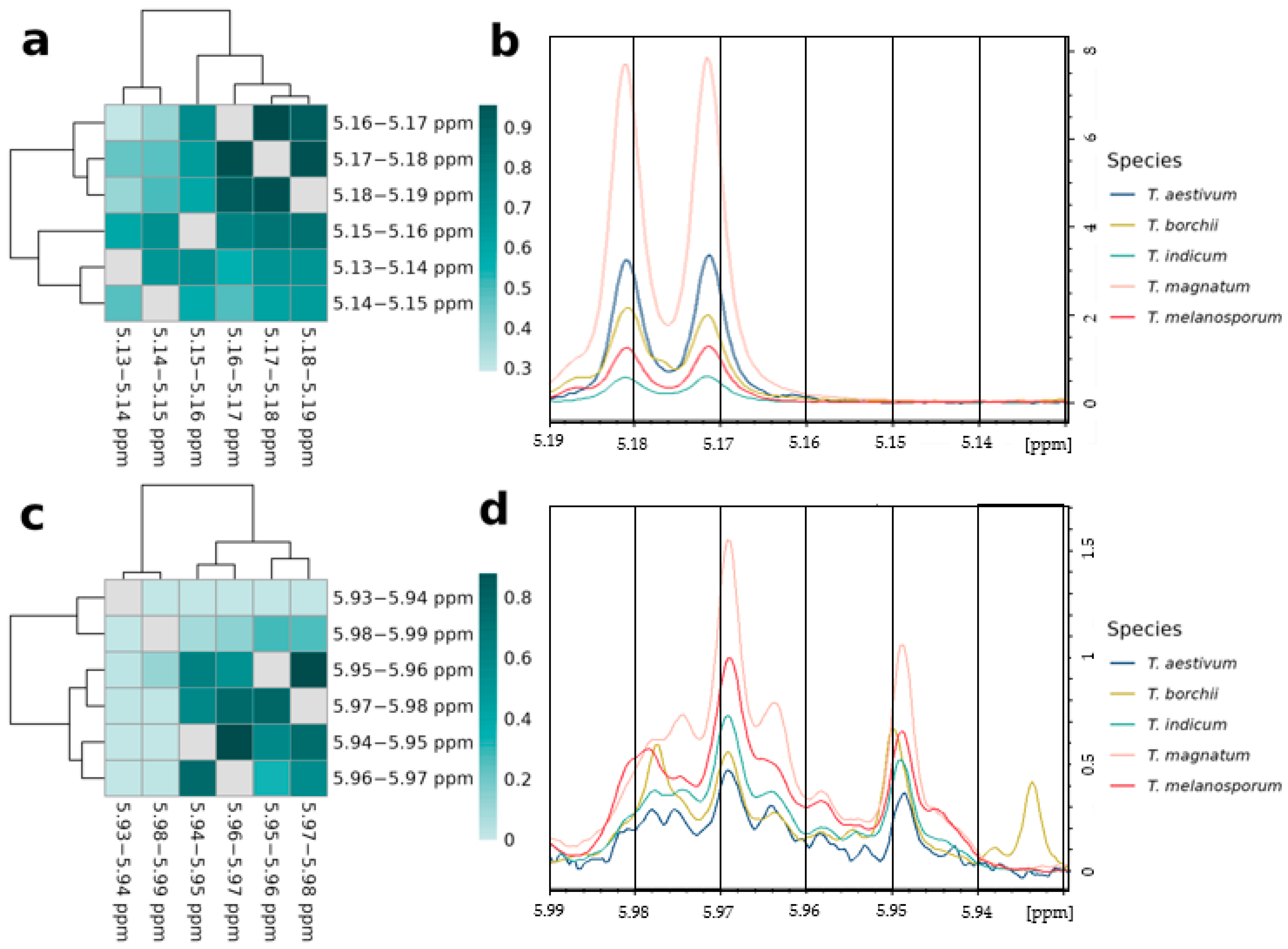
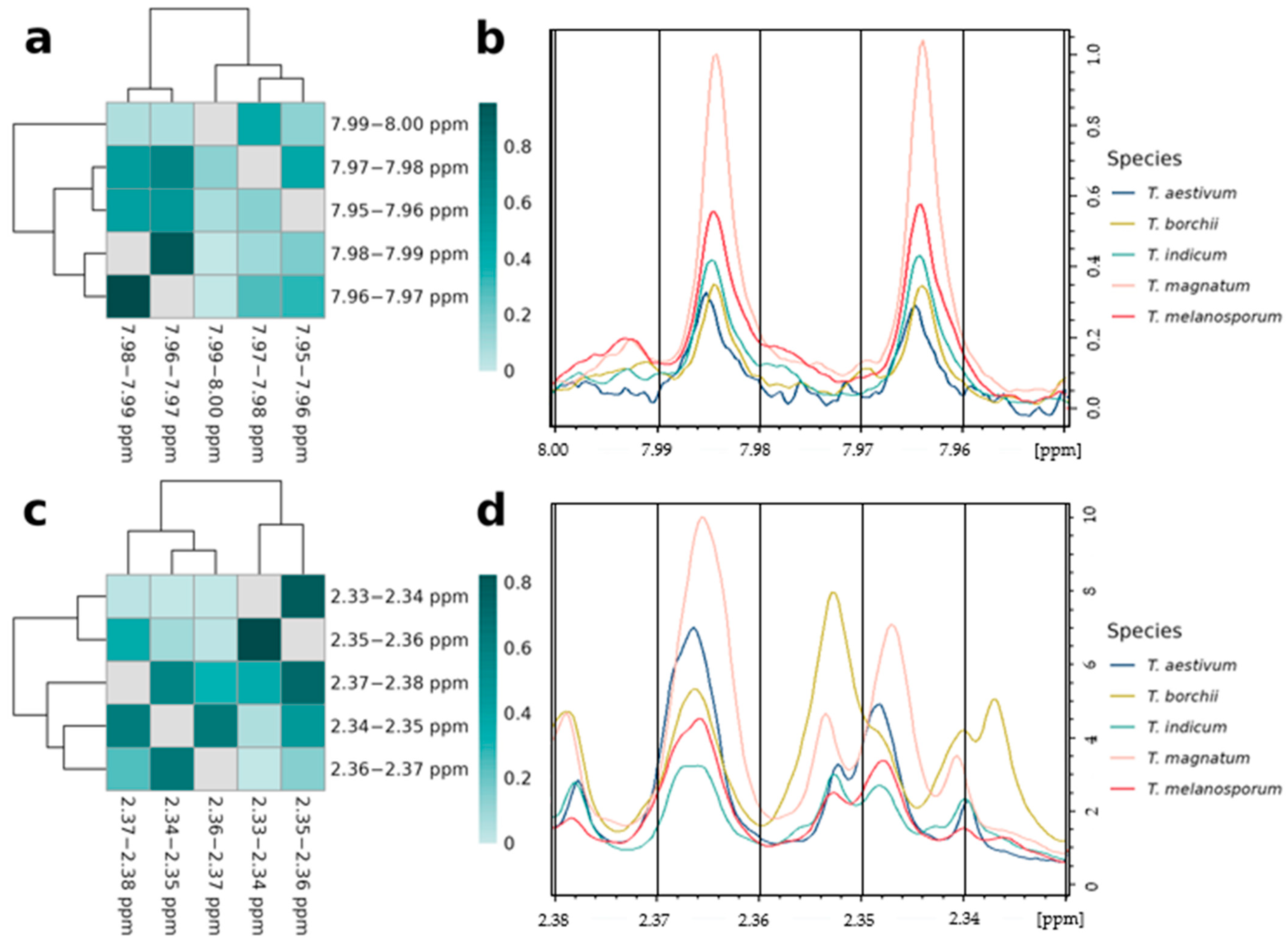
3.4.1. Relations of Variables Containing the Same Signals
3.4.2. Relations of Variables from the Same Metabolites
3.4.3. Relations of Variables from Different Metabolites
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wishart, D.S. Current Progress in Computational Metabolomics. Brief. Bioinform. 2007, 8, 279–293. [Google Scholar] [CrossRef]
- Fiehn, O. Metabolomics—The Link between Genotypes and Phenotypes. Plant Mol. Biol. 2002, 48, 155–171. [Google Scholar] [CrossRef]
- Mushtaq, M.Y.; Choi, Y.H.; Verpoorte, R.; Wilson, E.G. Extraction for Metabolomics: Access to the Metabolome. Phytochem. Anal. 2014, 25, 291–306. [Google Scholar] [CrossRef]
- Bachmann, R.; Klockmann, S.; Haerdter, J.; Fischer, M.; Hackl, T. 1H-NMR Spectroscopy for Determination of the Geographical Origin of Hazelnuts. J. Agric. Food Chem. 2018, 66, 11873–11879. [Google Scholar] [CrossRef]
- Shakiba, N.; Gerdes, A.; Holz, N.; Wenck, S.; Bachmann, R.; Schneider, T.; Seifert, S.; Fischer, M.; Hackl, T. Determination of the Geographical Origin of Hazelnuts (Corylus avellana L.) by Near-Infrared Spectroscopy (NIR) and a Low-Level Fusion with Nuclear Magnetic Resonance (NMR). Microchem. J. 2022, 174, 107066. [Google Scholar] [CrossRef]
- Creydt, M.; Hudzik, D.; Rurik, M.; Kohlbacher, O.; Fischer, M. Food Authentication: Small-Molecule Profiling as a Tool for the Geographic Discrimination of German White Asparagus. J. Agric. Food Chem. 2018, 66, 13328–13339. [Google Scholar] [CrossRef]
- Markley, J.L.; Brüschweiler, R.; Edison, A.S.; Eghbalnia, H.R.; Powers, R.; Raftery, D.; Wishart, D.S. The Future of NMR-Based Metabolomics. Curr. Opin. Biotechnol. 2017, 43, 34–40. [Google Scholar] [CrossRef]
- Bingol, K. Recent Advances in Targeted and Untargeted Metabolomics by NMR and MS/NMR Methods. High-Throughput 2018, 7, 9. [Google Scholar] [CrossRef]
- Nagana Gowda, G.A.; Raftery, D. Can NMR Solve Some Significant Challenges in Metabolomics? J. Magn. Reson. 2015, 260, 144–160. [Google Scholar] [CrossRef]
- Fan, T.W.-M.; Lane, A.N. Applications of NMR Spectroscopy to Systems Biochemistry. Prog. Nucl. Magn. Reson. Spectrosc. 2016, 92–93, 18–53. [Google Scholar] [CrossRef]
- Takis, P.G.; Ghini, V.; Tenori, L.; Turano, P.; Luchinat, C. Uniqueness of the NMR Approach to Metabolomics. TrAC Trends Anal. Chem. 2019, 120, 115300. [Google Scholar] [CrossRef]
- Hoch, J.C.; Baskaran, K.; Burr, H.; Chin, J.; Eghbalnia, H.R.; Fujiwara, T.; Gryk, M.R.; Iwata, T.; Kojima, C.; Kurisu, G.; et al. Biological Magnetic Resonance Data Bank. Nucleic Acids Res. 2023, 51, D368–D376. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Perez, I.; Posma, J.M.; Serrano-Contreras, J.I.; Boulangé, C.L.; Chan, Q.; Frost, G.; Stamler, J.; Elliott, P.; Lindon, J.C.; Holmes, E.; et al. Identifying Unknown Metabolites Using NMR-Based Metabolic Profiling Techniques. Nat. Protoc. 2020, 15, 2538–2567. [Google Scholar] [CrossRef]
- Bingol, K.; Brüschweiler, R. NMR/MS Translator for the Enhanced Simultaneous Analysis of Metabolomics Mixtures by NMR Spectroscopy and Mass Spectrometry: Application to Human Urine. J. Proteome Res. 2015, 14, 2642–2648. [Google Scholar] [CrossRef] [PubMed]
- Bingol, K.; Bruschweiler-Li, L.; Yu, C.; Somogyi, A.; Zhang, F.; Brüschweiler, R. Metabolomics Beyond Spectroscopic Databases: A Combined MS/NMR Strategy for the Rapid Identification of New Metabolites in Complex Mixtures. Anal. Chem. 2015, 87, 3864–3870. [Google Scholar] [CrossRef]
- Dai, D.; He, J.; Sun, R.; Zhang, R.; Aisa, H.A.; Abliz, Z. Nuclear Magnetic Resonance and Liquid Chromatography–Mass Spectrometry Combined with an Incompleted Separation Strategy for Identifying the Natural Products in Crude Extract. Anal. Chim. Acta 2009, 632, 221–228. [Google Scholar] [CrossRef] [PubMed]
- Watermann, S.; Bode, M.-C.; Hackl, T. Identification of Metabolites from Complex Mixtures by 3D Correlation of 1H NMR, MS and LC Data Using the SCORE-Metabolite-ID Approach. Sci. Rep. 2023, 13, 15834. [Google Scholar] [CrossRef]
- Cloarec, O.; Dumas, M.-E.; Craig, A.; Barton, R.H.; Trygg, J.; Hudson, J.; Blancher, C.; Gauguier, D.; Lindon, J.C.; Holmes, E.; et al. Statistical Total Correlation Spectroscopy: An Exploratory Approach for Latent Biomarker Identification from Metabolic 1H NMR Data Sets. Anal. Chem. 2005, 77, 1282–1289. [Google Scholar] [CrossRef] [PubMed]
- Crockford, D.J.; Holmes, E.; Lindon, J.C.; Plumb, R.S.; Zirah, S.; Bruce, S.J.; Rainville, P.; Stumpf, C.L.; Nicholson, J.K. Statistical Heterospectroscopy, an Approach to the Integrated Analysis of NMR and UPLC-MS Data Sets: Application in Metabonomic Toxicology Studies. Anal. Chem. 2006, 78, 363–371. [Google Scholar] [CrossRef]
- Ravanbakhsh, S.; Liu, P.; Bjordahl, T.C.; Mandal, R.; Grant, J.R.; Wilson, M.; Eisner, R.; Sinelnikov, I.; Hu, X.; Luchinat, C.; et al. Accurate, Fully-Automated NMR Spectral Profiling for Metabolomics. PLoS ONE 2015, 10, e0124219. [Google Scholar] [CrossRef]
- Emwas, A.-H.; Saccenti, E.; Gao, X.; McKay, R.T.; Dos Santos, V.A.P.M.; Roy, R.; Wishart, D.S. Recommended Strategies for Spectral Processing and Post-Processing of 1D 1H NMR Data of Biofluids with a Particular Focus on Urine. Metabolomics 2018, 14, 31. [Google Scholar] [CrossRef] [PubMed]
- Debik, J.; Sangermani, M.; Wang, F.; Madssen, T.S.; Giskeødegård, G.F. Multivariate Analysis of NMR-based Metabolomic Data. NMR Biomed. 2022, 35, e4638. [Google Scholar] [CrossRef] [PubMed]
- Wold, S.; Esbensen, K.; Geladi, P. Principal Component Analysis. Chemom. Intell. Lab. Syst. 1987, 2, 37–52. [Google Scholar] [CrossRef]
- Worley, B.; Powers, R. Multivariate Analysis in Metabolomics. Curr. Metabolomics 2012, 1, 92–107. [Google Scholar] [CrossRef]
- Bro, R.; Smilde, A.K. Principal Component Analysis. Anal. Methods 2014, 6, 2812–2831. [Google Scholar] [CrossRef]
- Boser, B.E.; Guyon, I.M.; Vapnik, V.N. A Training Algorithm for Optimal Margin Classifiers. In Proceedings of the Fifth Annual Workshop on Computational Learning Theory, Pittsburgh, PA, USA, 27–29 July 1992; pp. 144–152. [Google Scholar]
- Mendez, K.M.; Broadhurst, D.I.; Reinke, S.N. The Application of Artificial Neural Networks in Metabolomics: A Historical Perspective. Metabolomics 2019, 15, 142. [Google Scholar] [CrossRef]
- Breiman, L.; Friedman, J.H.; Olshen, R.A.; Stone, C.J. Classification and Regression Trees, 1st ed.; Routledge: London, UK, 2017; ISBN 978-1-315-13947-0. [Google Scholar]
- Breiman, L. Random Forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Kursa, M.B.; Rudnicki, W.R. Feature Selection with the Boruta Package. J. Stat. Softw. 2010, 36, 1–13. [Google Scholar] [CrossRef]
- Seifert, S.; Gundlach, S.; Szymczak, S. Surrogate Minimal Depth as an Importance Measure for Variables in Random Forests. Bioinformatics 2019, 35, 3663–3671. [Google Scholar] [CrossRef]
- Ishwaran, H.; Kogalur, U.B.; Chen, X.; Minn, A.J. Random Survival Forests for High-Dimensional Data: Random Survival Forests for High-Dimensional Data. Stat. Anal. Data Min. ASA Data Sci. J. 2011, 4, 115–132. [Google Scholar] [CrossRef]
- Voges, L.F.; Jarren, L.C.; Seifert, S. Exploitation of Surrogate Variables in Random Forests for Unbiased Analysis of Mutual Impact and Importance of Features. Bioinformatics 2023, 39, btad471. [Google Scholar] [CrossRef]
- Seifert, S. Application of Random Forest Based Approaches to Surface-Enhanced Raman Scattering Data. Sci. Rep. 2020, 10, 5436. [Google Scholar] [CrossRef] [PubMed]
- Živanović, V.; Seifert, S.; Drescher, D.; Schrade, P.; Werner, S.; Guttmann, P.; Szekeres, G.P.; Bachmann, S.; Schneider, G.; Arenz, C.; et al. Optical Nanosensing of Lipid Accumulation Due to Enzyme Inhibition in Live Cells. ACS Nano 2019, 13, 9363–9375. [Google Scholar] [CrossRef] [PubMed]
- Wenck, S.; Creydt, M.; Hansen, J.; Gärber, F.; Fischer, M.; Seifert, S. Opening the Random Forest Black Box of the Metabolome by the Application of Surrogate Minimal Depth. Metabolites 2022, 12, 5. [Google Scholar] [CrossRef] [PubMed]
- Lösel, H.; Brockelt, J.; Gärber, F.; Teipel, J.; Kuballa, T.; Seifert, S.; Fischer, M. Comparative Analysis of LC-ESI-IM-qToF-MS and FT-NIR Spectroscopy Approaches for the Authentication of Organic and Conventional Eggs. Metabolites 2023, 13, 882. [Google Scholar] [CrossRef]
- Mix, T.; Janneschütz, J.; Fischer, M.; Hackl, T. Differentiation of Truffle Species (Tuber spp.) by 1H NMR Spectroscopy and support vector machine. ChemRxiv 2023. preprint. [Google Scholar] [CrossRef]
- Mannina, L.; Sobolev, A.P.; Capitani, D. Applications of NMR Metabolomics to the Study of Foodstuffs: Truffle, Kiwifruit, Lettuce, and Sea Bass: General. Electrophoresis 2012, 33, 2290–2313. [Google Scholar] [CrossRef]
- Li, X.; Zhang, X.; Ye, L.; Kang, Z.; Jia, D.; Yang, L.; Zhang, B. LC-MS-Based Metabolomic Approach Revealed the Significantly Different Metabolic Profiles of Five Commercial Truffle Species. Front. Microbiol. 2019, 10, 2227. [Google Scholar] [CrossRef]
- Shaka, A.J.; Lee, C.J.; Pines, A. Iterative Schemes for Bilinear Operators; Application to Spin Decoupling. J. Magn. Reson. 1969 1988, 77, 274–293. [Google Scholar] [CrossRef]
- Dona, A.C.; Kyriakides, M.; Scott, F.; Shephard, E.A.; Varshavi, D.; Veselkov, K.; Everett, J.R. A Guide to the Identification of Metabolites in NMR-Based Metabonomics/Metabolomics Experiments. Comput. Struct. Biotechnol. J. 2016, 14, 135–153. [Google Scholar] [CrossRef]
- Wright, M.N.; Ziegler, A. Ranger: A Fast Implementation of Random Forests for High Dimensional Data in C++ and R. J. Stat. Softw. 2017, 77, 1–17. [Google Scholar] [CrossRef]
- Kucheryavskiy, S. Mdatools—R Package for Chemometrics. Chemom. Intell. Lab. Syst. 2020, 198, 103937. [Google Scholar] [CrossRef]
- Degenhardt, F.; Seifert, S.; Szymczak, S. Evaluation of Variable Selection Methods for Random Forests and Omics Data Sets. Brief. Bioinform. 2019, 20, 492–503. [Google Scholar] [CrossRef]
- Wickham, H. Ggplot2: Elegant Graphics for Data Analysis; Springer International Publishing: Cham, Switzerland, 2016; ISBN 978-3-319-24277-4. [Google Scholar]
- Kolde, R. Pheatmap: Pretty Heatmaps. 2019. Available online: https://cran.r-project.org/web/packages/pheatmap/pheatmap.pdf (accessed on 11 October 2023).
- Ward, J.H. Hierarchical Grouping to Optimize an Objective Function. J. Am. Stat. Assoc. 1963, 58, 236–244. [Google Scholar] [CrossRef]
- Martin, F.; Canet, D.; Marchal, J.P. 13C Nuclear Magnetic Resonance Study of Mannitol Cycle and Trehalose Synthesis during Glucose Utilization by the Ectomycorrhizal Ascomycete Cenococcum graniforme. Plant Physiol. 1985, 77, 499–502. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Merzendorfer, H. The Cellular Basis of Chitin Synthesis in Fungi and Insects: Common Principles and Differences. Eur. J. Cell Biol. 2011, 90, 759–769. [Google Scholar] [CrossRef]
- Genetet, I.; Martin, F.; Stewart, G.R. Nitrogen Assimilation in Mycorrhizas: Ammonium Assimilation in the N-Starved Ectomycorrhizal Fungus Cenococcum Graniforme. Plant Physiol. 1984, 76, 395–399. [Google Scholar] [CrossRef] [PubMed]
- Lam, H.-M.; Coschigano, K.T.; Oliveira, I.C.; Melo-Oliveira, R.; Coruzzi, G.M. The Molecular-Genetics of Nitrogen Assimilation into Amino Acids in Higher Plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1996, 47, 569–593. [Google Scholar] [CrossRef]
- Kenealy, W.; Zaady, E.; Du Preez, J.C.; Stieglitz, B.; Goldberg, I. Biochemical Aspects of Fumaric Acid Accumulation by Rhizopus arrhizus. Appl. Environ. Microbiol. 1986, 52, 128–133. [Google Scholar] [CrossRef] [PubMed]
- Roa Engel, C.A.; Straathof, A.J.J.; Zijlmans, T.W.; Van Gulik, W.M.; Van Der Wielen, L.A.M. Fumaric Acid Production by Fermentation. Appl. Microbiol. Biotechnol. 2008, 78, 379–389. [Google Scholar] [CrossRef]
- Citterio, B.; Malatesta, M.; Battistelli, S.; Marcheggiani, F.; Baffone, W.; Saltarelli, R.; Stocchi, V.; Gazzanelli, G. Possible Involvement of Pseudomonas fluorescens and Bacillaceae in Structural Modifications of Tuber borchii Fruit Bodies. Can. J. Microbiol. 2001, 47, 264–268. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.-X.; Rainey, P.B. Dual Involvement of CbrAB and NtrBC in the Regulation of Histidine Utilization in Pseudomonas fluorescens SBW25. Genetics 2008, 178, 185–195. [Google Scholar] [CrossRef]
- Gross, S.R. Genetic Regulatory Mechanisms in the Fungi. Annu. Rev. Genet. 1969, 3, 395–424. [Google Scholar] [CrossRef]
- Chen, T.H.H.; Murata, N. Enhancement of Tolerance of Abiotic Stress by Metabolic Engineering of Betaines and Other Compatible Solutes. Curr. Opin. Plant Biol. 2002, 5, 250–257. [Google Scholar] [CrossRef]
- Fernandes, J.D.S.; Martho, K.; Tofik, V.; Vallim, M.A.; Pascon, R.C. The Role of Amino Acid Permeases and Tryptophan Biosynthesis in Cryptococcus neoformans Survival. PLoS ONE 2015, 10, e0132369. [Google Scholar] [CrossRef] [PubMed]
- Spencer, B.; Hussey, E.C.; Orsi, B.A.; Scott, J.M. Mechanism of Choline O-Sulphate Utilization in Fungi. Biochem. J. 1968, 106, 461–469. [Google Scholar] [CrossRef]
- Ariño, J.; Velázquez, D.; Casamayor, A. Ser/Thr Protein Phosphatases in Fungi: Structure, Regulation and Function. Microb. Cell 2019, 6, 217–256. [Google Scholar] [CrossRef] [PubMed]



| T. aestivum | T. borchii | T. indicum | T. magnatum | T. melanosporum | |
|---|---|---|---|---|---|
| Amount | 28 | 7 | 12 | 21 | 12 |
| Color | black | white | black | white | black |
| Approach | Parameter | Description | Value |
|---|---|---|---|
| RF | ntree | number of trees | 10,000 |
| min.node.size | number of samples in terminal node | 1 | |
| mtry | number of candidate variables | 157 (p3/4) 1 | |
| case. weights | weights for sampling of training observations | chosen according to the size of the respective class | |
| SMD | s | Predefined number of surrogate splits | 42 (p ∙ 0.05) |
| Boruta | pValue | applied importance measure | impurity_corrected |
| importance | confidence level | 0.01 | |
| maxRuns | maximum number of importance source runs | 157 (p3/4) 1 |
| T. aestivum | T. borchii | T. indicum | T. magnatum | T. melanosporum | Sensitivity [%] | |
|---|---|---|---|---|---|---|
| T. aestivum | 28 | 0 | 0 | 0 | 0 | 100 |
| T. borchii | 0 | 7 | 0 | 0 | 0 | 100 |
| T. indicum | 0 | 0 | 12 | 0 | 0 | 100 |
| T. magnatum | 0 | 0 | 0 | 21 | 0 | 100 |
| T. melanosporum | 0 | 0 | 0 | 0 | 12 | 100 |
| Specificity [%] | 100 | 100 | 100 | 100 | 100 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wenck, S.; Mix, T.; Fischer, M.; Hackl, T.; Seifert, S. Opening the Random Forest Black Box of 1H NMR Metabolomics Data by the Exploitation of Surrogate Variables. Metabolites 2023, 13, 1075. https://doi.org/10.3390/metabo13101075
Wenck S, Mix T, Fischer M, Hackl T, Seifert S. Opening the Random Forest Black Box of 1H NMR Metabolomics Data by the Exploitation of Surrogate Variables. Metabolites. 2023; 13(10):1075. https://doi.org/10.3390/metabo13101075
Chicago/Turabian StyleWenck, Soeren, Thorsten Mix, Markus Fischer, Thomas Hackl, and Stephan Seifert. 2023. "Opening the Random Forest Black Box of 1H NMR Metabolomics Data by the Exploitation of Surrogate Variables" Metabolites 13, no. 10: 1075. https://doi.org/10.3390/metabo13101075
APA StyleWenck, S., Mix, T., Fischer, M., Hackl, T., & Seifert, S. (2023). Opening the Random Forest Black Box of 1H NMR Metabolomics Data by the Exploitation of Surrogate Variables. Metabolites, 13(10), 1075. https://doi.org/10.3390/metabo13101075





