Neurobiological Basis of Aversion-Resistant Ethanol Seeking in C. elegans
Abstract
1. Introduction
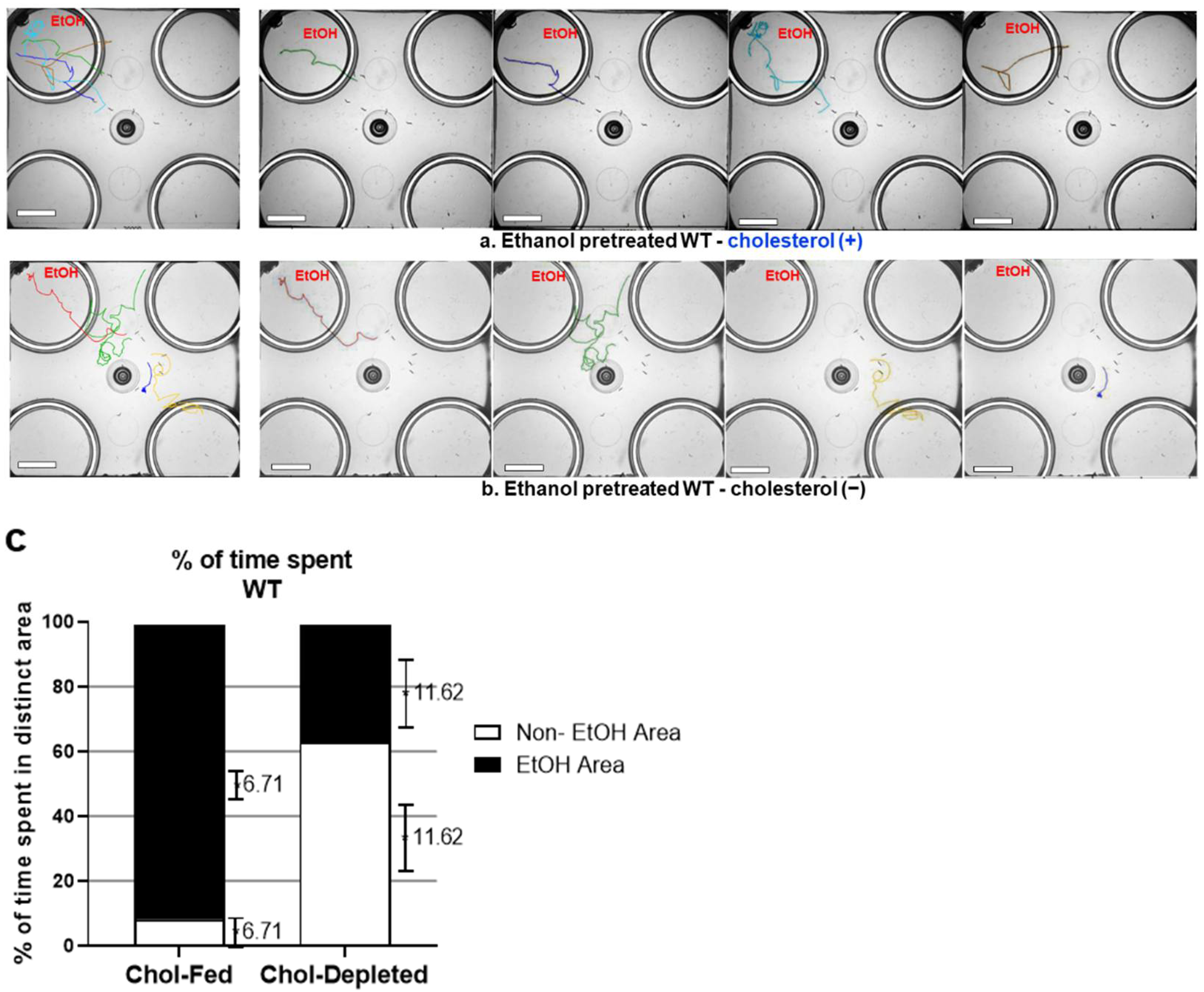
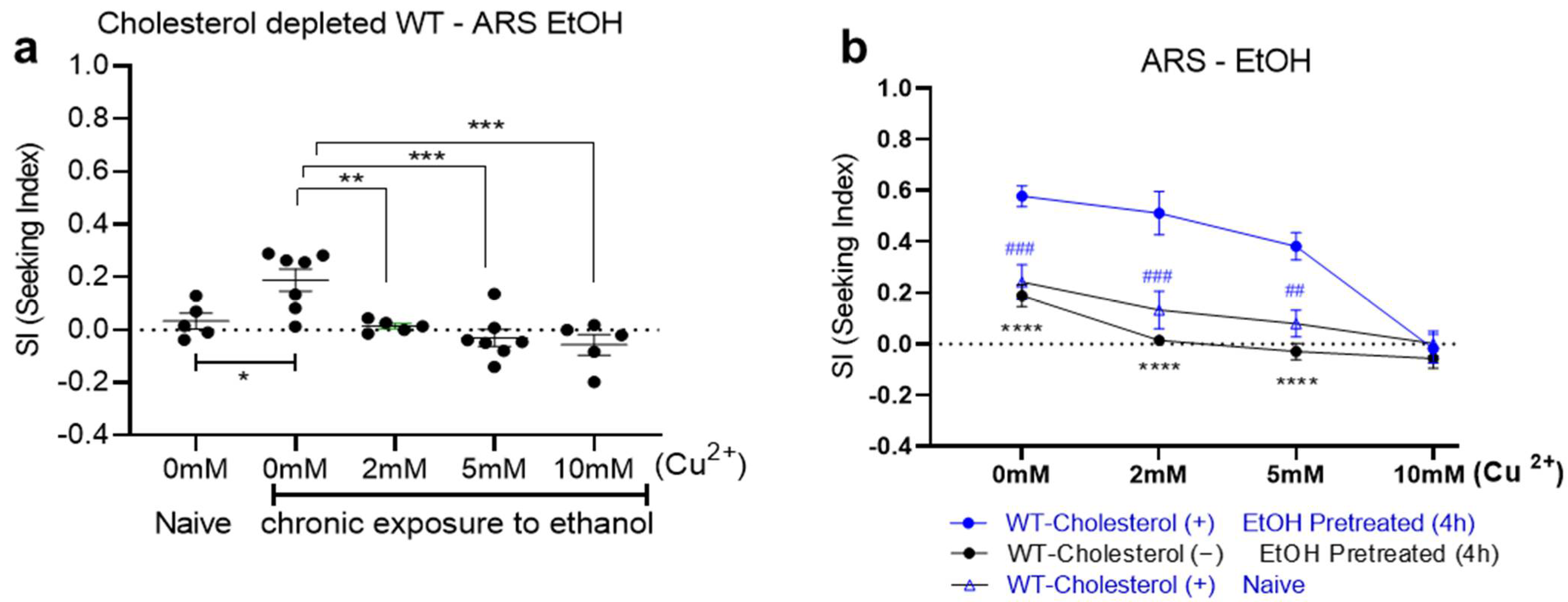
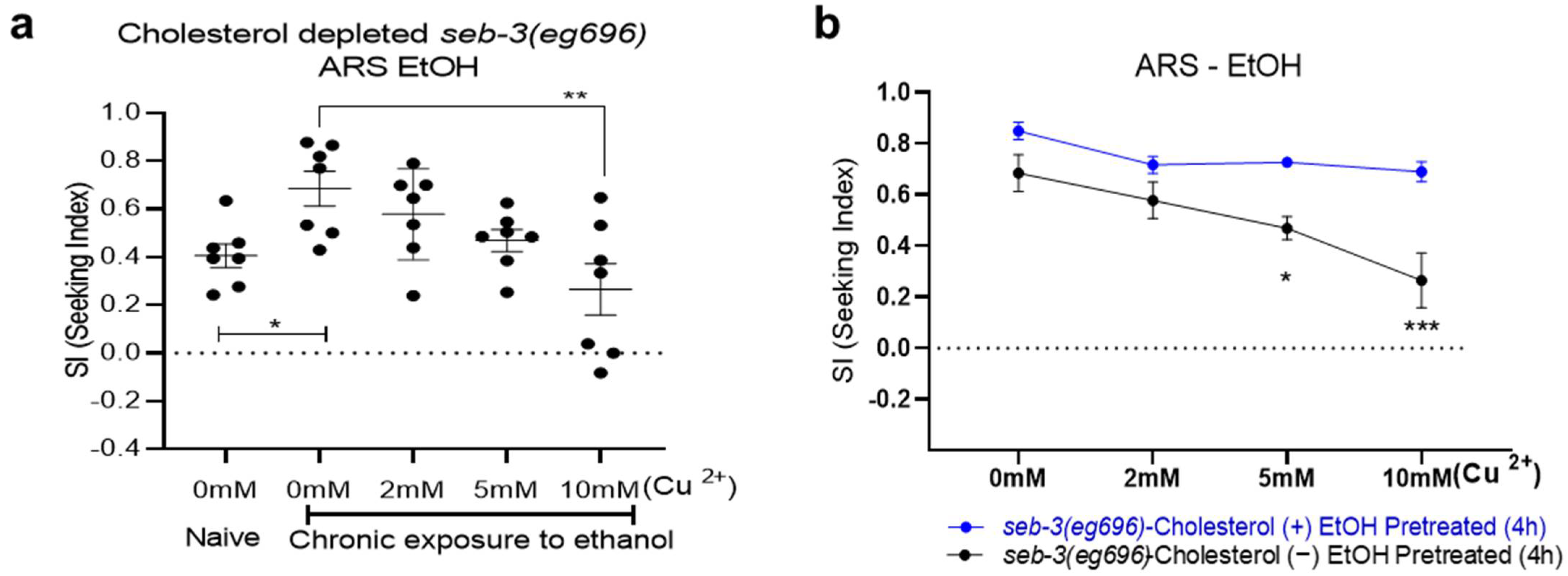
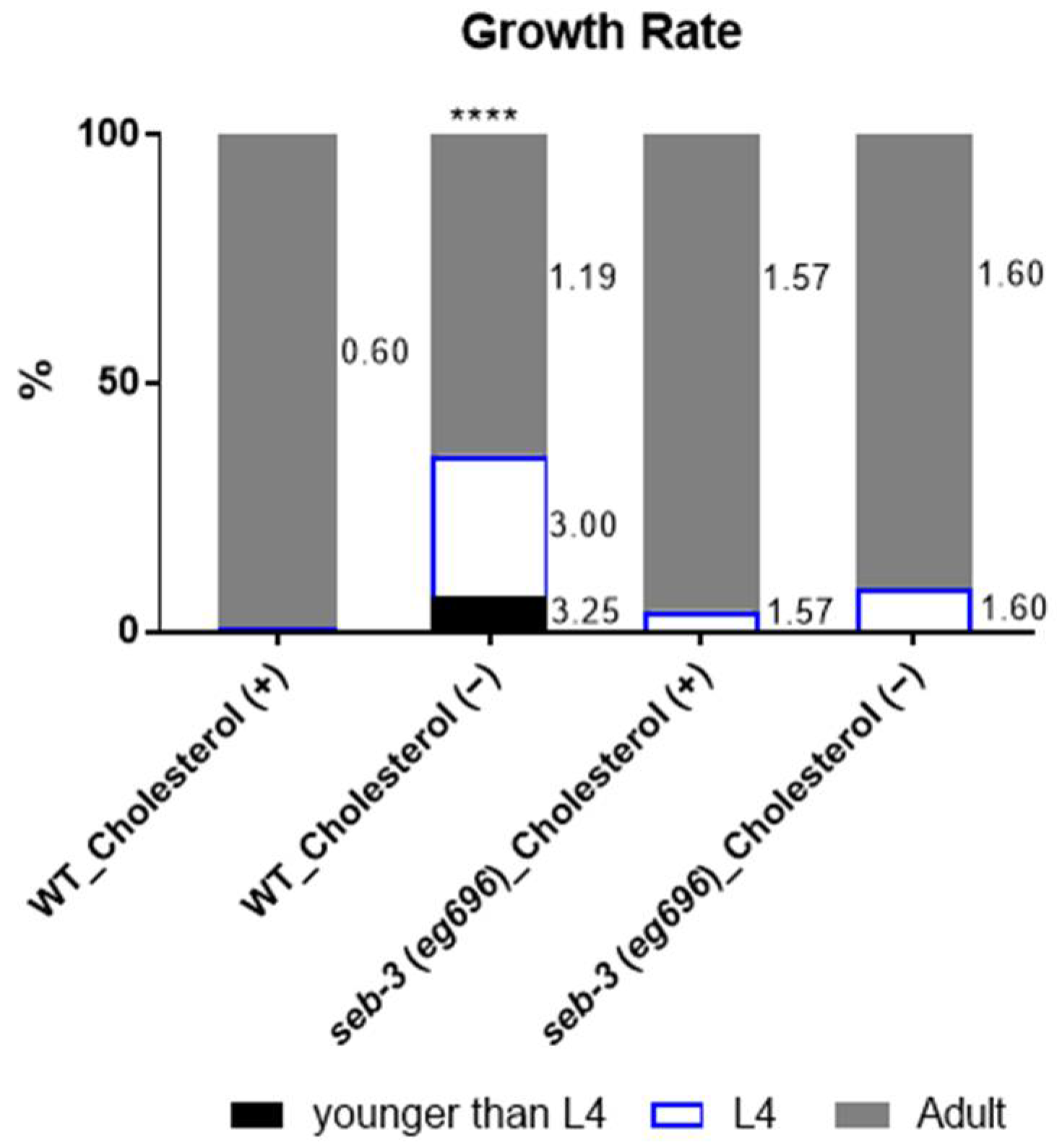
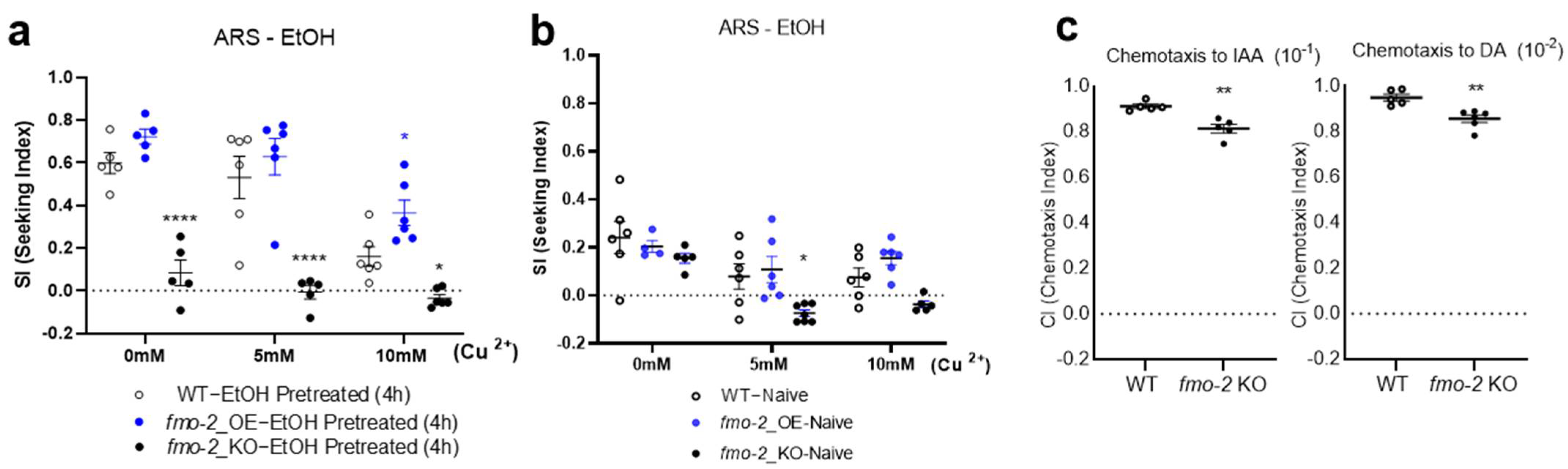
2. Results
3. Discussion
4. Materials and Methods
4.1. Cholesterol-Depleted Animals
4.2. Trajectory Analysis of C. elegans Locomotion in the Free-Moving Ethanol Preference Assay on a 4-Well Plate
4.3. Aversion-Resistant Seeking (ARS) of Ethanol Assay
4.4. Statistical Analysis
4.5. Gene Ontology Enrichment Analysis
4.6. Chemotaxis Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- World Health, O. Global Status Report on Alcohol and Health 2018; World Health Organization: Geneva, Switzerland, 2018. [Google Scholar]
- Esser, M.B.; Sherk, A.; Liu, Y.; Naimi, T.S.; Stockwell, T.; Stahre, M.; Kanny, D.; Landen, M.; Saitz, R.; Brewer, R.D. Deaths and Years of Potential Life Lost from Excessive Alcohol Use — United States, 2011–2015. MMWR Morb. Mortal. Wkly. Rep. 2020, 69, 1428–1433. [Google Scholar] [CrossRef] [PubMed]
- Rusyn, I.; Bataller, R. Alcohol and toxicity. J. Hepatol. 2013, 59, 387–388. [Google Scholar] [CrossRef] [PubMed]
- Rehm, J.; Baliunas, D.; Borges, G.L.; Graham, K.; Irving, H.; Kehoe, T.; Parry, C.D.; Patra, J.; Popova, S.; Poznyak, V.; et al. The relation between different dimensions of alcohol consumption and burden of disease: An overview. Addiction 2010, 105, 817–843. [Google Scholar] [CrossRef] [PubMed]
- Belin, D.; Everitt, B.J. Cocaine seeking habits depend upon dopamine-dependent serial connectivity linking the ventral with the dorsal striatum. Neuron 2008, 57, 432–441. [Google Scholar] [CrossRef] [PubMed]
- Fontenelle, L.F.; Oostermeijer, S.; Harrison, B.J.; Pantelis, C.; Yücel, M. Obsessive-compulsive disorder, impulse control disorders and drug addiction: Common features and potential treatments. Drugs 2011, 71, 827–840. [Google Scholar] [CrossRef]
- Baler, R.D.; Volkow, N.D. Drug addiction: The neurobiology of disrupted self-control. Trends Mol. Med. 2006, 12, 559–566. [Google Scholar] [CrossRef]
- Koob, G.F.; Volkow, N.D. Neurobiology of addiction: A neurocircuitry analysis. Lancet Psychiatry 2016, 3, 760–773. [Google Scholar] [CrossRef]
- Seif, T.; Chang, S.-J.J.; Simms, J.A.; Gibb, S.L.; Dadgar, J.; Chen, B.T.; Harvey, B.K.; Ron, D.; Messing, R.O.; Bonci, A.; et al. Cortical activation of accumbens hyperpolarization-active NMDARs mediates aversion-resistant alcohol intake. Nat. Neurosci. 2013, 16, 1094–1100. [Google Scholar] [CrossRef]
- Hopf, F.W.; Lesscher, H.M. Rodent models for compulsive alcohol intake. Alcohol 2014, 48, 253–264. [Google Scholar] [CrossRef]
- Salim, C.; Kan, A.K.; Batsaikhan, E.; Patterson, E.C.; Jee, C. Neuropeptidergic regulation of compulsive ethanol seeking in C. elegans. Sci. Rep. 2022, 12, 1804. [Google Scholar] [CrossRef]
- Flannery, B.A.; Roberts, A.J.; Cooney, N.; Swift, R.M.; Anton, R.F.; Rohsenow, D.J. The role of craving in alcohol use, dependence, and treatment. Alcohol Clin. Exp. Res. 2001, 25, 299–308. [Google Scholar] [CrossRef] [PubMed]
- Anton, R.F.; Moak, D.H.; Latham, P.K. The obsessive compulsive drinking scale: A new method of assessing outcome in alcoholism treatment studies. Arch. Gen. Psychiatry 1996, 53, 225–231. [Google Scholar] [CrossRef] [PubMed]
- Modell, J.G.; Glaser, F.B.; Cyr, L.; Mountz, J.M. Obsessive and compulsive characteristics of craving for alcohol in alcohol abuse and dependence. Alcohol. Clin. Exp. Res. 1992, 16, 272–274. [Google Scholar] [CrossRef] [PubMed]
- Radke, A.K.; Nakazawa, K.; Holmes, A. Cortical GluN2B deletion attenuates punished suppression of food reward-seeking. Psychopharmacology 2015, 232, 3753–3761. [Google Scholar] [CrossRef] [PubMed]
- Jee, C.; Lee, J.; Lim, J.P.; Parry, D.; Messing, R.O.; McIntire, S.L. SEB-3, a CRF receptor-like GPCR, regulates locomotor activity states, stress responses and ethanol tolerance in Caenorhabditis elegans. Genes Brain Behav. 2013, 12, 250–262. [Google Scholar] [CrossRef]
- Davies, A.G.; Pierce-Shimomura, J.T.; Kim, H.; VanHoven, M.K.; Thiele, T.R.; Bonci, A.; Bargmann, C.I.; McIntire, S.L. A central role of the BK potassium channel in behavioral responses to ethanol in C. elegans. Cell 2003, 115, 655–666. [Google Scholar] [CrossRef]
- Davies, A.G.; Bettinger, J.C.; Thiele, T.R.; Judy, M.E.; McIntire, S.L. Natural variation in the npr-1 gene modifies ethanol responses of wild strains of C. elegans. Neuron 2004, 42, 731–743. [Google Scholar] [CrossRef]
- Lee, J.; Jee, C.; McIntire, S.L. Ethanol preference in C. elegans. Genes Brain Behav. 2009, 8, 578–585. [Google Scholar] [CrossRef]
- Bettinger, J.C.; Leung, K.; Bolling, M.H.; Goldsmith, A.D.; Davies, A.G. Lipid environment modulates the development of acute tolerance to ethanol in Caenorhabditis elegans. PloS ONE 2012, 7, e35192. [Google Scholar] [CrossRef]
- Alaimo, J.T.; Davis, S.J.; Song, S.S.; Burnette, C.R.; Grotewiel, M.; Shelton, K.L.; Pierce-Shimomura, J.T.; Davies, A.G.; Bettinger, J.C. Ethanol Metabolism and Osmolarity Modify Behavioral Responses to Ethanol in C. elegans. Alcohol. Clin. Exp. Res. 2012, 36, 1840–1850. [Google Scholar] [CrossRef]
- Sun, L.; Wu, Q.; Liao, K.; Yu, P.; Cui, Q.; Rui, Q.; Wang, D. Contribution of heavy metals to toxicity of coal combustion related fine particulate matter (PM2.5) in Caenorhabditis elegans with wild-type or susceptible genetic background. Chemosphere 2016, 144, 2392–2400. [Google Scholar] [CrossRef] [PubMed]
- Hilliard, M.A.; Bargmann, C.I.; Bazzicalupo, P.C. elegans responds to chemical repellents by integrating sensory inputs from the head and the tail. Curr. Biol. CB 2002, 12, 730–734. [Google Scholar] [CrossRef] [PubMed]
- Deroche-Gamonet, V.; Belin, D.; Piazza, P.V. Evidence for addiction-like behavior in the rat. Science 2004, 305, 1014–1017. [Google Scholar] [CrossRef] [PubMed]
- Vengeliene, V.; Celerier, E.; Chaskiel, L.; Penzo, F.; Spanagel, R. Compulsive alcohol drinking in rodents. Addict. Biol. 2009, 14, 384–396. [Google Scholar] [CrossRef]
- Barbier, E.; Johnstone, A.L.; Khomtchouk, B.B.; Tapocik, J.D.; Pitcairn, C.; Rehman, F.; Augier, E.; Borich, A.; Schank, J.R.; Rienas, C.A.; et al. Dependence-induced increase of alcohol self-administration and compulsive drinking mediated by the histone methyltransferase PRDM2. Mol. Psychiatry 2017, 22, 1746–1758. [Google Scholar] [CrossRef]
- Vendruscolo, L.F.; Barbier, E.; Schlosburg, J.E.; Misra, K.K.; Whitfield, T.W., Jr.; Logrip, M.L.; Rivier, C.; Repunte-Canonigo, V.; Zorrilla, E.P.; Sanna, P.P.; et al. Corticosteroid-dependent plasticity mediates compulsive alcohol drinking in rats. J. Neurosci. 2012, 32, 7563–7571. [Google Scholar] [CrossRef]
- Funk, D.; Coen, K.; Tamadon, S.; Lê, A.D. Effect of chronic alcohol vapor exposure on reinstatement of alcohol seeking induced by U50,488. Neuropharmacology 2019, 148, 210–219. [Google Scholar] [CrossRef]
- Hopf, F.W.; Chang, S.J.; Sparta, D.R.; Bowers, M.S.; Bonci, A. Motivation for alcohol becomes resistant to quinine adulteration after 3 to 4 months of intermittent alcohol self-administration. Alcohol. Clin. Exp. Res. 2010, 34, 1565–1573. [Google Scholar] [CrossRef]
- Sterken, M.G.; van Wijk, M.H.; Quamme, E.C.; Riksen, J.A.G.; Carnell, L.; Mathies, L.D.; Davies, A.G.; Kammenga, J.E.; Bettinger, J.C. Transcriptional analysis of the response of C. elegans to ethanol exposure. Sci. Rep. 2021, 11, 10993. [Google Scholar] [CrossRef]
- Hamilton, J.A.; Hillard, C.J.; Spector, A.A.; Watkins, P.A. Brain uptake and utilization of fatty acids, lipids and lipoproteins: Application to neurological disorders. J. Mol. Neurosci. 2007, 33, 2–11. [Google Scholar] [CrossRef]
- Bozek, K.; Wei, Y.; Yan, Z.; Liu, X.; Xiong, J.; Sugimoto, M.; Tomita, M.; Pääbo, S.; Sherwood, C.C.; Hof, P.R.; et al. Organization and evolution of brain lipidome revealed by large-scale analysis of human, chimpanzee, macaque, and mouse tissues. Neuron 2015, 85, 695–702. [Google Scholar] [CrossRef] [PubMed]
- Hieb, W.F.; Rothstein, M. Sterol requirement for reproduction of a free-living nematode. Science 1968, 160, 778–780. [Google Scholar] [CrossRef] [PubMed]
- Matyash, V.; Entchev, E.V.; Mende, F.; Wilsch-Bräuninger, M.; Thiele, C.; Schmidt, A.W.; Knölker, H.J.; Ward, S.; Kurzchalia, T.V. Sterol-derived hormone(s) controls entry into diapause in Caenorhabditis elegans by consecutive activation of DAF-12 and DAF-16. PLoS Biol. 2004, 2, e280. [Google Scholar] [CrossRef] [PubMed]
- Rauthan, M.; Pilon, M. The mevalonate pathway in C. elegans. Lipids Health Dis. 2011, 10, 243. [Google Scholar] [CrossRef]
- Satouchi, K.; Hirano, K.; Sakaguchi, M.; Takehara, H.; Matsuura, F. Phospholipids from the free-living nematode Caenorhabditis elegans. Lipids 1993, 28, 837–840. [Google Scholar] [CrossRef]
- Kniazeva, M.; Crawford, Q.T.; Seiber, M.; Wang, C.Y.; Han, M. Monomethyl branched-chain fatty acids play an essential role in Caenorhabditis elegans development. PLoS Biol. 2004, 2, E257. [Google Scholar] [CrossRef]
- Watts, J.L.; Browse, J. Genetic dissection of polyunsaturated fatty acid synthesis in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 2002, 99, 5854–5859. [Google Scholar] [CrossRef]
- Merris, M.; Wadsworth, W.G.; Khamrai, U.; Bittman, R.; Chitwood, D.J.; Lenard, J. Sterol effects and sites of sterol accumulation in Caenorhabditis elegans developmental requirement for 4α-methyl sterols. J. Lipid Res. 2003, 44, 172–181. [Google Scholar] [CrossRef]
- Unal, C.T.; Beverley, J.A.; Willuhn, I.; Steiner, H. Long-lasting dysregulation of gene expression in corticostriatal circuits after repeated cocaine treatment in adult rats: Effects on zif 268 and homer 1a. Eur. J. Neurosci. 2009, 29, 1615–1626. [Google Scholar] [CrossRef]
- Zhou, Z.; Enoch, M.-A.A.; Goldman, D. Gene expression in the addicted brain. Int. Rev. Neurobiol. 2014, 116, 251–273. [Google Scholar] [CrossRef]
- Ribeiro, E.A.; Scarpa, J.R.; Garamszegi, S.P.; Kasarskis, A.; Mash, D.C.; Nestler, E.J. Gene Network Dysregulation in Dorsolateral Prefrontal Cortex Neurons of Humans with Cocaine Use Disorder. Sci. Rep. 2017, 7, 5412. [Google Scholar] [CrossRef] [PubMed]
- Raudvere, U.; Kolberg, L.; Kuzmin, I.; Arak, T.; Adler, P.; Peterson, H.; Vilo, J. g:Profiler: A web server for functional enrichment analysis and conversions of gene lists (2019 update). Nucleic Acids Res. 2019, 47, W191–W198. [Google Scholar] [CrossRef] [PubMed]
- Huang da, W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef] [PubMed]
- Huang da, W.; Sherman, B.T.; Lempicki, R.A. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009, 37, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Papadopoulos, J.S.; Agarwala, R. COBALT: Constraint-based alignment tool for multiple protein sequences. Bioinformatics 2007, 23, 1073–1079. [Google Scholar] [CrossRef]
- Kim, W.; Underwood, R.S.; Greenwald, I.; Shaye, D.D. OrthoList 2: A New Comparative Genomic Analysis of Human and Caenorhabditis elegans Genes. Genetics 2018, 210, 445–461. [Google Scholar] [CrossRef]
- Krueger, S.K.; Williams, D.E. Mammalian flavin-containing monooxygenases: Structure/function, genetic polymorphisms and role in drug metabolism. Pharmacol. Ther. 2005, 106, 357–387. [Google Scholar] [CrossRef]
- Veeravalli, S.; Omar, B.A.; Houseman, L.; Hancock, M.; Gonzalez Malagon, S.G.; Scott, F.; Janmohamed, A.; Phillips, I.R.; Shephard, E.A. The phenotype of a flavin-containing monooyxgenase knockout mouse implicates the drug-metabolizing enzyme FMO1 as a novel regulator of energy balance. Biochem. Pharmacol. 2014, 90, 88–95. [Google Scholar] [CrossRef]
- Gonzalez Malagon, S.G.; Melidoni, A.N.; Hernandez, D.; Omar, B.A.; Houseman, L.; Veeravalli, S.; Scott, F.; Varshavi, D.; Everett, J.; Tsuchiya, Y.; et al. The phenotype of a knockout mouse identifies flavin-containing monooxygenase 5 (FMO5) as a regulator of metabolic ageing. Biochem. Pharmacol. 2015, 96, 267–277. [Google Scholar] [CrossRef]
- Leiser, S.F.; Miller, H.; Rossner, R.; Fletcher, M.; Leonard, A.; Primitivo, M.; Rintala, N.; Ramos, F.J.; Miller, D.L.; Kaeberlein, M. Cell nonautonomous activation of flavin-containing monooxygenase promotes longevity and health span. Science 2015, 350, 1375–1378. [Google Scholar] [CrossRef]
- Hoppe, J.M.; Frick, A.; Åhs, F.; Linnman, C.; Appel, L.; Jonasson, M.; Lubberink, M.; Långström, B.; Frans, Ö.; von Knorring, L.; et al. Association between amygdala neurokinin-1 receptor availability and anxiety-related personality traits. Transl. Psychiatry 2018, 8, 168. [Google Scholar] [CrossRef] [PubMed]
- Warrier, M.; Shih, D.M.; Burrows, A.C.; Ferguson, D.; Gromovsky, A.D.; Brown, A.L.; Marshall, S.; McDaniel, A.; Schugar, R.C.; Wang, Z.; et al. The TMAO-Generating Enzyme Flavin Monooxygenase 3 Is a Central Regulator of Cholesterol Balance. Cell Rep. 2015, 10, 326–338. [Google Scholar] [CrossRef] [PubMed]
- De Felipe, C.; Herrero, J.F.; O’Brien, J.A.; Palmer, J.A.; Doyle, C.A.; Smith, A.J.; Laird, J.M.; Belmonte, C.; Cervero, F.; Hunt, S.P. Altered nociception, analgesia and aggression in mice lacking the receptor for substance P. Nature 1998, 392, 394–397. [Google Scholar] [CrossRef] [PubMed]
- Bargmann, C.I.; Hartwieg, E.; Horvitz, H.R. Odorant-selective genes and neurons mediate olfaction in C. elegans. Cell 1993, 74, 515–527. [Google Scholar] [CrossRef] [PubMed]
- Sengupta, P.; Colbert, H.A.; Bargmann, C.I. The C. elegans gene odr-7 encodes an olfactory-specific member of the nuclear receptor superfamily. Cell 1994, 79, 971–980. [Google Scholar] [CrossRef]
- Bargmann, C.I.; Horvitz, H.R. Chemosensory neurons with overlapping functions direct chemotaxis to multiple chemicals in C. elegans. Neuron 1991, 7, 729–742. [Google Scholar] [CrossRef]
- Hilliard, M.A.; Bergamasco, C.; Arbucci, S.; Plasterk, R.H.; Bazzicalupo, P. Worms taste bitter: ASH neurons, QUI-1, GPA-3 and ODR-3 mediate quinine avoidance in Caenorhabditis elegans. EMBO J. 2004, 23, 1101–1111. [Google Scholar] [CrossRef]
- Bargmann, C.I. Chemosensation in C. elegans. WormBook Online Rev. C. elegans Biol. 2006. [Google Scholar] [CrossRef]
- Hernandez, D.; Janmohamed, A.; Chandan, P.; Phillips, I.R.; Shephard, E.A. Organization and evolution of the flavin-containing monooxygenase genes of human and mouse: Identification of novel gene and pseudogene clusters. Pharmacogenetics 2004, 14, 117–130. [Google Scholar] [CrossRef]
- Overby, L.H.; Carver, G.C.; Philpot, R.M. Quantitation and kinetic properties of hepatic microsomal and recombinant flavin-containing monooxygenases 3 and 5 from humans. Chem. Biol. Interact. 1997, 106, 29–45. [Google Scholar] [CrossRef]
- Cherrington, N.J.; Cao, Y.; Cherrington, J.W.; Rose, R.L.; Hodgson, E. Physiological factors affecting protein expression of flavin-containing monooxygenases 1, 3 and 5. Xenobiotica 1998, 28, 673–682. [Google Scholar] [CrossRef] [PubMed]
- Shih, D.M.; Wang, Z.; Lee, R.; Meng, Y.; Che, N.; Charugundla, S.; Qi, H.; Wu, J.; Pan, C.; Brown, J.M.; et al. Flavin containing monooxygenase 3 exerts broad effects on glucose and lipid metabolism and atherosclerosis. J. Lipid Res. 2015, 56, 22–37. [Google Scholar] [CrossRef] [PubMed]
- Ashrafi, K.; Chang, F.Y.; Watts, J.L.; Fraser, A.G.; Kamath, R.S.; Ahringer, J.; Ruvkun, G. Genome-wide RNAi analysis of Caenorhabditis elegans fat regulatory genes. Nature 2003, 421, 268–272. [Google Scholar] [CrossRef] [PubMed]
- Richard, D.; Lin, Q.; Timofeeva, E. The corticotropin-releasing factor family of peptides and CRF receptors: Their roles in the regulation of energy balance. Eur. J. Pharmacol. 2002, 440, 189–197. [Google Scholar] [CrossRef]
- Alsebaaly, J.; Dugast, E.; Favot, L.; Khabbaz, L.; Solinas, M.; Thiriet, N. Persistent Neuroadaptations in the Expression of Genes Involved in Cholesterol Homeostasis Induced by Chronic, Voluntary Alcohol Intake in Rats. Front. Mol. Neurosci. 2018, 11, 457. [Google Scholar] [CrossRef]
- Cho, W.; Kang, J.L.; Park, Y.M. Corticotropin-Releasing Hormone (CRH) Promotes Macrophage Foam Cell Formation via Reduced Expression of ATP Binding Cassette Transporter-1 (ABCA1). PLoS ONE 2015, 10, e0130587. [Google Scholar] [CrossRef]
- Oram, J.F.; Vaughan, A.M. ATP-Binding cassette cholesterol transporters and cardiovascular disease. Circ. Res. 2006, 99, 1031–1043. [Google Scholar] [CrossRef] [PubMed]
- Klappe, K.; Hummel, I.; Hoekstra, D.; Kok, J.W. Lipid dependence of ABC transporter localization and function. Chem. Phys. Lipids 2009, 161, 57–64. [Google Scholar] [CrossRef]
- Landry, Y.D.; Denis, M.; Nandi, S.; Bell, S.; Vaughan, A.M.; Zha, X. ATP-binding cassette transporter A1 expression disrupts raft membrane microdomains through its ATPase-related functions. J. Biol. Chem. 2006, 281, 36091–36101. [Google Scholar] [CrossRef]
- Cutler, R.G.; Kelly, J.; Storie, K.; Pedersen, W.A.; Tammara, A.; Hatanpaa, K.; Troncoso, J.C.; Mattson, M.P. Involvement of oxidative stress-induced abnormalities in ceramide and cholesterol metabolism in brain aging and Alzheimer’s disease. Proc. Natl. Acad. Sci. USA 2004, 101, 2070–2075. [Google Scholar] [CrossRef]
- Zhou, H.; Liu, R. ER stress and hepatic lipid metabolism. Front. Genet. 2014, 5, 112. [Google Scholar] [CrossRef] [PubMed]
- Chuang, J.C.; Cui, H.; Mason, B.L.; Mahgoub, M.; Bookout, A.L.; Yu, H.G.; Perello, M.; Elmquist, J.K.; Repa, J.J.; Zigman, J.M.; et al. Chronic social defeat stress disrupts regulation of lipid synthesis. J. Lipid Res. 2010, 51, 1344–1353. [Google Scholar] [CrossRef] [PubMed]
- Brenner, S. The genetics of Caenorhabditis elegans. Genetics 1974, 77, 71–94. [Google Scholar] [CrossRef] [PubMed]
- Corsi, A.K.; Wightman, B.; Chalfie, M. A Transparent Window into Biology: A Primer on Caenorhabditis elegans. Genetics 2015, 200, 387–407. [Google Scholar] [CrossRef]
- Bandyopadhyay, J.; Lee, J.; Lee, J.; Lee, J.I.; Yu, J.-R.R.; Jee, C.; Cho, J.-H.H.; Jung, S.; Lee, M.H.; Zannoni, S.; et al. Calcineurin, a calcium/calmodulin-dependent protein phosphatase, is involved in movement, fertility, egg laying, and growth in Caenorhabditis elegans. Mol. Biol. Cell 2002, 13, 3281–3293. [Google Scholar] [CrossRef]
| Altered | Gene ID | Gene | FoldChange | FDR | p_Value | Description | Human Orthologue |
|---|---|---|---|---|---|---|---|
| Up | WBGene00001477 | fmo-2 | 20.97 | 0.05 | 0.04 | Flavin containing monooxygenase | FMO5 (ENSG00000131781), FMO3 (ENSG00000007933) |
| Down | WBGene00003995 | pgp-1 | 5.04 | 0.04 | 0.04 | ABC transporter family (ABCB) | ABCB1 (ENSG00000085563), ABCB11 (ENSG00000073734), ABCB4 (ENSG00000005471) |
| Down | WBGene00004002 | pgp-8 | 2.65 | 0.05 | 0.04 | ABCB1(ENSG00000085563), ABCB11 (ENSG00000073734), ABCB5 (ENSG00000004846) | |
| Down | WBGene00004003 | pgp-9 | 2.42 | 0.01 | 0.00 | ABCB1(ENSG00000085563), ABCB11 (ENSG00000073734), ABCB4 (ENSG00000005471) | |
| Down | WBGene00001817 | haf-7 | 2.05 | 0.01 | 0.00 | ABCB9 (ENSG00000150967), TAP1 (ENSG00000168394) | |
| Down * | WBGene00004004 | pgp-10 | 2.34 | 0.05 | 0.02 | ABCB1 (ENSG00000085563), ABCB4 (ENSG00000005471) | |
| Down * | WBGene00004005 | pgp-11 | 2.26 | 0.05 | 0.02 | ABCB1 (ENSG00000085563), ABCB11 (ENSG00000073734), ABCB5 (ENSG00000004846) | |
| Down * | WBGene00015479 | wht-1 | 1.60 | 0.05 | 0.03 | ABC transporter family (ABCG) | ABCG1 (ENSG00000160179), ABCG4 (ENSG00000172350) |
| Down * | WBGene00000023 | abt-5 | 2.53 | 0.05 | 0.01 | ABC transporter family (ABCA) | ABCA1 (ENSG00000085563), ABCA3 (ENSG00000167972), ABCA12 (ENSG00000144452), ABCA1 (ENSG00000179869) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jee, C.; Batsaikhan, E.; Salim, C. Neurobiological Basis of Aversion-Resistant Ethanol Seeking in C. elegans. Metabolites 2023, 13, 62. https://doi.org/10.3390/metabo13010062
Jee C, Batsaikhan E, Salim C. Neurobiological Basis of Aversion-Resistant Ethanol Seeking in C. elegans. Metabolites. 2023; 13(1):62. https://doi.org/10.3390/metabo13010062
Chicago/Turabian StyleJee, Changhoon, Enkhzul Batsaikhan, and Chinnu Salim. 2023. "Neurobiological Basis of Aversion-Resistant Ethanol Seeking in C. elegans" Metabolites 13, no. 1: 62. https://doi.org/10.3390/metabo13010062
APA StyleJee, C., Batsaikhan, E., & Salim, C. (2023). Neurobiological Basis of Aversion-Resistant Ethanol Seeking in C. elegans. Metabolites, 13(1), 62. https://doi.org/10.3390/metabo13010062






