Metabolite Signature of Simvastatin Treatment Involves Multiple Metabolic Pathways
Abstract
1. Introduction
2. Materials and Methods
2.1. Participants
2.2. Clinical and Laboratory Measurements
2.3. Calculations
2.4. Metabolomics Analysis
2.5. Statistical Analysis
3. Results
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Stamler, J.; Wentworth, D.; Neaton, J.D. Is the relationship between serum cholesterol and risk of premature death from coronary heart disease continuous and graded? Findings in 356,222 primary screenees of the Multiple Risk Factor Intervention Trial (MRFIT). JAMA 1986, 256, 2823–2828. [Google Scholar] [CrossRef] [PubMed]
- Castelli, W.P.; Garrison, R.J.; Wilson, P.W.; Abbott, R.D.; Kalousdian, S.; Kannel, W.B. Incidence of coronary heart disease and lipoprotein cholesterol levels: The Framingham Study. JAMA 1986, 256, 2835–2858. [Google Scholar] [CrossRef]
- Mackay, J.; Mensah, G.A.; Greenlund, K. The Atlas of Heart Disease and Stroke; World Health Organization: Geneva, Switzerland, 2004. [Google Scholar]
- Stone, N.J.; Robinson, J.G.; Lichtenstein, A.H.; Bairey Merz, C.N.; Blum, C.B.; Eckel, R.H.; Goldberg, A.C.; Gordon, D.G.; Levy, D.; Lloyd-Jones, D.M.; et al. ACC/AHA guideline on the treatment of blood cholesterol to reduce atherosclerotic cardiovascular risk in adults: A report of the American College of Cardiology/American Heart Association Task Force on Practice Guidelines. J. Am. Coll. Cardiol. 2014, 63, 2889–2934. [Google Scholar] [CrossRef] [PubMed]
- Collins, R.; Reith, C.; Emberson, J.; Armitage, J.; Baigent, C.; Blackwell, L.; Blumenthal, R.; Danesh, J.; Smith, G.D.; DeMets, D.; et al. Interpretation of the evidence for the efficacy and safety of statin therapy. Lancet 2016, 388, 2532–2561. [Google Scholar] [CrossRef]
- Statins for millions more? Lancet 2014, 383, 669. [CrossRef]
- Cederberg, H.; Stančáková, A.; Yaluri, N.; Modi, S.; Kuusisto, J.; Laakso, M. Increased risk of diabetes with statin treatment is associated with impaired insulin sensitivity and insulin secretion: A 6 year follow-up study of the METSIM cohort. Diabetologia 2015, 58, 1109–1117. [Google Scholar] [CrossRef] [PubMed]
- Swerdlow, D.I.; Preiss, D.; Kuchenbaecker, K.B.; Holmes, M.V.; Engmann, J.E.L.; Shah, T.; Sofat, R.; Stender, S.; Johnson, P.C.D.; Robert, A.; et al. HMG-coenzyme A reductase inhibition, type 2 diabetes, and bodyweight: Evidence from genetic analysis and randomised trials. Lancet 2015, 385, 351–361. [Google Scholar] [CrossRef]
- Liu, G.; Shi, M.; Mosley, J.D.; Weng, C.; Zhang, Y.; Ta, M.; Lee, M.; Jarvik, G.P.; Hakonarson, H.; Namjou-Khaleset, B.; et al. A Mendelian Randomization approach using 3-HMG-coenzyme-A reductase gene variation to evaluate the association of statin-induced low-density lipoprotein cholesterol lowering with noncardiovascular disease phenotypes. JAMA Netw. Open 2021, 4, e2112820. [Google Scholar] [CrossRef]
- Jiang, J.-I.; Jiang, D.-J.; Tang, Y.-H.; Li, N.-S.; Deng, H.-W.; Li, Y.J. Effect of simvastatin on endothelium-dependent vaso-relaxation and endogenous nitric oxide synthase inhibitor. Acta Pharmacol. Sin. 2004, 25, 893–901. [Google Scholar]
- Giurgea, A.G.; Margeta, C.; Maca, T.; Rezaie-Majd, A.; Bucek, R.A.; Manavi, M.; Afarideh, R.; Minar, E.; Baghestanian, M. Simvastatin reduces serum level of vascular endothelial growth factor in hypercholesterolemic patients. J. Cardiovasc. Pharmacol. 2006, 47, 30–36. [Google Scholar] [CrossRef]
- Ludwig, S.; Dharmalingam, S.; Erickson-Nesmith, S.; Ren, S.; Zhu, F.; Ma, G.M.; Zhao, R.; Fenton, J.W.; Ofosu, F.A.; Velthuis, H.T.; et al. Impact of simvastatin on hemostatic andfibrinolytic regulators in type 2 diabetes mellitus. Diabetes Res. Clin. Pract. 2005, 70, 110–118. [Google Scholar] [CrossRef] [PubMed]
- Shiomi, M.; Yamada, S.; Ito, T. Atheroma stabilizing effects of simvastatin due to depression of macrophages or lipid accumulation in the atheromatous plaques of coronary plaque-prone WHHL rabbits. Aherosclerosis 1998, 101, 2711–2719. [Google Scholar] [CrossRef] [PubMed]
- Hernández-Perera, O.; Pérez-Sala, D.; Navarro-Antolín, J.; Sánchez-Pascuala, R.; Hernández, G.; Díaz, C.; Lamas, L. Effects of the 3-hydroxy-3-methylglutaryl-CoA reductase inhibitors, atorvastatin and simvastatin, on the expression of endothelin-1 and endothelial nitric oxide synthase in vascular endothelial cells. J. Clin. Investig. 2005, 178, 287–294. [Google Scholar] [CrossRef] [PubMed]
- Kanda, H.; Yokota, K.; Kohno, C.; Sawada, T.; Sato, K.; Yamaguchi, M.; Komagata, Y.; Shimada, K.; Yamamoto, K.; Mimura, T. Effects of low-dosage simvastatin on rheumatoid arthritis through reduction of Th1/Th2 and CD4/CD8 ratios. Mod. Rheumatol. 2007, 17, 364–368. [Google Scholar] [CrossRef]
- Kaddurah-Daouk, R.; Krishnan, K.R. Metabolomics: A global biochemical approach to the study of central nervous system diseases. Neuropsychopharmacology 2009, 34, 173–186. [Google Scholar] [CrossRef]
- Kaddurah-Daouk, R.; Kristal, B.S.; Weinshilboum, R.M. Metabolomics: A global biochemical approach to drug response and disease. Annu. Rev. Pharmacol. Toxicol. 2008, 48, 653–683. [Google Scholar] [CrossRef]
- Würtz, P.; Wang, Q.; Soininen, P.; Kangas, A.J.; Fatemifar, G.; Tynkkynen, T.; Tiainen, M.; Perola, M.; Tillin, T.; Hughes, A.D.; et al. Metabolomic Profiling of Statin Use and Genetic Inhibition of HMG-CoA Reductase. J. Am. Coll. Cardiol. 2016, 67, 1200–1210. [Google Scholar] [CrossRef]
- Nagana Gowda, G.A.; Djukovic, D. Overview of mass spectrometry-based metabolomics: Opportunities and challenges. Methods Mol. Biol. 2014, 1198, 3–12. [Google Scholar]
- Laakso, M.; Kuusisto, J.; Stančáková, A.; Kuulasmaa, T.; Pajukanta, P.; Lusis, A.J.; Collins, F.S.; Mohlke, K.L.; Boehnke, M. The Metabolic Syndrome in Men study: A resource for studies of metabolic and cardiovascular diseases. J. Lipid Res. 2017, 58, 481–493. [Google Scholar] [CrossRef]
- Stancáková, A.; Javorský, M.; Kuulasmaa, T.; Haffner, S.M.; Kuusisto, J.; Laakso, M. Changes in insulin sensitivity and insulin release in relation to glycemia and glucose tolerance in 6,414 Finnish men. Diabetes 2009, 58, 1212–1221. [Google Scholar] [CrossRef]
- American Diabetes Association. Diagnosis and classification of diabetes mellitus. Diabetes Care 2013, 36, 67–74. [Google Scholar] [CrossRef] [PubMed]
- Fernandes Silva, L.; Vangipurapu, J.; Kuulasmaa, T.; Laakso, M. An intronic variant in the GCKR gene is associated with multiple lipids. Sci. Rep. 2019, 9, 10240. [Google Scholar] [CrossRef] [PubMed]
- Trupp, M.; Zhu, H.; Wikoff, W.R.; Baillie, R.A.; Zeng, Z.B.; Karp, P.D.; Fiehn, O.; Krauss, R.M.; Kaddurah-Daouket, R. Metabolomics reveals amino acids contribute to variation in response to simvastatin treatment. PLoS ONE 2012, 7, e38386. [Google Scholar] [CrossRef] [PubMed]
- Scandinavian Simvastatin Survival Study Group. Randomized trial of cholesterol lowering in 4444 patients with coronary heart disease: The Scandinavian Simvastatin Survival Study (4S). Lancet 1994, 344, 1383–1389. [Google Scholar]
- Maltese, W.A.; Aprille, J.R. Relation of mevalonate synthesis to mitochondrial ubiquinone content and respiratory function in cultured neuroblastoma cells. J. Biol. Chem. 1985, 260, 11524–11529. [Google Scholar] [CrossRef]
- Su, B.; Ryan, R.O. Metabolic biology of 3-methylglutaconic acid-uria: A new perspective. J. Inherit. Metab. Dis. 2014, 37, 359–368. [Google Scholar] [CrossRef]
- Loria, P.; Bertolotti, M.; Cassinadri, M.T.; Dilengite, M.A.; Bozzoli, M.; Carubbi, F.; Concari, M.; Guicciardi, M.E.; Carulli, N. Short-term effects of simvastatin on bile acid synthesis and bile lipid secretion in human subjects. Hepatology 1994, 19, 882–888. [Google Scholar] [CrossRef]
- Schooling, C.M.; Au Yeung, S.L.; Freeman, G.; Cowling, B.J. The effect of statins on testosterone in men and women, a systematic review and meta-analysis of randomized controlled trials. BMC Med. 2013, 11, 57. [Google Scholar] [CrossRef]
- Phillips, P.S.; Haas, R.H.; Bannykh, S.; Hathaway, S.; Gray, N.L.; Kimura, B.J.; Vladutiu, G.D.; England, J.D.F. Statin-associated myopathy with normal creatine kinase levels. Ann. Intern. Med. 2002, 137, 581–585. [Google Scholar] [CrossRef]
- Leipnitz, G.; Seminotti, B.; Amaral, A.U.; de Bortoli, G.; Solano, A.; Schuck, P.F.; Wyse, A.T.; Wannmacher, C.M.; Latini, A.; Wajner, M. Induction of oxidative stress by the metabolites accumulating in 3-methylglutaconic aciduria in cerebral cortex of young rats. Life Sci. 2008, 82, 652–662. [Google Scholar] [CrossRef]
- Newgard, C.B.; An, J.; Bain, J.R.; Muehlbauer, M.J.; Stevens, R.D.; Lien, L.F.; Haqq, A.M.; Shah, S.H.; Arlotto, M.; Slentz, C.A.; et al. A branched-chain amino acid-related metabolic signature that differentiates obese and lean humans and contributes to insulin resistance. Cell Metab. 2009, 9, 311–326. [Google Scholar] [CrossRef] [PubMed]
- Mihalik, S.J.; Goodpaster, B.H.; Kelley, D.E.; Chace, D.H.; Vockley, J.; Toledo, F.G.S.; DeLanye, J.P. Increased levels of plasma acylcarnitines in obesity and type 2 diabetes and identification of a marker of glucolipotoxicity. Obesity 2010, 18, 1695–1700. [Google Scholar] [CrossRef] [PubMed]
- Laakso, M.; Kuusisto, J. Diabetes secondary to treatment with statins. Curr. Diabetes Rep. 2017, 17, 10. [Google Scholar] [CrossRef] [PubMed]
- Waterman, I.J.; Zammit, V.A. Differential effects of fenofibrate or simvastatin treatment of rats on hepatic microsomal overt and latent diacylglycerol acyltransferase activities. Diabetes 2002, 51, 1708–1713. [Google Scholar] [CrossRef][Green Version]
- Trub, A.G.; Wagner, G.R.; Anderson, K.A.; Crown, S.B.; Zhang, G.-F.; Thompson, J.W.; Ilkayeva, O.R.; Stevens, R.D.; Grimsrud, P.A.; Kulkarni, R.A.; et al. Statin therapy inhibits fatty acid synthase via dynamic protein modifications. Nat. Commun. 2022, 13, 2542. [Google Scholar] [CrossRef]
- Gbelcová, H.; Svéda, M.; Laubertová, L.; Varga, I.; Vítek, L.; Kolář, M.; Strnad, H.; Zelenka, J.; Böhmer, D.; Ruml, T. The effect of simvastatin on lipid droplets accumulation in human embryonic kidney cells and pancreatic cancer cells. Lipids Health Dis. 2013, 12, 126. [Google Scholar] [CrossRef]
- Watkins, P.A. Fatty acyl-CoA synthetases. In Encyclopedia of Biological Chemistry, 2nd ed.; Lennarz, W.J., M. Lane, M.D., Eds.; Academic Press: Cambridge, MA, USA, 2013; pp. 290–295. [Google Scholar]
- Grevengoed, T.J.; Klett, E.L.; Coleman, R.A. Acyl-CoA metabolism and partitioning. Annu. Rev. Nutr. 2014, 34, 1–30. [Google Scholar] [CrossRef]
- Yanagita, T.; Yamamoto, K.; Ishida, S.; Sonda, K.; Morito, F.; Saku, K.; T Sakaie, T. Effects of simvastatin, a cholesterol synthesis inhibitor, on phosphatidylcholine synthesis in HepG2 cells. Clin. Ther. 1994, 16, 200–208. [Google Scholar]
- Li, L.O.; Klett, E.L.; Coleman, R.A. Acyl-CoA synthesis, lipid metabolism and lipotoxicity. Biochim. Biophys. Acta (BBA)-Mol. Cell Biol. Lipids 2010, 1801, 246–251. [Google Scholar] [CrossRef]
- Momin, A.A.; Park, H.; Portz, B.J.; Haynes, C.A.; Shaner, R.L.; Kelly, S.L.; Jordan, I.K.; Merrill, J. A method for visualization of “omic” datasets for sphingolipid metabolism to predict potentially interesting differences. J. Lipid Res. 2011, 52, 1073–1083. [Google Scholar] [CrossRef]
- Wilken, D.R.; McMacken, M.L.; Rodriquez, A. Choline and betaine aldehyde oxidation by rat liver mitochondria. Biochim. Biophys. Acta 1970, 216, 305–317. [Google Scholar] [CrossRef]
- Friesen, R.W.; Novak, E.M.; Hasman, D.; Innis, S.M. Relationship of dimethylglycine, choline, and betaine with oxoproline in plasma of pregnant women and their newborn infants. J. Nutr. 2007, 37, 2641–2646. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Goto, S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef] [PubMed]
- Priolo, C.; Khabibullin, D.; Reznik, E.; Filippakis, H.; Ogórek, B.; Kavanagh, T.R.; Nijmeh, J.; Herbert, Z.T.; Asara, J.M.; Kwiatkowski, D.J.; et al. Impairment of gamma-glutamyl transferase 1 activity in the metabolic pathogenesis of chromophobe renal cell carcinoma. Proc. Natl. Acad. Sci. USA 2018, 115, E6274–E6282. [Google Scholar] [CrossRef] [PubMed]
- Griffith, O.W.; Bridges, R.J.; Meister, A. Evidence that the gamma-glutamyl cycle functions in vivo using intracellular glutathione: Effects of amino acids and selective inhibition of enzymes. Proc. Natl. Acad. Sci. USA 1978, 75, 5405–5408. [Google Scholar] [CrossRef] [PubMed]
- Wishart, D.S.; Feunang, Y.D.; Marcu, A.; Guo, A.C.; Liang, K.; Vázquez-Fresno, R.; Sajed, T.; Johnson, D.; Li, C.; Karu, N.; et al. HMDB 4.0: The human metabolome database for 2018. Nucleic Acids Res. 2018, 46, D608–D617. [Google Scholar] [CrossRef]
- Majumdar, R.; Yori, A.; Rush, P.W.; Raymond, K.; Gavrilov, D.; Tortorelli, S.; Matern, D.; Rinaldo, P.; Feldman, G.L.; Oglesbee, D. Allelic spectrum of formiminotransferase-cyclodeaminase gene variants in individuals with formiminoglutamic aciduria. Mol. Genet. Genom. Med. 2017, 5, 795–799. [Google Scholar] [CrossRef] [PubMed]
- Grace, H.; McGregor, A.D.; Campbell, S.K.; Fey, S.K.; Tumanov, F.S.; Sumpton, D.; Blanco, G.R.; Mackay, G.; Nixon, C.; Vazquez, A.; et al. Targeting the metabolic response to statin-mediated oxidative stress produces a synergistic antitumor response. Cancer Res. 2020, 80, 175–188. [Google Scholar]
- Liang, X.; Zhang, L.; Natarajan, S.K.; Becker, D.F. Proline mechanisms of stress survival. Antioxid. Redox Signal. 2013, 19, 998–1011. [Google Scholar] [CrossRef]
- Smirnoff, N.; Cumbes, Q.J. Hydroxyl radical scavenging activity of compatible solutes. Phytochemistry 1989, 28, 1057–1060. [Google Scholar] [CrossRef]
- Mor, A.; Tankiewicz-Kwedlo, A.; Krupa, A.; Pawlak, D. Role of kynurenine pathway in oxidative stress during neurodegenerative disorders. Cells 2021, 10, 1603. [Google Scholar] [CrossRef] [PubMed]
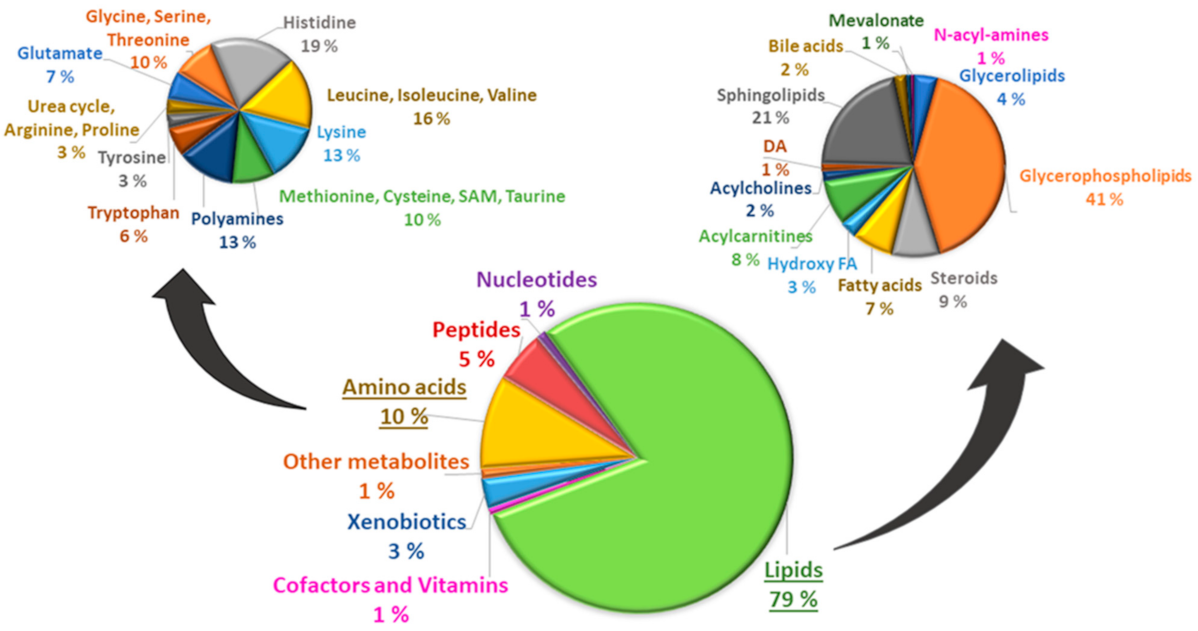
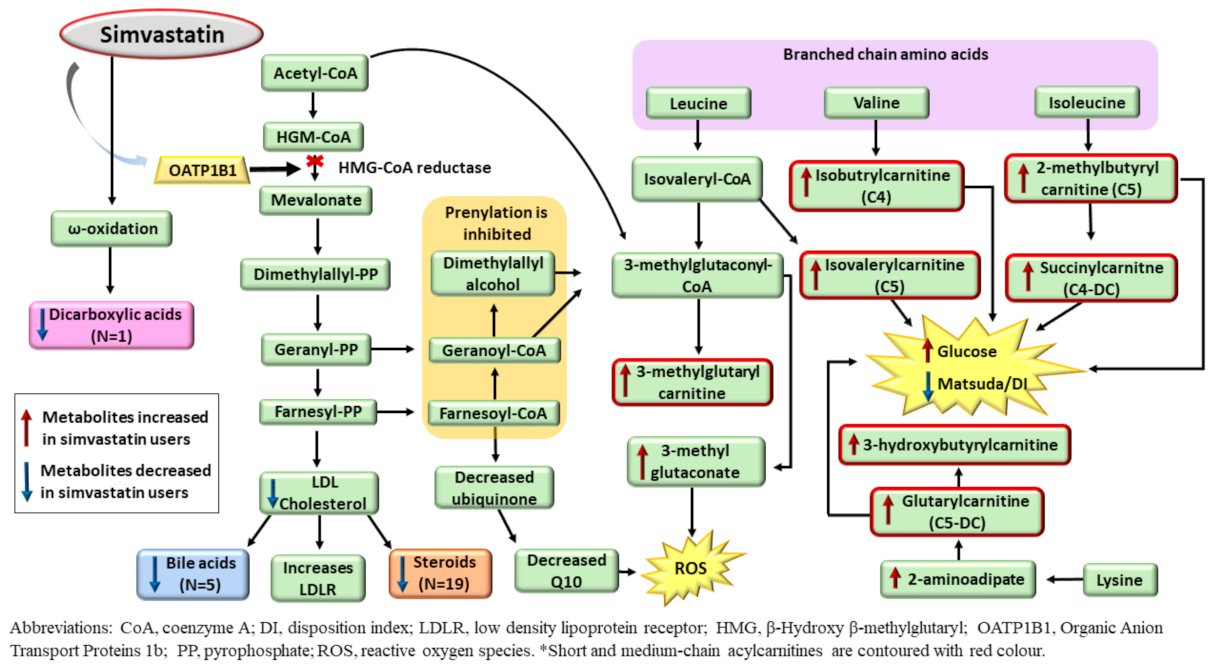
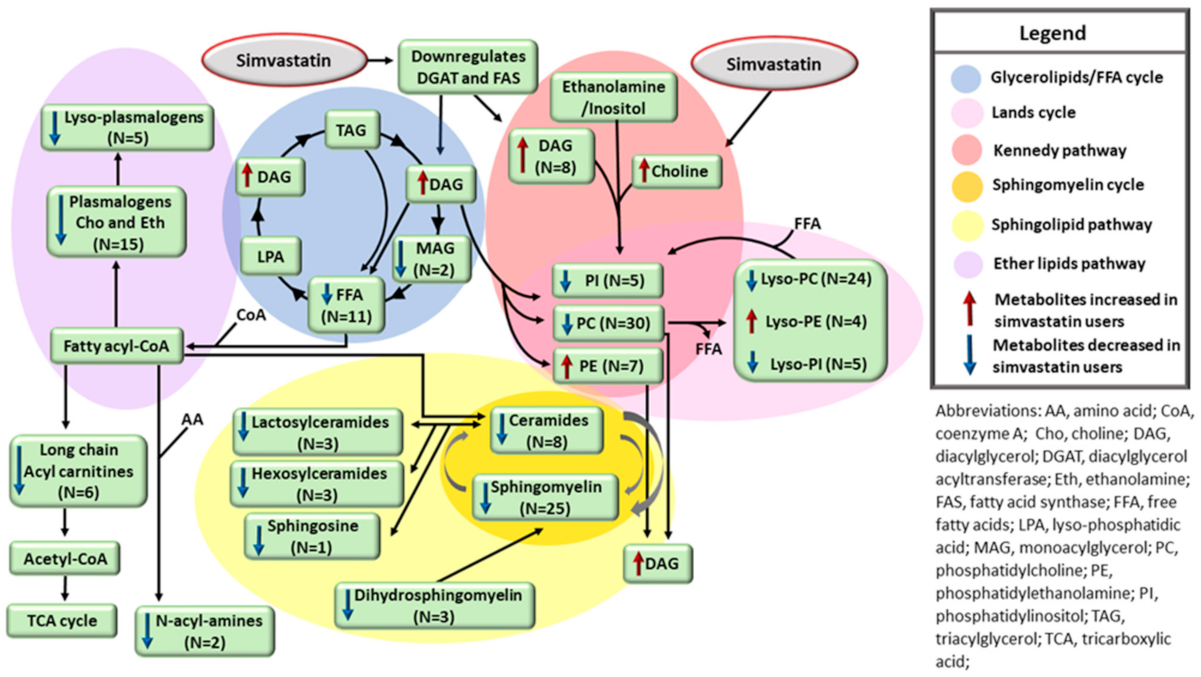
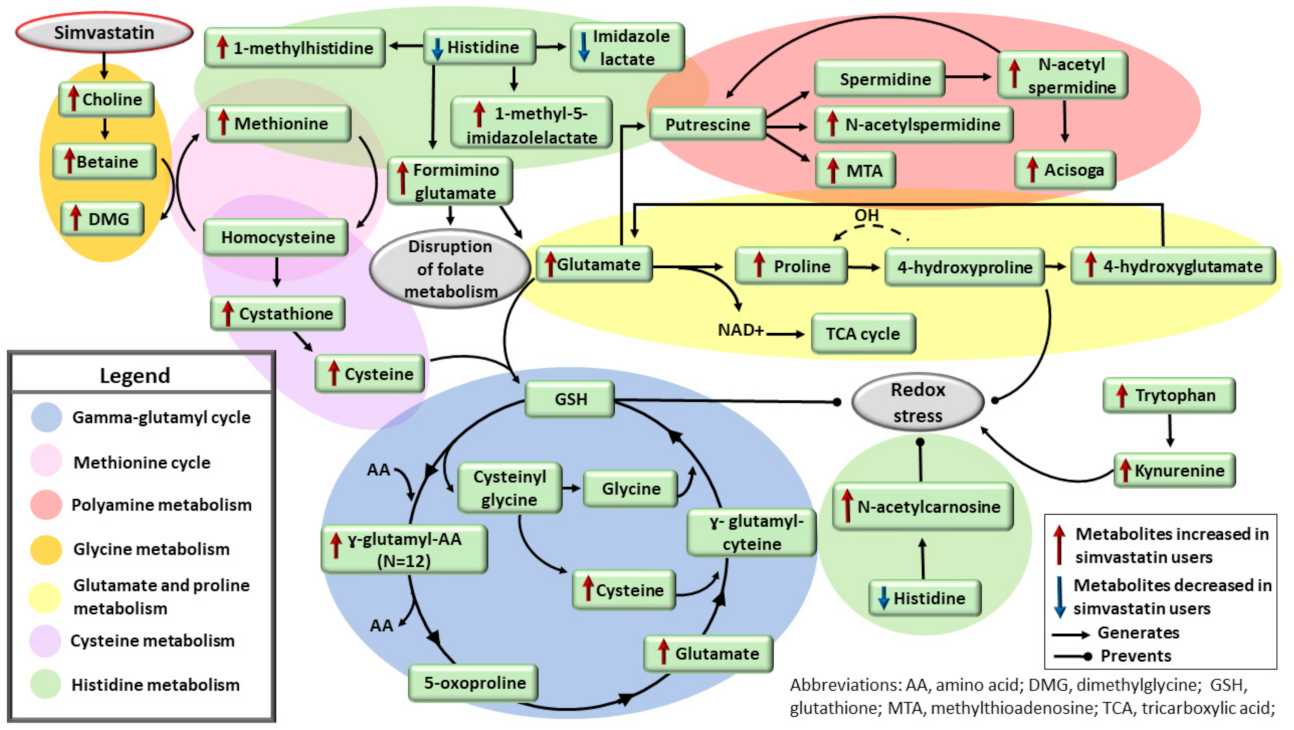
| Clinical and Laboratory Characteristics | Participants on Simvastatin Treatment * | Participants Not on Simvastatin Treatment * | p Value ** | ||||
|---|---|---|---|---|---|---|---|
| n | Mean | SD | n | Mean | SD | ||
| Age | 1332 | 59.74 | 7,08 | 6200 | 56.62 | 6.92 | <0.001 |
| Body mass index | 1331 | 27.43 | 3.96 | 6198 | 26.60 | 3.74 | <0.001 |
| Waist (cm) | 1331 | 99.11 | 10.85 | 6197 | 96.84 | 10.55 | <0.001 |
| Systolic blood pressure | 1332 | 137.88 | 15.82 | 6200 | 136.81 | 16.28 | NS |
| Fasting plasma glucose (mmol/L) | 1332 | 6.42 | 1.74 | 6200 | 5.93 | 1.65 | <0.001 |
| 2 h plasma glucose (mmol/L) | 1332 | 5.79 | 0.48 | 6200 | 5.69 | 0.48 | <0.001 |
| Fasting plasma insulin (mU/L) | 1331 | 9.53 | 6.93 | 6197 | 7.82 | 5.52 | <0.001 |
| Matsuda ISI (mg/dl, mU/L) | 1320 | 5.71 | 3.44 | 6167 | 7.33 | 4.29 | <0.001 |
| Disposition index | 1320 | 153.22 | 65.16 | 6167 | 166.90 | 73.62 | <0.001 |
| LDL cholesterol (mmol/L) | 1332 | 2.71 | 0.71 | 6197 | 3.57 | 0.82 | <0.001 |
| Total triglycerides (mmol/L) | 1332 | 1.41 | 0.72 | 6200 | 1.38 | 1.00 | NS |
| Smokers % | 13.0% | 16.6% | - | NS | |||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fernandes Silva, L.; Ravi, R.; Vangipurapu, J.; Laakso, M. Metabolite Signature of Simvastatin Treatment Involves Multiple Metabolic Pathways. Metabolites 2022, 12, 753. https://doi.org/10.3390/metabo12080753
Fernandes Silva L, Ravi R, Vangipurapu J, Laakso M. Metabolite Signature of Simvastatin Treatment Involves Multiple Metabolic Pathways. Metabolites. 2022; 12(8):753. https://doi.org/10.3390/metabo12080753
Chicago/Turabian StyleFernandes Silva, Lilian, Rowmika Ravi, Jagadish Vangipurapu, and Markku Laakso. 2022. "Metabolite Signature of Simvastatin Treatment Involves Multiple Metabolic Pathways" Metabolites 12, no. 8: 753. https://doi.org/10.3390/metabo12080753
APA StyleFernandes Silva, L., Ravi, R., Vangipurapu, J., & Laakso, M. (2022). Metabolite Signature of Simvastatin Treatment Involves Multiple Metabolic Pathways. Metabolites, 12(8), 753. https://doi.org/10.3390/metabo12080753







