Analyzing and Resolving Infeasibility in Flux Balance Analysis of Metabolic Networks
Abstract
:1. Introduction
2. Preliminaries
2.1. Definitions
2.2. Classical MFA
2.3. Conservation Relations
2.4. Correcting Measured Rates to Make Inconsistent Systems Feasible
3. Results
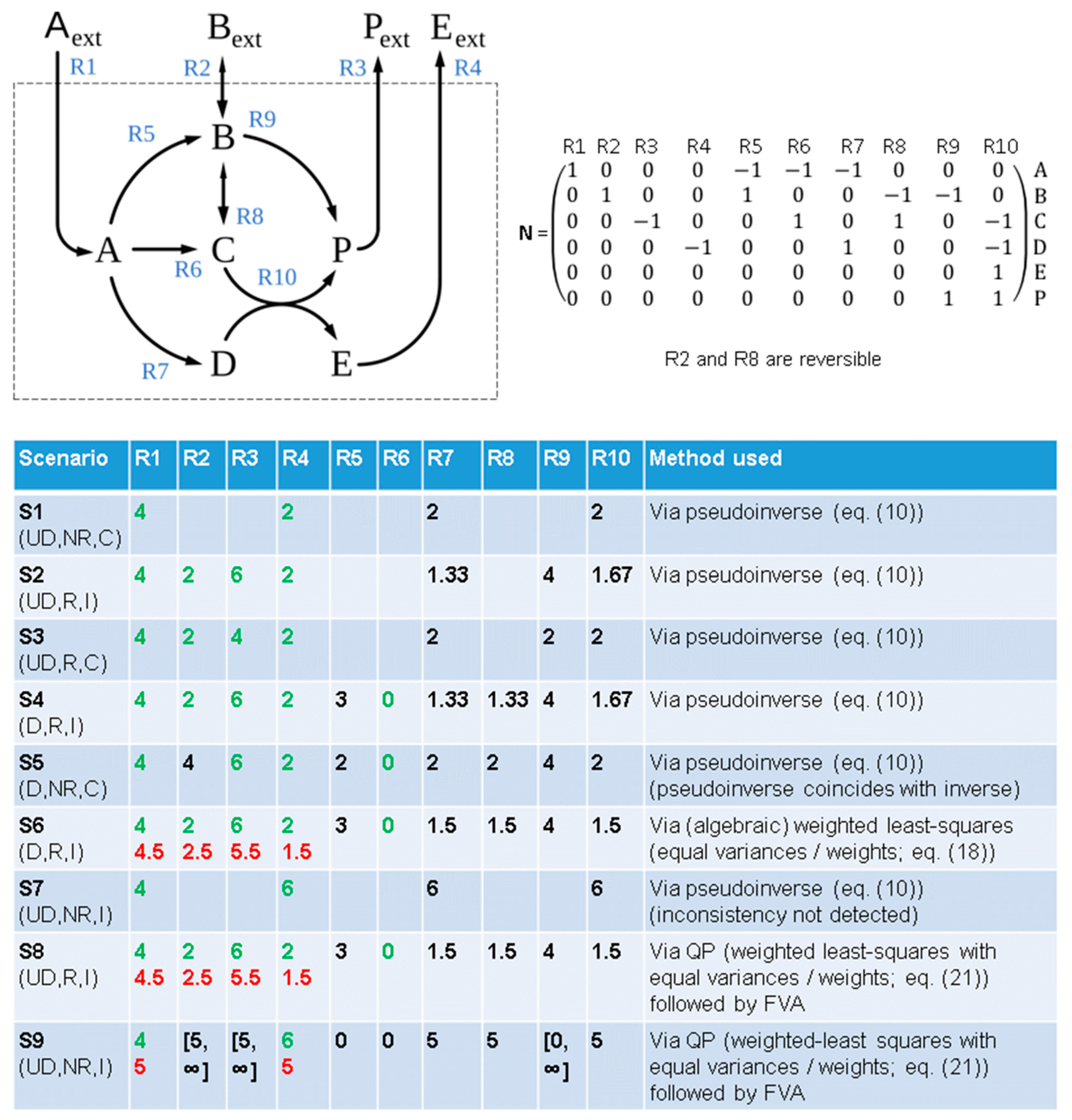
3.1. Infeasibility in FBA Scenarios with Known Fluxes
3.2. Weighted Least-Squares Solution via Quadratic Optimization
- W1: If the variances of measurements ()are available, these could be used for the weights: .
- W2: If the measurements variances are unavailable, one could set so that a deviation from a flux of larger magnitude weighs less than the same deviation from a flux of lower magnitude. This assumes that the measurement noise is correlated with the magnitude of the measured value.
- W3: As the simplest approach, one could choose equal weights for all corrections: .
3.3. Using Linear Optimization to Correct Flux Measurements in Infeasible FBA Systems
3.4. Allowing Minimal Corrections of Other Constraints to Make Infeasible FBA Systems Feasible
3.5. A Practical Guide for How to Proceed with Infeasible FBA Problems
- (1)
- Detect (in)feasibility: infeasibility of a metabolic flux (balance) scenario can or will be detected by an associated error message of the LP solver when performing an FBA optimization or an FVA. If this infeasibility is not a consequence of fixing some reaction rates, i.e., if the base system (1)–(3) is already infeasible, use the methods explained in the previous section for resolving inconsistencies in these constraints.
- (2)
- If not yet performed, identify all reactions with a fixed rate (e.g., by searching for reactions in the model where lower and upper bound are identical).
- (3)
- It is recommended to check via Equation (15) whether there are algebraic redundancies in the defined scenario (solely related to the steady-state constraint) and, if so, to find out which of the fixed reaction rates induce this redundancy (via the redundancy matrix (12)). This can be very helpful to find possible sources of mutually inconsistent rates (but it does not provide a (complete) explanation if constraints (2) and (3) are also involved in the inconsistency).
- (4)
- Decide on which optimization approach (QP (21) or LP (24)) and which of the three weighting schemes, W1–W3, are to be applied. Recommendation: if the measurements variances are known and if a suitable QP solver is available, then use a QP with ; otherwise, choose the LP approach with . In the latter case, the may be manually (re)adjusted to increase knowledge about the (un)certainty of the fixed rates. Generally, large weights ( > 1000) should be used for rates fixed at zero (.
- (5)
- Compute the corrections with the respective optimization approach and analyze them to identify the given rates that were assigned the largest changes and have thus caused the largest inconsistencies.
- (6)
- Apply the corrections and compute with the balanced (now feasible) system the solutions for the original FBA/FVA problem. In particular, FVA can be used to identify unknown rates that are uniquely determined from the measured rates (FVA delivers identical lower and upper bounds for those rates). Generally, when performing FBA or FVA in the corrected system, one may face numerical problems: the precision of the calculated (and applied) corrections might not be sufficient enough for the solver used in the subsequent FBA/FVA optimizations, again resulting in an infeasibility message of the solver. If this happens to be the case, it is advised to use a slightly lower precision for the subsequent optimizations. In the example applications described in Section 3.7, we did not encounter such a problem.
3.6. Implementation in CellNetAnalyzer and CNApy
3.7. Relevant Examples from Core and Genome-Scale Models of Escherichia coli
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bordbar, A.; Monk, J.M.; King, Z.A.; Palsson, B.O. Constraint-based models predict metabolic and associated cellular functions. Nat. Rev. Genet. 2014, 15, 107–120. [Google Scholar] [CrossRef]
- Lewis, N.E.; Nagarajan, H.; Palsson, B.O. Constraining the metabolic genotype–phenotype relationship using a phylogeny of in silico methods. Nat. Rev. Microbiol. 2012, 10, 291–305. [Google Scholar] [CrossRef] [Green Version]
- Raman, K.; Chandra, N. Flux balance analysis of biological systems: Applications and challenges. Brief. Bioinform. 2009, 10, 435–449. [Google Scholar] [CrossRef]
- Orth, J.D.; Thiele, I.; Palsson, B.Ø. What is flux balance analysis? Nat. Biotechnol. 2010, 28, 245–248. [Google Scholar] [CrossRef]
- Aiba, S.; Matsuoka, M. Identification of metabolic model: Citrate production from glucose by Candida lipolytica. Biotechnol. Bioeng. 1979, 21, 1373–1386. [Google Scholar] [CrossRef]
- Vallino, J.J.; Stephanopoulos, G. Metabolic flux distributions in Corynebacterium glutamicum during growth and lysine overproduction. Biotechnol. Bioeng. 1993, 41, 633–646. [Google Scholar] [CrossRef]
- Stephanopoulos, G.N.; Aristidou, A.A.; Nielsen, J. Metabolic Engineering: Principles and Methodology; Academic Press: SanDiego, CA, USA, 1993. [Google Scholar]
- Antoniewicz, M.R. A guide to metabolic flux analysis in metabolic engineering: Methods, tools and applications. Meta. Eng. 2021, 63, 2–12. [Google Scholar] [CrossRef]
- van der Heijden, R.T.; Heijnen, J.J.; Hellinga, C.; Romein, B.; Luyben, K.C. Linear constraint relations in biochemical reaction systems: I. Classification of the calculability and the balanceability of conversion rates. Biotechnol. Bioeng. 1994, 43, 3–10. [Google Scholar] [CrossRef]
- van der Heijden, R.T.; Romein, B.; Heijnen, J.J.; Hellinga, C.; Luyben, K.C. Linear constraint relations in biochemical reaction systems: II. Diagnosis and estimation of gross errors. Biotechnol. Bioeng. 1994, 43, 11–20. [Google Scholar] [CrossRef]
- Klamt, S.; Schuster, S.; Gilles, E.D. Calculability analysis in underdetermined metabolic networks illustrated by a model of the central metabolism in purple nonsulfur bacteria. Biotechnol. Bioeng. 2002, 77, 734–751. [Google Scholar] [CrossRef]
- Sánchez, B.J.; Zhang, C.; Nilsson, A.; Lahtvee, P.J.; Kerkhoven, E.J.; Nielsen, J. Improving the phenotype predictions of a yeast genome-scale metabolic model by incorporating enzymatic constraints. Mol. Syst. Bio. 2017, 13, 935. [Google Scholar] [CrossRef]
- Bekiaris, P.S.; Klamt, S. Automatic construction of metabolic models with enzyme constraints. BMC Bioinform. 2020, 21, 19. [Google Scholar] [CrossRef]
- Strang, G. Linear Algebra and Its Applications; Academic Press: New York, NY, USA, 1980. [Google Scholar]
- Boyd, S.; Vandenberghe, L. Convex Optimization; Cambridge University Press: Cambridge, UK, 2004. [Google Scholar]
- Mahadevan, R.; Schilling, C.H. The effects of alternate optimal solutions in constraint-based genome-scale metabolic models. Metab. Eng. 2003, 5, 264–276. [Google Scholar] [CrossRef]
- Strutz, T. Data Fitting and Uncertainty—A Practical Introduction to Weighted Least Squares and Beyond; Springer Vieweg: Wiesbaden, Germany, 2016. [Google Scholar]
- Klamt, S.; Saez-Rodriguez, J.; Gilles, E.D. Structural and functional analysis of cellular networks with CellNetAnalyzer. BMC Syst. Biol. 2008, 1, 2. [Google Scholar] [CrossRef] [Green Version]
- von Kamp, A.; Thiele, S.; Hädicke, O.; Klamt, S. Use of CellNetAnalyzer in biotechnology and metabolic engineering. J. Biotechnol. 2017, 261, 221–228. [Google Scholar] [CrossRef]
- Thiele, S.; von Kamp, A.; Bekiaris, P.S.; Schneider, P.; Klamt, S. CNApy: A CellNetAnalyzer GUI in Python for analyzing and designing metabolic networks. Bioinformatics 2022, 38, 1467–1469. [Google Scholar] [CrossRef]
- Hädicke, O.; Klamt, S. EColiCore2: A reference model of the central metabolism of Escherichia coli and the relationships to its genome-scale parent model. Sci. Rep. 2017, 7, 39647. [Google Scholar] [CrossRef] [Green Version]
- Boecker, S.; Slaviero, G.; Schramm, T.; Szymanski, W.; Steuer, R.; Link, H.; Klamt, S. Deciphering the physiological response of Escherichia coli under high ATP demand. Mol. Syst. Biol. 2021, 17, e10504. [Google Scholar] [CrossRef]
- Orth, J.D.; Conrad, T.M.; Na, J.; Lerman, J.A.; Nam, H.; Feist, A.M.; Palsson, B.O. A comprehensive genome-scale reconstruction of Escherichia coli metabolism. Mol. Syst. Biol. 2011, 7, 535. [Google Scholar] [CrossRef]
- Abbate, T.; Dewasme, L.; Wouwer, A.V.; Bogaerts, P. Adaptive flux variability analysis of HEK cell cultures. Comput. Chem. Eng. 2020, 133, 106633. [Google Scholar] [CrossRef]
- Segrè, D.; Vitkup, D.; Church, G.M. Analysis of optimality in natural and perturbed metabolic networks. Proc. Natl. Acad. Sci. USA 2002, 99, 15112–15117. [Google Scholar] [CrossRef] [Green Version]
- Gevorgyan, A.; Poolman, M.G.; Fell, D.A. Detection of stoichiometric inconsistencies in biomolecular models. Bioinformatics 2008, 24, 2245–2251. [Google Scholar] [CrossRef] [Green Version]
- Murty, K.G.; Kabadi, S.N.; Chandrasekaran, R. Infeasibility analysis for linear systems, a survey. Arab. J. Sci. Technol. 2000, 25, 3–18. [Google Scholar]
- COBRA Toolbox Function Solving the Cardinality Optimization Problem. Available online: https://github.com/opencobra/cobratoolbox/blob/master/src/base/solvers/cardOpt/sparseLP/optimizeCardinality.m (accessed on 15 June 2022).
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Klamt, S.; von Kamp, A. Analyzing and Resolving Infeasibility in Flux Balance Analysis of Metabolic Networks. Metabolites 2022, 12, 585. https://doi.org/10.3390/metabo12070585
Klamt S, von Kamp A. Analyzing and Resolving Infeasibility in Flux Balance Analysis of Metabolic Networks. Metabolites. 2022; 12(7):585. https://doi.org/10.3390/metabo12070585
Chicago/Turabian StyleKlamt, Steffen, and Axel von Kamp. 2022. "Analyzing and Resolving Infeasibility in Flux Balance Analysis of Metabolic Networks" Metabolites 12, no. 7: 585. https://doi.org/10.3390/metabo12070585
APA StyleKlamt, S., & von Kamp, A. (2022). Analyzing and Resolving Infeasibility in Flux Balance Analysis of Metabolic Networks. Metabolites, 12(7), 585. https://doi.org/10.3390/metabo12070585






