Screening of Candidate Bioactive Secondary Plant Metabolite Ion-Features from Moringa oleifera Accessions Associated with High and Low Enteric Methane Inhibition from Ruminants
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials and Preparation of Crude Extracts
2.2. Determination of In Vitro Total Gas, Methane, and Organic Matter Digestibility
2.3. UPLC-MS Data Analysis
2.4. Statistical Analysis
3. Results
3.1. In Vitro Methane Inhibition
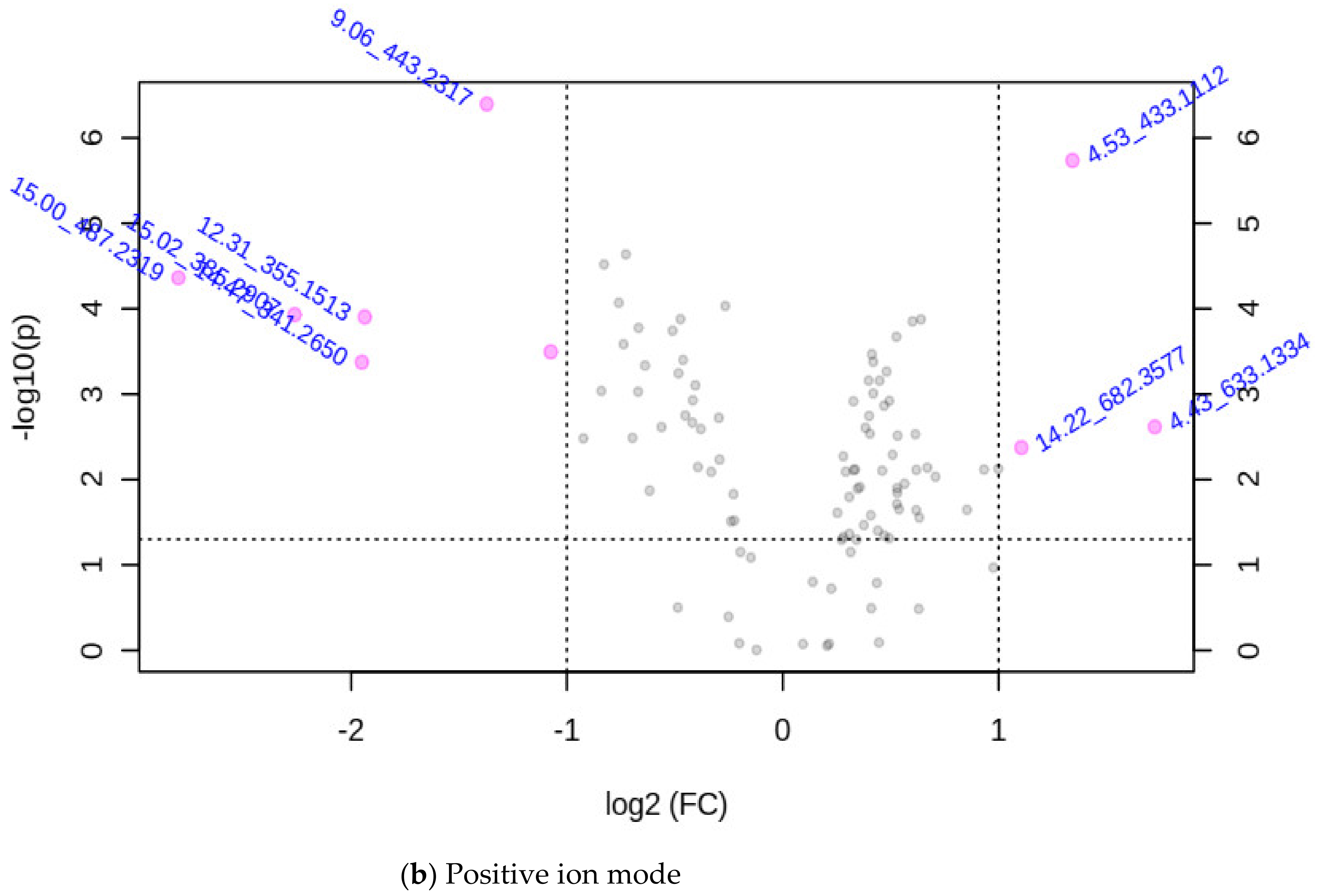
3.2. Characterizing the m/z Ion-Features of Moringa Accessions with Methane Inhibition
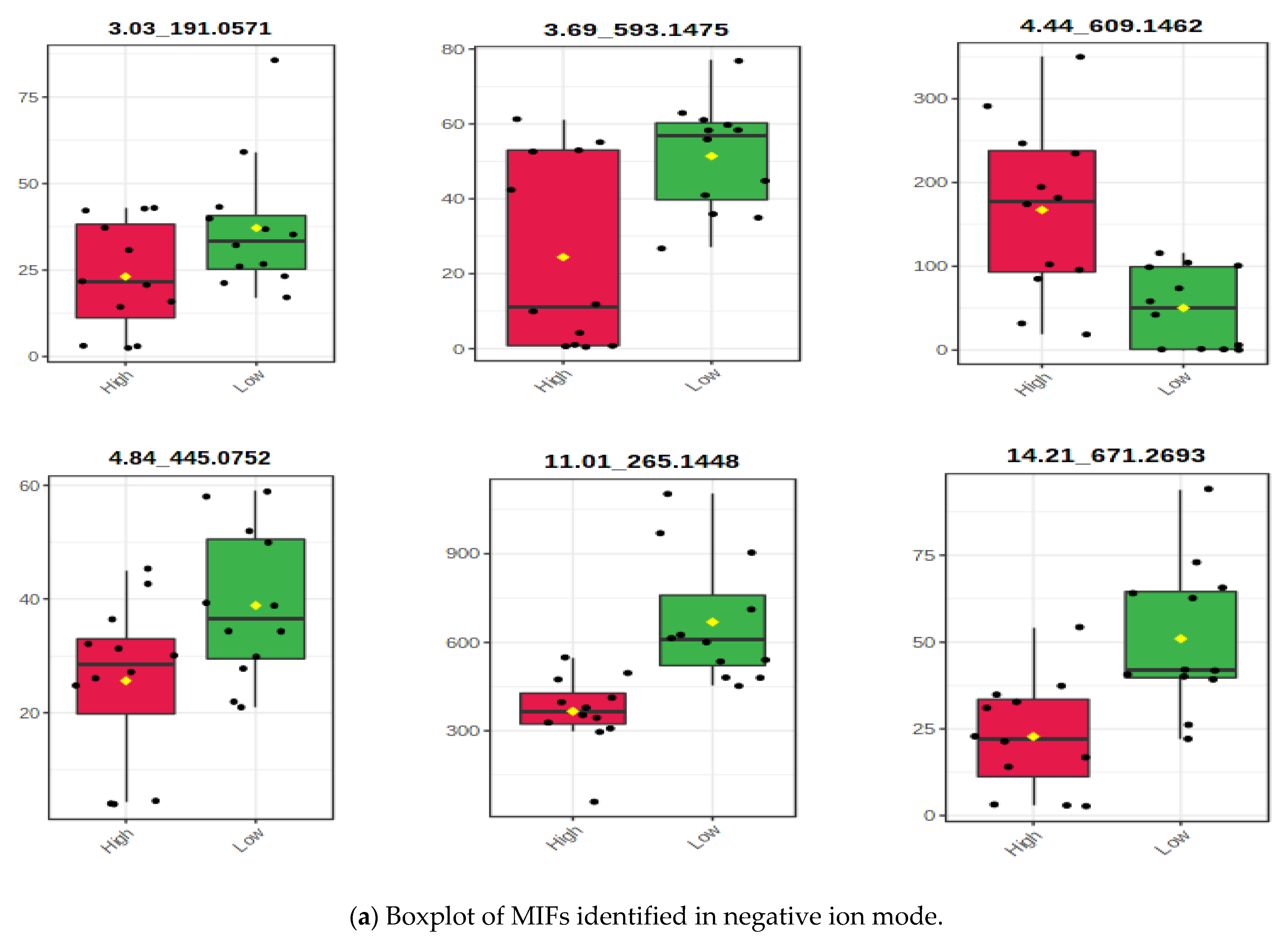
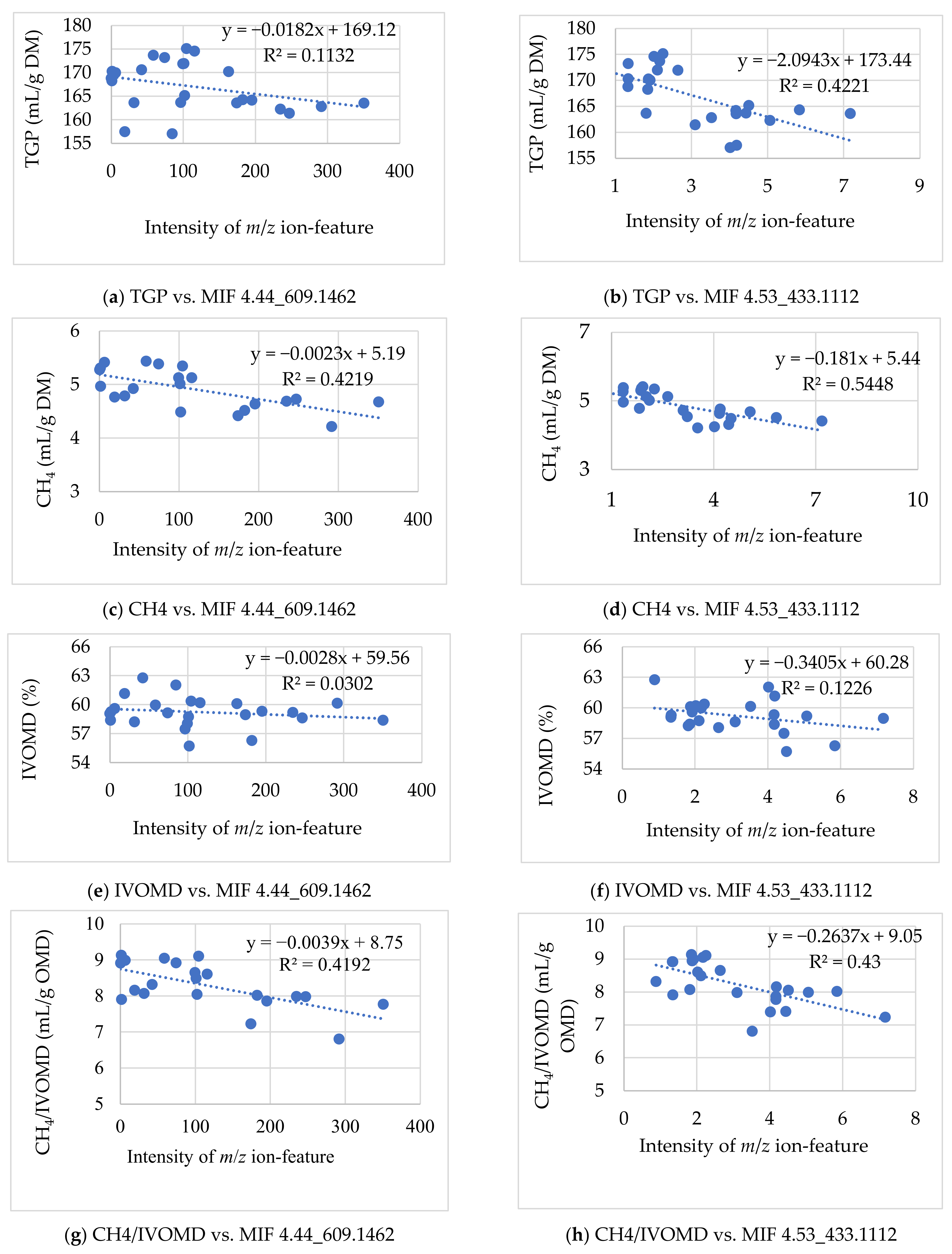
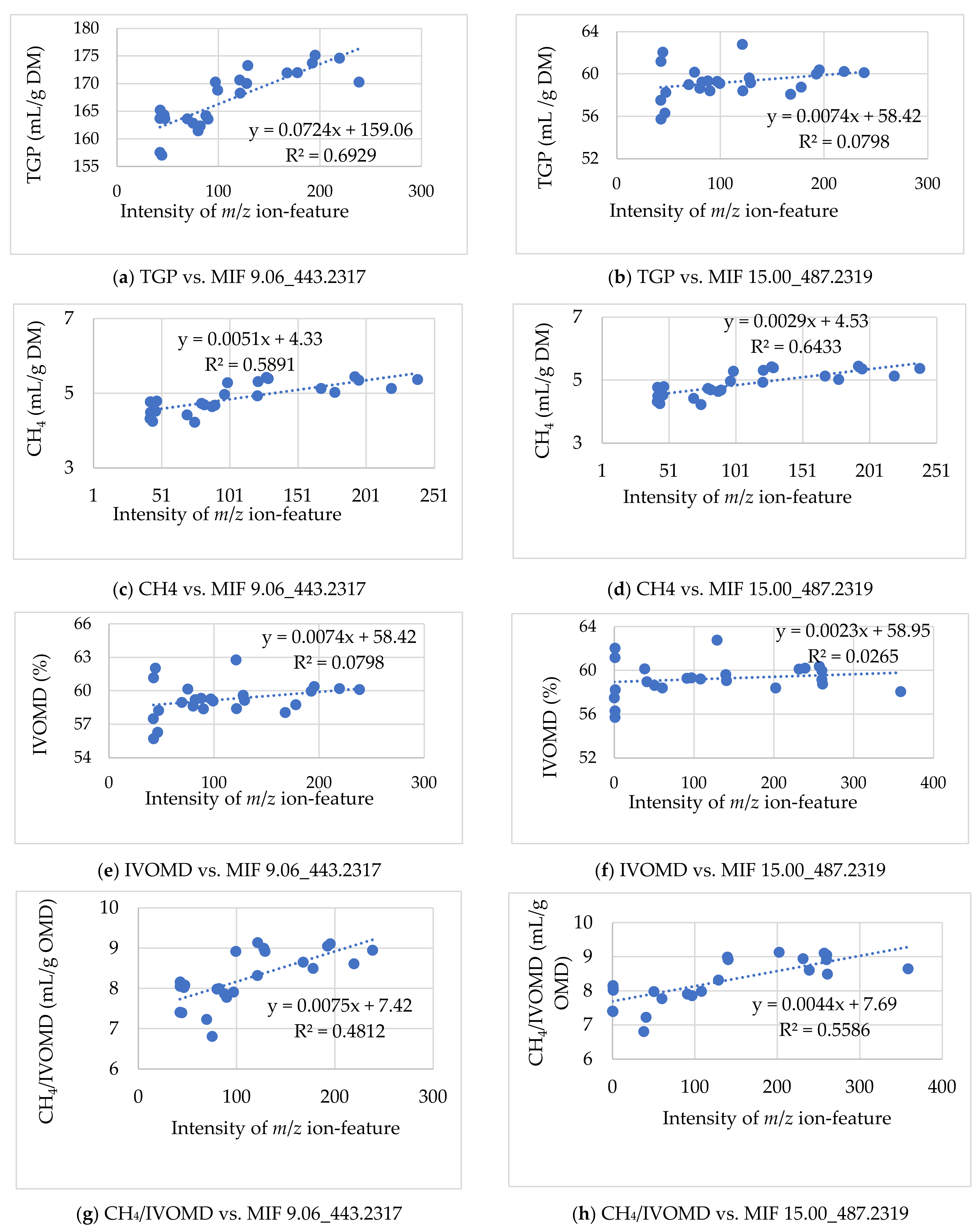
3.2.1. Associating the m/z Ion-Features Contribution to High Methane Inhibition
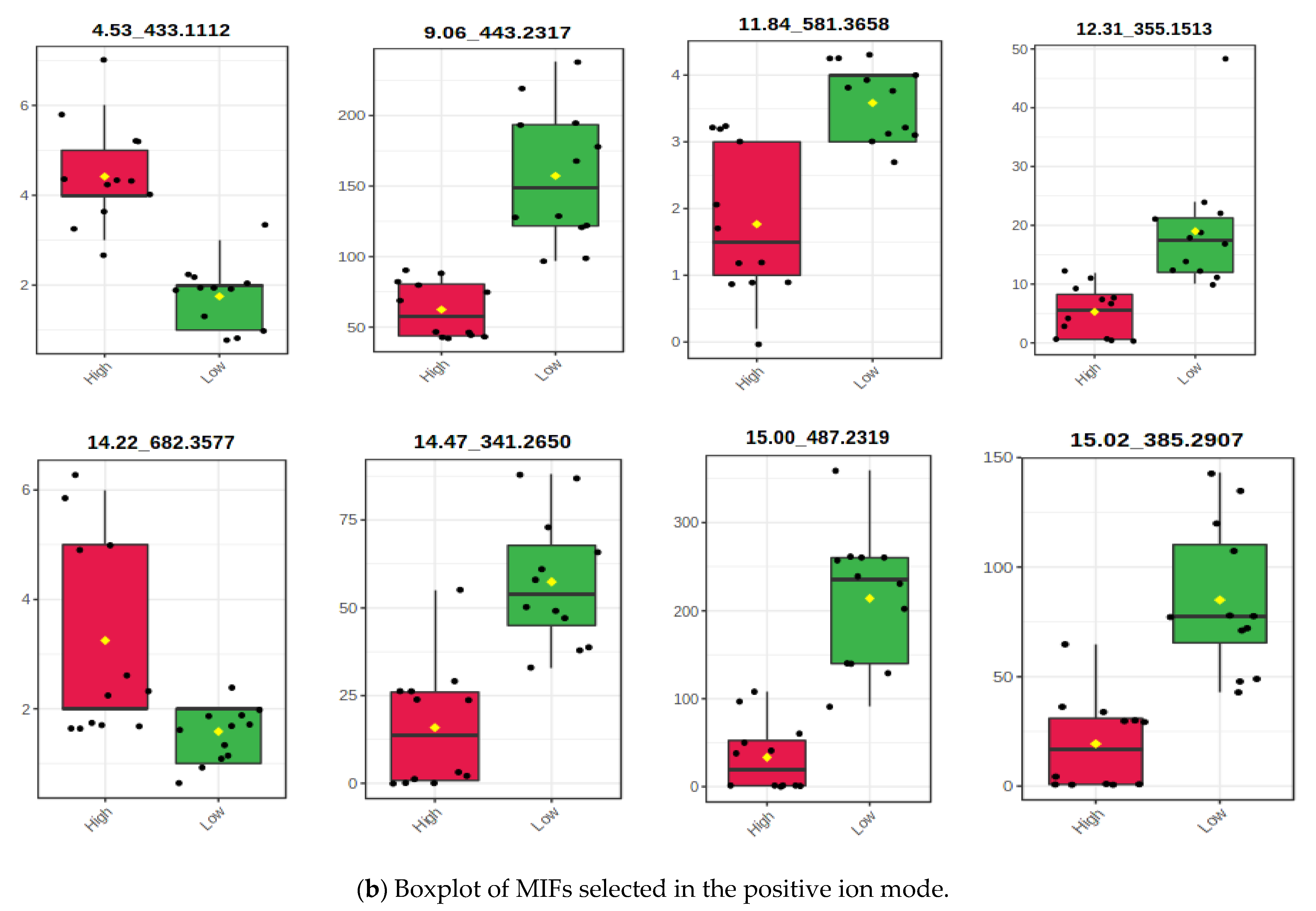
3.2.2. Associating the m/z Ion-Features with Their Contribution to Low Methane Inhibition
4. Discussion
5. Practical Applications and Future Research Perspectives
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Paul, E.A.; Sheehan, J.; Antle, J.M.; Paustian, K. Agriculture’s Role in Greenhouse Gas Mitigation. In Arlington, US: Prepared for the Pew Center on Global Climate Change; Report No.: 4; Food & Agriculture Org: Rome, Italy, 2006; Available online: www.pewclimate.org (accessed on 24 January 2019).
- Steinfeld, H.; Gerber, P.; Wassenaar, T.D.; Castel, V.; Rosales, M.; Rosales, M.; de Haan, C. Livestock’s Long Shadow: Environmental Issues and Options; Food & Agriculture Org: Rome, Italy, 2006. [Google Scholar]
- Bodas, R.; Prieto, N.; García-González, R.; Andrés, S.; Giráldez, F.J.; López, S. Manipulation of rumen fermentation and methane production with plant secondary metabolites. Anim. Feed Sci. Technol. 2012, 176, 78–93. [Google Scholar] [CrossRef]
- Hill, J.; McSweeney, C.; Wright, A.-D.G.; Bishop-Hurley, G.; Kalantar-zadeh, K. Measuring methane production from ruminants. Trends Biotechnol. 2016, 34, 26–35. [Google Scholar] [CrossRef]
- Chapman, C.; Chester-Jones, H.; Ziegler, D.; Clapper, J.; Erickson, P. Effects of cinnamaldehyde or monensin on performance of weaned Holstein dairy heifers. J. Dairy Sci. 2017, 100, 1712–1719. [Google Scholar] [CrossRef] [Green Version]
- McCann, J.C.; Elolimy, A.A.; Loor, J.J. Rumen microbiome, probiotics, and fermentation additives. Vet. Clin. Food Anim. Pract. 2017, 33, 539–553. [Google Scholar] [CrossRef]
- Medjekal, S.; Bodas, R.; Bousseboua, H.; López, S. Evaluation of three medicinal plants for methane production potential, fiber digestion and rumen fermentation in vitro. Energy Procedia 2017, 119, 632–641. [Google Scholar] [CrossRef]
- Neubauer, V.; Petri, R.; Humer, E.; Kröger, I.; Mann, E.; Reisinger, N.; Wagner, M.; Zebeli, Q. High-grain diets supplemented with phytogenic compounds or autolyzed yeast modulate ruminal bacterial community and fermentation in dry cows. J. Dairy Sci. 2018, 101, 2335–2349. [Google Scholar] [CrossRef]
- Demİrtaş, A.; Öztürk, H.; Pİşkİn, İ. Overview of plant extracts and plant secondary metabolites as alternatives to antibiotics for modification of ruminal fermentation. Ank. Üniversitesi Vet. Fakültesi Derg. 2018, 65, 213–217. [Google Scholar]
- Rira, M.; Chentli, A.; Boufenera, S.; Bousseboua, H. Effects of plants containing secondary metabolites on ruminal methanogenesis of sheep in vitro. Energy Procedia 2015, 74, 15–24. [Google Scholar] [CrossRef] [Green Version]
- Ku-Vera, J.C.; Jiménez-Ocampo, R.; Valencia-Salazar, S.S.; Montoya-Flores, M.D.; Molina-Botero, I.C.; Arango, J.; Gómez-Bravo, C.A.; Aguilar-Pérez, C.F.; Solorio-Sánchez, F.J. Role of secondary plant metabolites on enteric methane mitigation in ruminants. Front. Vet. Sci. 2020, 7, 584. [Google Scholar] [CrossRef]
- Jayanegara, A.; Yogianto, Y.; Wina, E.; Sudarman, A.; Kondo, M.; Obitsu, T.; Kreuzer, M. Combination effects of plant extracts rich in tannins and saponins as feed additives for mitigating In Vitro ruminal methane and ammonia formation. Animals 2020, 10, 1531. [Google Scholar] [CrossRef]
- Kingston-Smith, A.H.; Edwards, J.E.; Huws, S.A.; Kim, E.J.; Abberton, M. Plant-based strategies towards minimising ‘livestock’s long shadow’. Proc. Nutr. Soc. 2010, 69, 613–620. [Google Scholar] [CrossRef] [Green Version]
- Kim, E.T.; Kim, C.-H.; Min, K.-S.; Lee, S.S. Effects of plant extracts on microbial population, methane emission and ruminal fermentation characteristics in in vitro. Asian-Australas. J. Anim. Sci. 2012, 25, 806. [Google Scholar] [CrossRef]
- Pal, K.; Patra, A.; Sahoo, A.; Kumawat, P. Evaluation of several tropical tree leaves for methane production potential, degradability and rumen fermentation in vitro. Phytochemistry 2015, 180, 98–105. [Google Scholar] [CrossRef]
- Akanmu, A.M.; Hassen, A. The use of certain medicinal plant extracts reduced in vitro methane production while improving in vitro organic matter digestibility. Anim. Prod. Sci. 2017, 58, 900–908. [Google Scholar] [CrossRef] [Green Version]
- Jafari, S.; Meng, G.Y.; Rajion, M.A.; Jahromi, M.F.; Ebrahimi, M. Manipulation of rumen microbial fermentation by polyphenol rich solvent fractions from papaya leaf to reduce green-house gas methane and biohydrogenation of C18 PUFA. J. Agric. Food Chem. 2016, 64, 4522–4530. [Google Scholar] [CrossRef]
- Patra, A.K.; Saxena, J. Dietary phytochemicals as rumen modifiers: A review of the effects on microbial populations. Antonie Van Leeuwenhoek 2009, 96, 363–375. [Google Scholar] [CrossRef]
- Erb, M.; Kliebenstein, D.J. Plant secondary metabolites as defenses, regulators, and primary metabolites: The blurred functional trichotomy. Plant Physiol. 2020, 184, 39–52. [Google Scholar] [CrossRef]
- Francis, G.; Kerem, Z.; Makkar, H.P.; Becker, K. The biological action of saponins in animal systems: A review. Br. J. Nutr. 2002, 88, 587–605. [Google Scholar] [CrossRef]
- Faehnrich, B.; Franz, C.; Nemaz, P.; Kaul, H.-P. Medicinal plants and their secondary metabolites—State of the art and trends in breeding, analytics and use in feed supplementation—With special focus on German chamomile. J. Appl. Bot. Food Qual. 2021, 94, 61–74. [Google Scholar]
- Leone, A.; Spada, A.; Battezzati, A.; Schiraldi, A.; Aristil, J.; Bertoli, S. Cultivation, genetic, ethnopharmacology, phytochemistry and pharmacology of Moringa oleifera leaves: An overview. Int. J. Mol. Sci. 2015, 16, 12791–12835. [Google Scholar] [CrossRef]
- Sudha, R.; Philip, X.C.; Suriyakumari, K. Phytochemical constituents of leaves of Moringa oleifera grow in Cuddalore District, Tamil Nadu, India. SBV J. Basic Clin. App. Health Sci. 2020, 3, 164–167. [Google Scholar] [CrossRef]
- Fidrianny, I.; Kanapa, I.; Singgih, M. Phytochemistry and pharmacology of moringa tree: An overview. Biointerface Res. Appl. Chem. 2021, 11, 10776–10789. [Google Scholar]
- Bhalla, N.; Ingle, N.; Patri, S.V.; Haranath, D. Phytochemical analysis of Moringa oleifera leaves extracts by GC-MS and free radical scavenging potency for industrial applications. Saudi J. Biol. Sci. 2021, 28, 6915–6928. [Google Scholar] [CrossRef]
- Amaglo, N.K.; Bennett, R.N.; Curto, R.B.L.; Rosa, E.A.; Turco, V.L.; Giuffrida, A.; Curto, A.L.; Crea, F.; Timpo, G.M. Profiling selected phytochemicals and nutrients in different tissues of the multipurpose tree Moringa oleifera L., grown in Ghana. Food Chem. 2010, 122, 1047–1054. [Google Scholar] [CrossRef]
- Leone, A.; Fiorillo, G.; Criscuoli, F.; Ravasenghi, S.; Santagostini, L.; Fico, G.; Spadafranca, A.; Battezzati, A.; Schiraldi, A.; Pozzi, F. Nutritional characterization and phenolic profiling of Moringa oleifera leaves grown in Chad, Sahrawi Refugee Camps, and Haiti. Int. J. Mol. Sci. 2015, 16, 18923–18937. [Google Scholar] [CrossRef] [Green Version]
- López, S.; García-González, R.; Fernández, M.; Bodas, R.; González, J. Medicinal plants as feed additives in animal nutrition. In Natural Products I; Studium Press LLC: Houston, TX, USA, 2007; pp. 309–333. [Google Scholar]
- Akula, R.; Ravishankar, G.A. Influence of abiotic stress signals on secondary metabolites in plants. Plant Signal. Behav. 2011, 6, 1720–1731. [Google Scholar] [CrossRef]
- Rodrigues, N.P.; Salva, T.d.J.G.; Bragagnolo, N. Influence of coffee genotype on bioactive compounds and the in vitro capacity to scavenge reactive oxygen and nitrogen species. J. Agric. Food Chem. 2015, 63, 4815–4826. [Google Scholar] [CrossRef]
- Skrovankova, S.; Sumczynski, D.; Mlcek, J.; Jurikova, T.; Sochor, J. Bioactive compounds and antioxidant activity in different types of berries. Int. J. Mol. Sci. 2015, 16, 24673–24706. [Google Scholar] [CrossRef] [Green Version]
- Lin, H.; Zhu, H.; Tan, J.; Wang, H.; Wang, Z.; Li, P.; Zhao, C.; Liu, J. Comparative analysis of chemical constituents of Moringa oleifera leaves from China and India by ultra-performance liquid chromatography coupled with quadrupole-time-of-flight mass spectrometry. Molecules 2019, 24, 942. [Google Scholar] [CrossRef] [Green Version]
- Teclegeorgish, Z.W.; Aphane, Y.M.; Mokgalaka, N.S.; Steenkamp, P.; Tembu, V.J. Nutrients, secondary metabolites and anti-oxidant activity of Moringa oleifera leaves and Moringa-based commercial products. S. Afr. J. Bot. 2021, 142, 409–420. [Google Scholar] [CrossRef]
- Oskoueian, E.; Abdullah, N.; Oskoueian, A. Effects of flavonoids on rumen fermentation activity, methane production, and microbial population. BioMed Res. Int. 2013, 2013, 349129. [Google Scholar] [CrossRef] [Green Version]
- Sinz, S.; Kunz, C.; Liesegang, A.; Braun, U.; Marquardt, S.; Soliva, C.R.; Kreuzer, M. In vitro bioactivity of various pure flavonoids in ruminal fermentation, with special reference to methane formation. Czech J. Anim. Sci. 2018, 63, 293–304. [Google Scholar] [CrossRef]
- Dall’Agnol, R.; Ferraz, A.; Bernardi, A.; Albring, D.; Nör, C.; Sarmento, L.; Lamb, L.; Hass, M.; Von Poser, G.; Schapoval, E. Antimicrobial activity of some Hypericum species. Phytomedicine 2003, 10, 511–516. [Google Scholar] [CrossRef]
- Patra, A.; Kamra, D.; Agarwal, N. Effect of plant extracts on in vitro methanogenesis, enzyme activities and fermentation of feed in rumen liquor of buffalo. Anim. Feed Sci. Technol. 2006, 128, 276–291. [Google Scholar] [CrossRef]
- Tavendale, M.H.; Meagher, L.P.; Pacheco, D.; Walker, N.; Attwood, G.T.; Sivakumaran, S. Methane production from in vitro rumen incubations with Lotus pedunculatus and Medicago sativa, and effects of extractable condensed tannin fractions on methanogenesis. Anim. Feed Sci. Technol. 2005, 123, 403–419. [Google Scholar] [CrossRef]
- Cushnie, T.T.; Lamb, A.J. Antimicrobial activity of flavonoids. Int. J. Antimicrob. Agents 2005, 26, 343–356. [Google Scholar] [CrossRef]
- Benchaar, C.; Calsamiglia, S.; Chaves, A.; Fraser, G.; Colombatto, D.; McAllister, T.; Beauchemin, K. A review of plant-derived essential oils in ruminant nutrition and production. Anim. Feed Sci. Technol. 2008, 145, 209–228. [Google Scholar] [CrossRef]
- Singh, B.N.; Singh, B.; Singh, R.; Prakash, D.; Dhakarey, R.; Upadhyay, G.; Singh, H. Oxidative DNA damage protective activity, antioxidant and anti-quorum sensing potentials of Moringa oleifera. Food Chem. Toxicol. 2009, 47, 1109–1116. [Google Scholar] [CrossRef]
- Zhang, M.; Hettiarachchy, N.S.; Horax, R.; Kannan, A.; Praisoody, A.; Muhundan, A. Phytochemicals, antioxidant and antimicrobial activity of Hibiscus sabdariffa, Centella asiatica, Moringa oleifera and Murraya koenigii leaves. J. Med. Plants Res. 2011, 5, 6672–6680. [Google Scholar] [CrossRef]
- Haque, M.N. Dietary manipulation: A sustainable way to mitigate methane emissions from ruminants. J. Anim. Sci. Technol. 2018, 60, 15. [Google Scholar] [CrossRef] [Green Version]
- Melesse, A. Comparative assessment on chemical compositions and feeding values of leaves of Moringa stenopetala and Moringa oleifera using in vitro gas production method. Ethiop. J. Sci. Technol. 2011, 2, 31–41. Available online: https://www.semanticscholar.org/paper/Comparative-assessment-on-chemical-compositions-and-Melesse/d5f5045527c98159a5c112d8b1641d5754a85313 (accessed on 5 February 2018).
- Delgado, D.C.; Galindo, J.; González, R.; González, N.; Scull, I.; Dihigo, L.; Cairo, J.; Aldama, A.I.; Moreira, O. Feeding of tropical trees and shrub foliages as a strategy to reduce ruminal methanogenesis: Studies conducted in Cuba. Trop. Anim. Health Prod. 2012, 44, 1097–1104. [Google Scholar] [CrossRef] [PubMed]
- Ghamkhar, K.; Rochfort, S.; Banik, B.K.; Revell, C. Candidate metabolites for methane mitigation in the forage legume biserrula. Agron. Sustain. Dev. 2018, 38, 30. [Google Scholar] [CrossRef] [Green Version]
- Addisu, Z.; Hassen, A.; Apostolides, Z.; Tjelele, J. Relationships between Agronomic Traits of Moringa Accessions and In Vitro Gas Production Characteristics of a Test Feed Incubated with the Respective Moringa Leaf Extracts. Ph.D. Thesis, University of Pretoria, Pretoria, South Africa, 2022. in press. [Google Scholar]
- Eloff, J. Which extractant should be used for the screening and isolation of antimicrobial components from plants? J. Ethnopharmacol. 1998, 60, 1–8. [Google Scholar] [CrossRef]
- Goering, H.K.; Van Soest, P.J. Forage Fiber Analyses: Apparatus, Reagents, Procedures, and Some Applications; Agricultural Research iService; US Department of Agriculture: Washington, DC, USA, 1970.
- Mould, F.L.; Morgan, R.; Kliem, K.E.; Krystallidou, E. A review and simplification of the in vitro incubation medium. Anim. Feed Sci. Technol. 2005, 123–124, 155–172. [Google Scholar] [CrossRef]
- Theodorou, M.K.; Williams, B.A.; Dhanoa, M.S.; McAllan, A.B.; France, J. A simple gas production method using a pressure transducer to determine the fermentation kinetics of ruminant feeds. Anim. Feed Sci. Technol. 1994, 48, 185–197. [Google Scholar] [CrossRef]
- Ankom, T. Pressure to Gas Production Conversion, User Manual Service Procedure 005. Calculation of Gas Volume in mL at 39 °C with Pressure Measured in Psi; ANKOM Technology Macedon: New York, NY, USA, 2014; Available online: https://www.manualsdir.com/manuals/655546/ankom-rf.html?page=46 (accessed on 12 September 2018).
- Tilley, J.; Terry, D.R. A two-stage technique for the in vitro digestion of forage crops. Grass Forage Sci. 1963, 18, 104–111. [Google Scholar] [CrossRef]
- Engels, E.; Van der Merwe, F. Application of an in vitro technique to South African forages with special reference to the effect to certain factors on the results. S. Afr. J. Agric. Sci. 1967, 10, 983–995. [Google Scholar]
- Yang, J.; Zhao, X.; Lu, X.; Lin, X.; Xu, G. A data preprocessing strategy for metabolomics to reduce the mask effect in data analysis. Front. Mol. Biosci. 2015, 2, 4. [Google Scholar] [CrossRef] [Green Version]
- Vinaixa, M.; Samino, S.; Saez, I.; Duran, J.; Guinovart, J.J.; Yanes, O. A guideline to univariate statistical analysis for LC/MS-based untargeted metabolomics-derived data. Metabolites 2012, 2, 775–795. [Google Scholar] [CrossRef]
- SAS Institute. SAS/ACCESS® 9.4 Interface to ADABAS: Reference; SAS Institute Inc.: Cary, NC, USA, 2013. [Google Scholar]
- Isah, T. Stress and defense responses in plant secondary metabolites production. Biol. Res. 2019, 52, 39. [Google Scholar] [CrossRef] [Green Version]
- Yang, L.; Wen, K.-S.; Ruan, X.; Zhao, Y.-X.; Wei, F.; Wang, Q. Response of plant secondary metabolites to environmental factors. Molecules 2018, 23, 762. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Kong, D.; Fu, Y.; Sussman, M.R.; Wu, H. The effect of developmental and environmental factors on secondary metabolites in medicinal plants. Plant Physiol. Biochem. 2020, 148, 80–89. [Google Scholar] [CrossRef]
- Al_husnan, L.A.; Alkahtani, M.D. Impact of Moringa aqueous extract on pathogenic bacteria and fungi in vitro. Ann. Agric. Sci. 2016, 61, 247–250. [Google Scholar] [CrossRef]
- Cieslak, A.; Szumacher-Strabel, M.; Stochmal, A.; Oleszek, W. Plant components with specific activities against rumen methanogens. Animal 2013, 7 (Suppl. 2), 253–265. [Google Scholar] [CrossRef]
- Othman, L.; Sleiman, A.; Abdel-Massih, R.M. Antimicrobial activity of polyphenols and alkaloids in middle eastern plants. Front. Microbiol. 2019, 10, 911. [Google Scholar] [CrossRef]
- Lelario, F.; Scrano, L.; De Franchi, S.; Bonomo, M.; Salzano, G.; Milan, S.; Milella, L.; Bufo, S. Identification and antimicrobial activity of most representative secondary metabolites from different plant species. Chem. Biol. Technol. Agric. 2018, 5, 1–12. [Google Scholar] [CrossRef]
- Macheboeuf, D.; Morgavi, D.; Papon, Y.; Mousset, J.-L.; Arturo-Schaan, M. Dose–response effects of essential oils on in vitro fermentation activity of the rumen microbial population. Anim. Feed Sci. Technol. 2008, 145, 335–350. [Google Scholar] [CrossRef]
- Gokulan, K.; Kolluru, P.; Cerniglia, C.E.; Khare, S. Dose-dependent effects of Aloin on the intestinal bacterial community structure, short chain fatty acids metabolism and intestinal epithelial cell permeability. Front. Microbiol. 2019, 10, 474. [Google Scholar] [CrossRef]
- Burrell-Saward, H.; Harris, A.J.; de LaFlor, R.; Sallam, H.; Alavijeh, M.S.; Ward, T.H.; Croft, S.L. Dose-dependent effect and pharmacokinetics of fexinidazole and its metabolites in a mouse model of human African trypanosomiasis. Int. J. Antimicrob. Agents 2017, 50, 203–209. [Google Scholar] [CrossRef]
- Goel, G.; Makkar, H.P. Methane mitigation from ruminants using tannins and saponins. Trop. Anim. Health Prod. 2012, 44, 729–739. [Google Scholar] [CrossRef]
- Jouany, J.-P.; Morgavi, D. Use of ‘natural’ products as alternatives to antibiotic feed additives in ruminant production. Animal 2007, 1, 1443–1466. [Google Scholar] [CrossRef] [Green Version]
- Cardoso-Gutierrez, E.; Aranda-Aguirre, E.; Robles-Jimenez, L.; Castelán-Ortega, O.; Chay-Canul, A.; Foggi, G.; Angeles-Hernandez, J.; Vargas-Bello-Pérez, E.; González-Ronquillo, M. Effect of tannins from tropical plants on methane production from ruminants: A systematic review. Vet. Anim. Sci. 2021, 14, 100214. [Google Scholar] [CrossRef]
- Rossi, C.A.S.; Grossi, S.; Dell’Anno, M.; Compiani, R.; Rossi, L. Effect of a Blend of Essential Oils, Bioflavonoids and Tannins on In Vitro Methane Production and In Vivo Production Efficiency in Dairy Cows. Animals 2022, 12, 728. [Google Scholar] [CrossRef]
- Zhou, R.; Wu, J.; Lang, X.; Liu, L.; Casper, D.P.; Wang, C.; Zhang, L.; Wei, S. Effects of oregano essential oil on in vitro ruminal fermentation, methane production, and ruminal microbial community. J. Dairy Sci. 2020, 103, 2303–2314. [Google Scholar] [CrossRef]
- Lee, S.-R.; Cho, Y.; Ju, H.K.; Kim, E. Theoretical Methane Emission Estimation from Volatile Fatty Acids in Bovine Rumen Fluid. Appl. Sci. 2021, 11, 7730. [Google Scholar] [CrossRef]
- Mitsumori, M.; Sun, W. Control of rumen microbial fermentation for mitigating methane emissions from the rumen. Asian-Australas. J. Anim. Sci. 2008, 21, 144. [Google Scholar] [CrossRef]
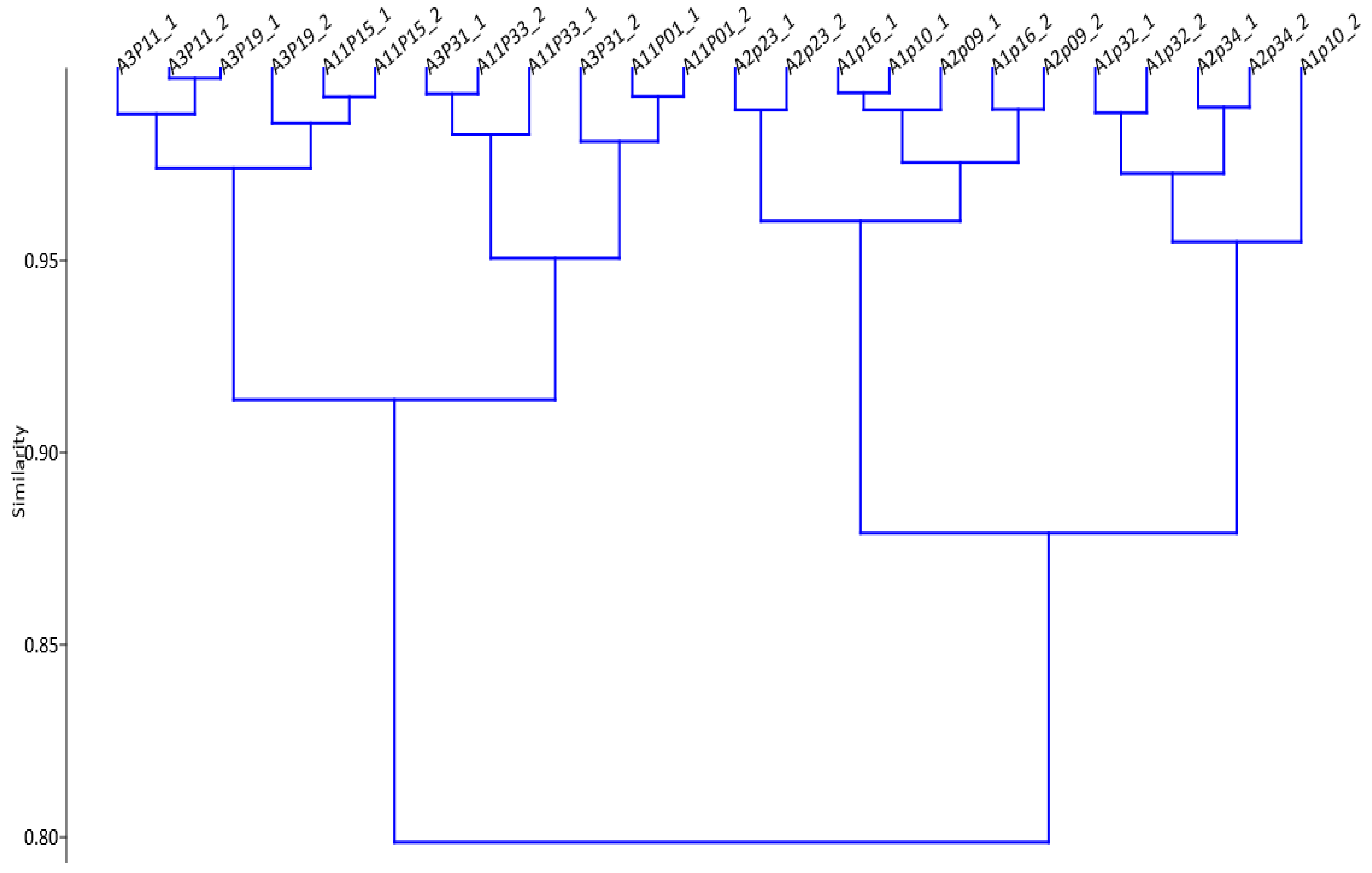
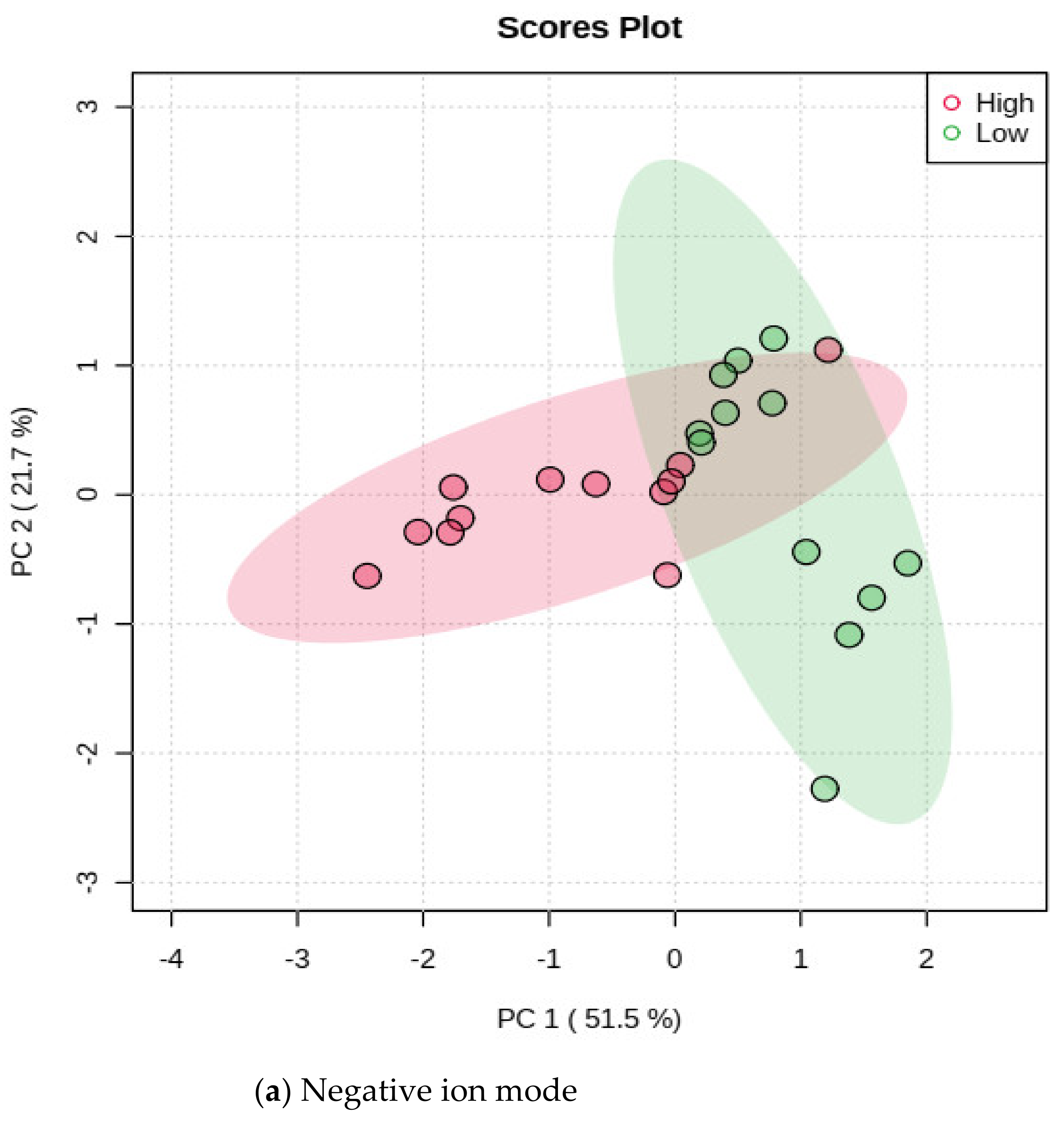
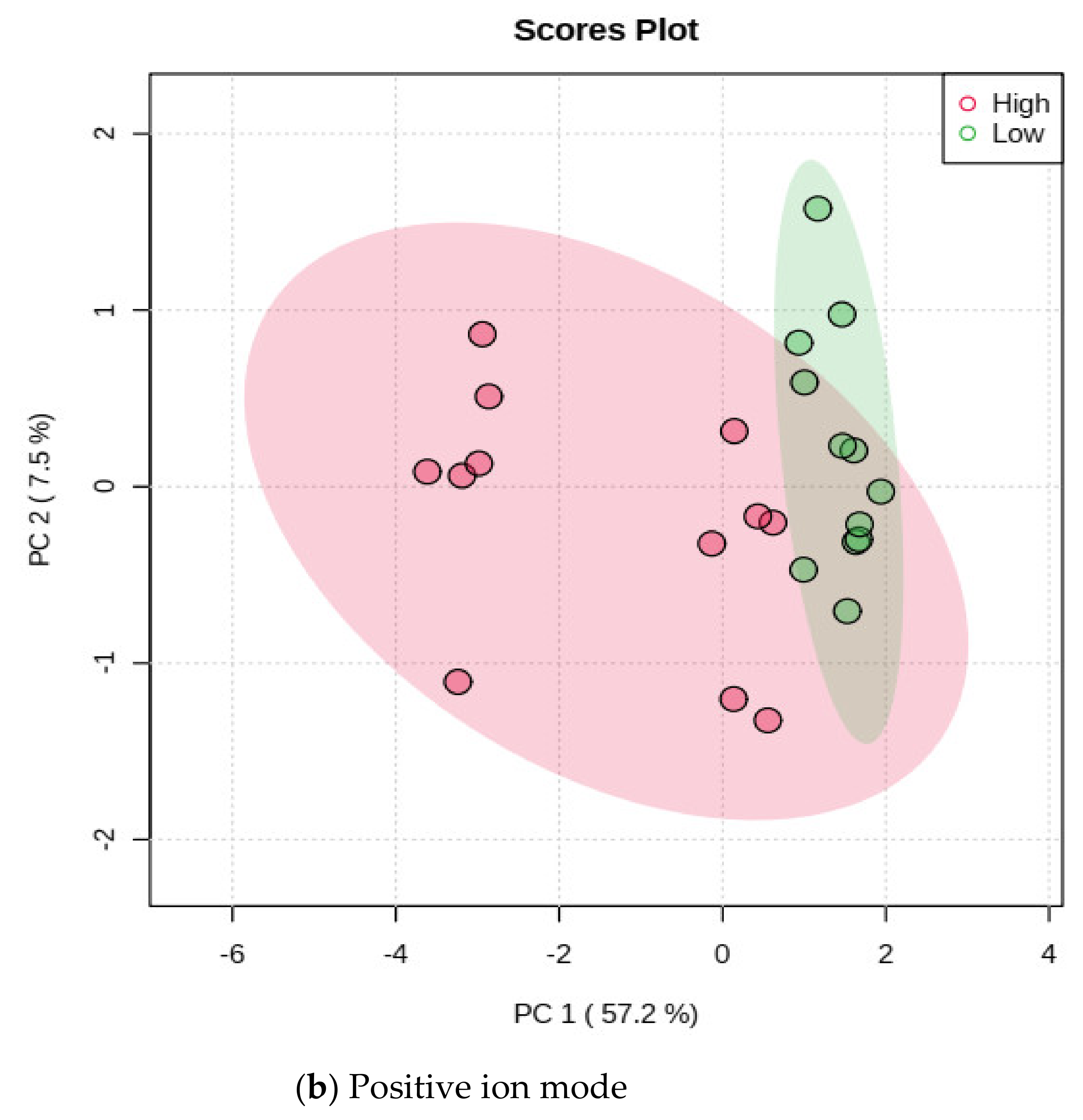
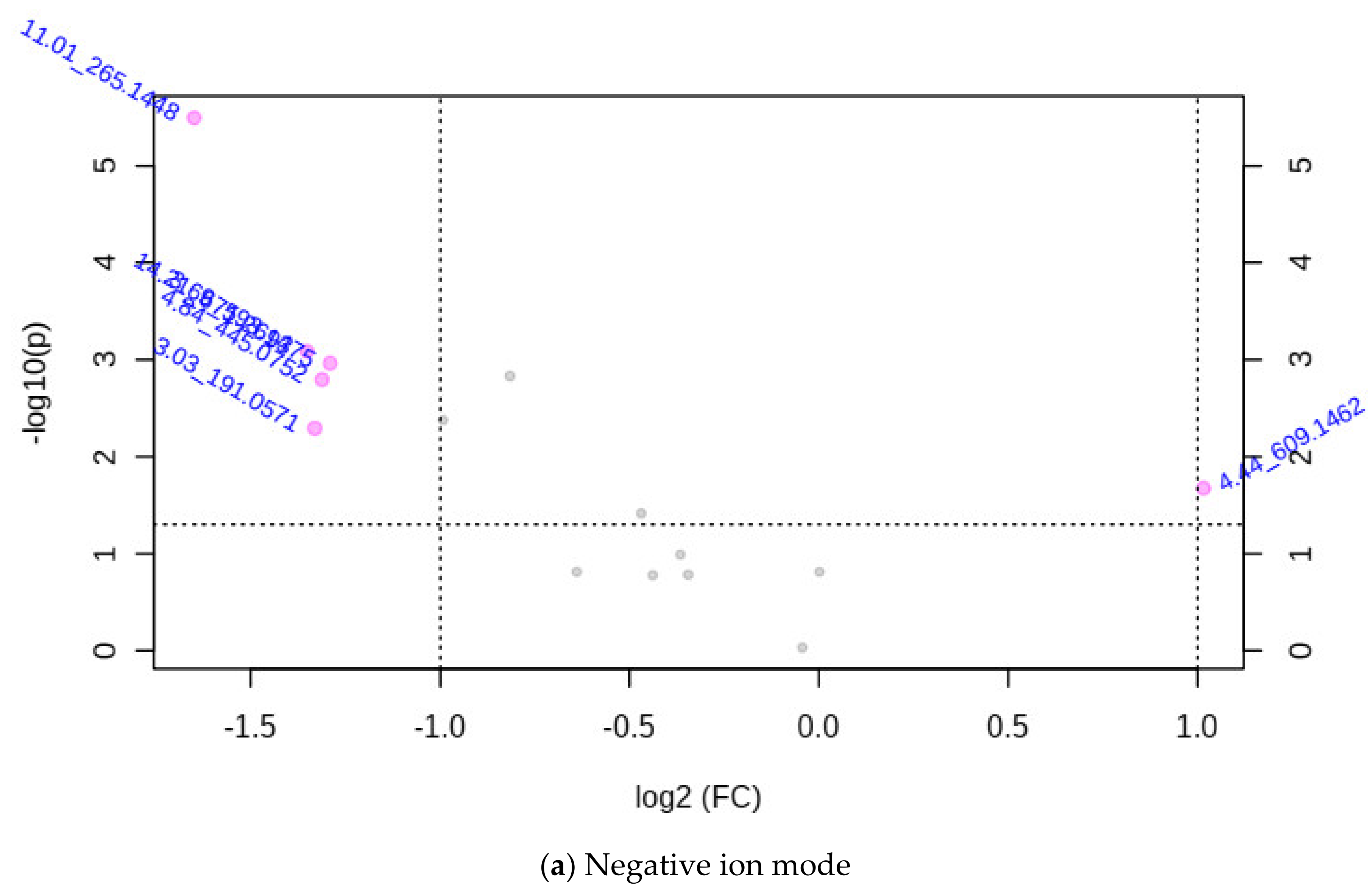
| Accession | CO | Sample Code | TGP | TCH4P | % IVOMD | % CH4 Inhibition | CH4 IVOMD | CH4 IG |
|---|---|---|---|---|---|---|---|---|
| Bulk (A1) | Kenya | A01p10_1 | 173.71 | 5.37 | 59.99 | 15.72 | 8.95 | low |
| A01p10_2 | 170.03 | 5.13 | 59.62 | 19.42 | 8.61 | low | ||
| A01p16_1 | 171.97 | 5.35 | 58.76 | 15.97 | 9.11 | low | ||
| A01p16_2 | 170.28 | 5.44 | 60.13 | 14.55 | 9.05 | low | ||
| A01p32_1 | 168.82 | 5.02 | 59.09 | 21.20 | 8.49 | low | ||
| A01p32_2 | 168.28 | 5.06 | 58.41 | 20.64 | 8.65 | low | ||
| Mean ± SEM | 170.51 ± 1.23 B | 5.23 ± 0.13 B | 59.33 ± 0.38 A | 17.92 ± 2.01 B | 8.81 ± 0.18 B | |||
| 07229 (A2) | Kenya | A02p09_1 | 175.14 | 5.39 | 60.39 | 15.42 | 8.92 | low |
| A02p09_2 | 174.59 | 5.42 | 60.22 | 14.96 | 8.99 | low | ||
| A02p23_1 | 171.93 | 5.31 | 58.07 | 16.70 | 9.14 | low | ||
| A02p23_2 | 173.25 | 5.28 | 59.16 | 17.15 | 8.92 | low | ||
| A02p34_1 | 170.29 | 4.93 | 59.29 | 22.54 | 8.32 | low | ||
| A02p34_2 | 170.65 | 4.97 | 62.78 | 22.03 | 7.91 | low | ||
| Mean ± SEM | 172.64 ± 1.55 B | 5.21 ± 0.15 B | 59.99 ± 0.88 A | 18.13 ± 2.35 B | 8.70 ± 0.36 B | |||
| 07633 (A3) | Mali | A03P11_1 | 163.61 | 4.64 | 58.98 | 27.20 | 7.86 | high |
| A03P11_2 | 162.81 | 4.68 | 60.16 | 26.57 | 7.77 | high | ||
| A03P19_1 | 161.43 | 4.69 | 58.64 | 26.42 | 7.99 | high | ||
| A03P19_2 | 162.31 | 4.73 | 59.22 | 25.77 | 7.98 | high | ||
| A03P31_1 | 157.05 | 4.22 | 62.05 | 33.69 | 6.81 | high | ||
| A03P31_2 | 157.53 | 4.42 | 61.18 | 30.55 | 7.23 | high | ||
| Mean ± SEM | 160.79 ± 2.20 C | 4.56 ± 0.14 C | 60.04 ± 0.99 A | 28.37 ± 2.20 A | 7.61 ± 0.37 C | |||
| Pretoria (A11) | South Africa | A11P01_1 | 164.31 | 4.52 | 56.3 | 29.10 | 8.02 | high |
| A11P01_2 | 165.19 | 4.49 | 55.73 | 29.56 | 8.05 | high | ||
| A11P15_1 | 163.57 | 4.77 | 58.4 | 25.17 | 8.16 | high | ||
| A11P15_2 | 164.19 | 4.79 | 59.34 | 24.75 | 8.08 | high | ||
| A11P33_1 | 163.65 | 4.32 | 58.25 | 32.26 | 7.41 | high | ||
| A11P33_2 | 163.71 | 4.25 | 57.51 | 33.22 | 7.40 | high | ||
| Mean ± SEM | 164.10 ± 0.40 BC | 4.52 ± 0.16 C | 57.59 ± 1.02 B | 29.01 ± 2.48 A | 7.85 ± 0.28 C | |||
| Control | 203.84 ± 5.32 A | 6.37 ± 0.18 A | 54.68 ± 0.38 C | 11.25 ± 0.40 A | ||||
| m/z Ion-Features (MIFs) | |||
|---|---|---|---|
| Negative Ion Mode | Positive Ion Mode | ||
| 4.44_609.1462 | 4.53_433.1112 | 14.22_682.3577 | |
| Detected mass | 609.1462 | 433.1112 | 682.3577 |
| Actual mass | 609.1462 | 432.1112 | 681.3577 |
| RT | 4.44 | 4.53 | 14.22 |
| ALI | 63.89 | 1.81 | 1.54 |
| AHI | 167.66 | 4.10 | 2.83 |
| Fold change | 2.02 | 2.54 | 2.13 |
| p-value | 0.021 | 4.7 × 10−7 | 0.0047 |
| Pearson correlation of the selected MIFs with TCH4P, TGP and IVOMD (2-tailed) | |||
| TCH4P | −0.48853 * | −0.73828 ** | −0.50434 * |
| TGP | −0.63255 ** | −0.79327 ** | −0.36979 * |
| IVOMD | 0.08915 | −0.29749 | −0.45888 * |
| CH4/IVOMD | −0.58925 ** | −0.65649 ** | −0.36577 |
| m/z Ion-Features | |||||||
|---|---|---|---|---|---|---|---|
| Negative Ion Mode | Positive Ion Mode | ||||||
| 11.01_265.1448 | 9.06_443.2317 | 11.84_581.3658 | 12.31_355.1513 | 14.47_341.2650 | 15.00_487.2319 | 15.02_385.2907 | |
| Detected mass | 265.1448 | 443.2317 | 581.3558 | 355.1513 | 341.2650 | 487.2319 | 385.2907 |
| Actual mass | 265.1448 | 442.2317 | 580.3658 | 354.1513 | 340.2650 | 486.2319 | 384.2907 |
| RT | 11.01 | 9.06 | 11.84 | 12.31 | 14.47 | 15.00 | 15.02 |
| ALI | 668.19 | 157.22 | 3.53 | 18.91 | 57.35 | 214.01 | 84.97 |
| AHI | 394.07 | 58.52 | 1.81 | 5.15 | 15.87 | 33.12 | 19.11 |
| Fold change | 0.32 | 0.39 | 0.48 | 0.26 | 0.26 | 0.14 | 0.21 |
| p-value | 3.2 × 10−6 | 4.75 × 10−7 | 2.9 × 10−4 | 1.1 × 10−4 | 4.2 × 10−4 | 4.27 × 10−6 | 1.2 × 10−4 |
| Pearson correlation of the selected MIFs with TCH4P, TGP and IVOMD (2-tailed) | |||||||
| TCH4P | 0.615 ** | 0.768 ** | 0.653 ** | 0.693 ** | 0.632 ** | 0.802 ** | 0.741 ** |
| TGP | 0.662 ** | 0.648 ** | 0.509 * | 0.580 ** | 0.520 ** | 0.675 ** | 0.637 ** |
| IVOMD | 0.154 | 0.264 | 0.439 | 0.294 | 0.325 | 0.177 | 0.317 |
| CH4/IVOMD | 0.578 ** | 0.694 ** | 0.523 ** | 0.611 ** | 0.531 ** | 0.747 ** | 0.655 ** |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zeru, A.E.; Hassen, A.; Apostolides, Z.; Tjelele, J. Screening of Candidate Bioactive Secondary Plant Metabolite Ion-Features from Moringa oleifera Accessions Associated with High and Low Enteric Methane Inhibition from Ruminants. Metabolites 2022, 12, 501. https://doi.org/10.3390/metabo12060501
Zeru AE, Hassen A, Apostolides Z, Tjelele J. Screening of Candidate Bioactive Secondary Plant Metabolite Ion-Features from Moringa oleifera Accessions Associated with High and Low Enteric Methane Inhibition from Ruminants. Metabolites. 2022; 12(6):501. https://doi.org/10.3390/metabo12060501
Chicago/Turabian StyleZeru, Addisu Endalew, Abubeker Hassen, Zeno Apostolides, and Julius Tjelele. 2022. "Screening of Candidate Bioactive Secondary Plant Metabolite Ion-Features from Moringa oleifera Accessions Associated with High and Low Enteric Methane Inhibition from Ruminants" Metabolites 12, no. 6: 501. https://doi.org/10.3390/metabo12060501
APA StyleZeru, A. E., Hassen, A., Apostolides, Z., & Tjelele, J. (2022). Screening of Candidate Bioactive Secondary Plant Metabolite Ion-Features from Moringa oleifera Accessions Associated with High and Low Enteric Methane Inhibition from Ruminants. Metabolites, 12(6), 501. https://doi.org/10.3390/metabo12060501








