Short Linear Motifs Orchestrate Functioning of Human Proteins during Embryonic Development, Redox Regulation, and Cancer
Abstract
1. Introduction
2. Results
2.1. Human Proteins Aligned to Human AFP Segments
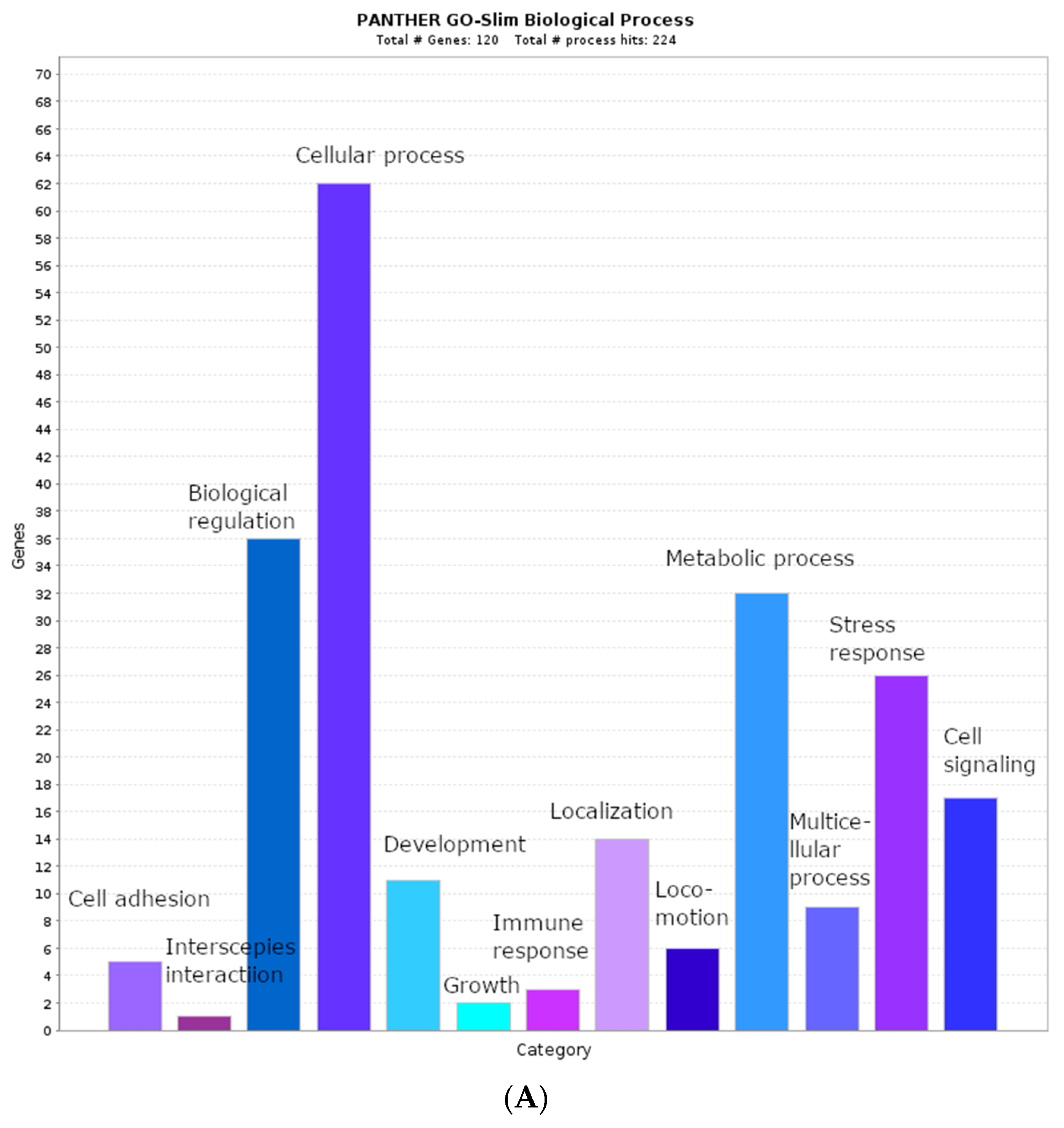
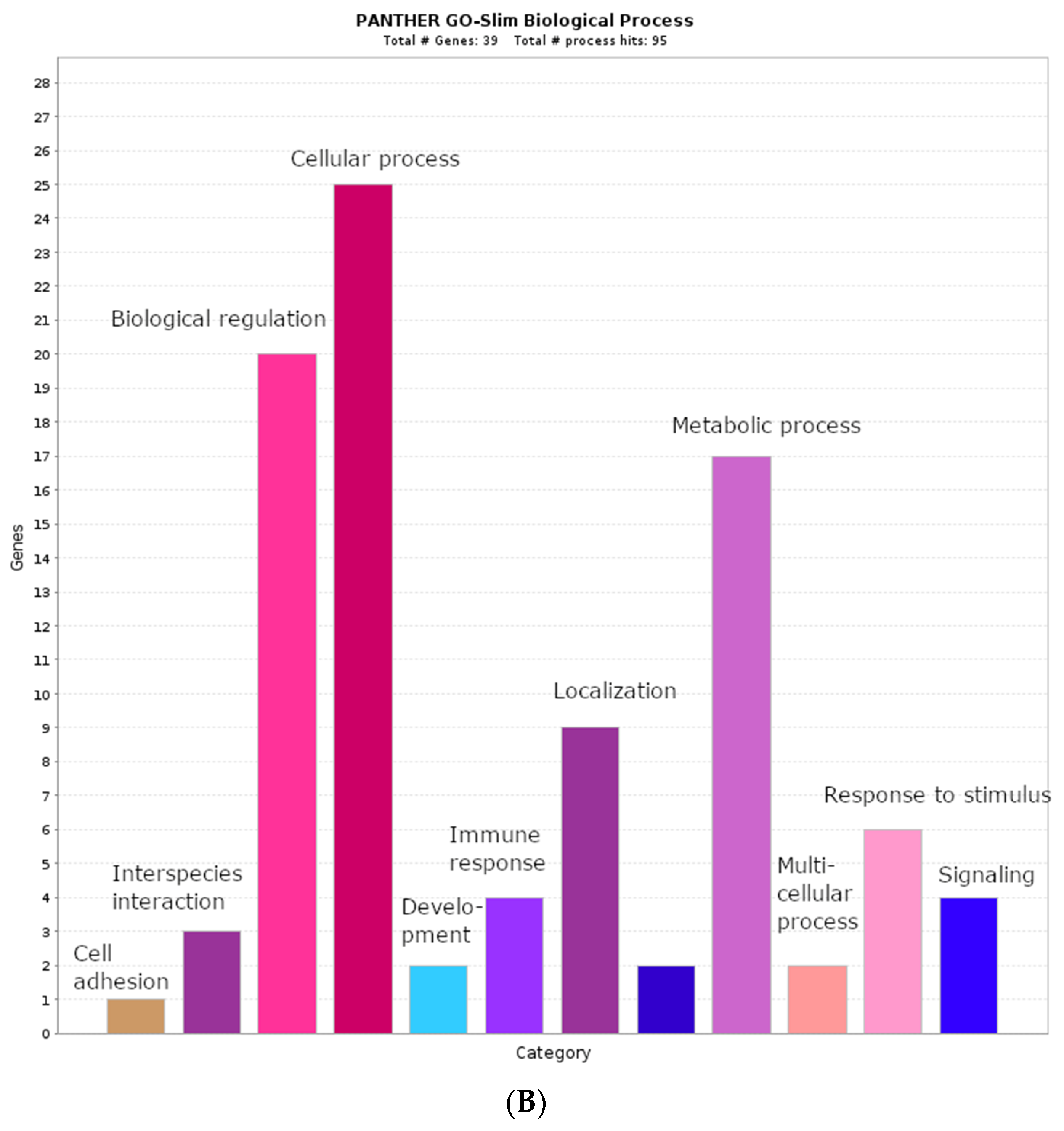
2.2. Biological Process Categories
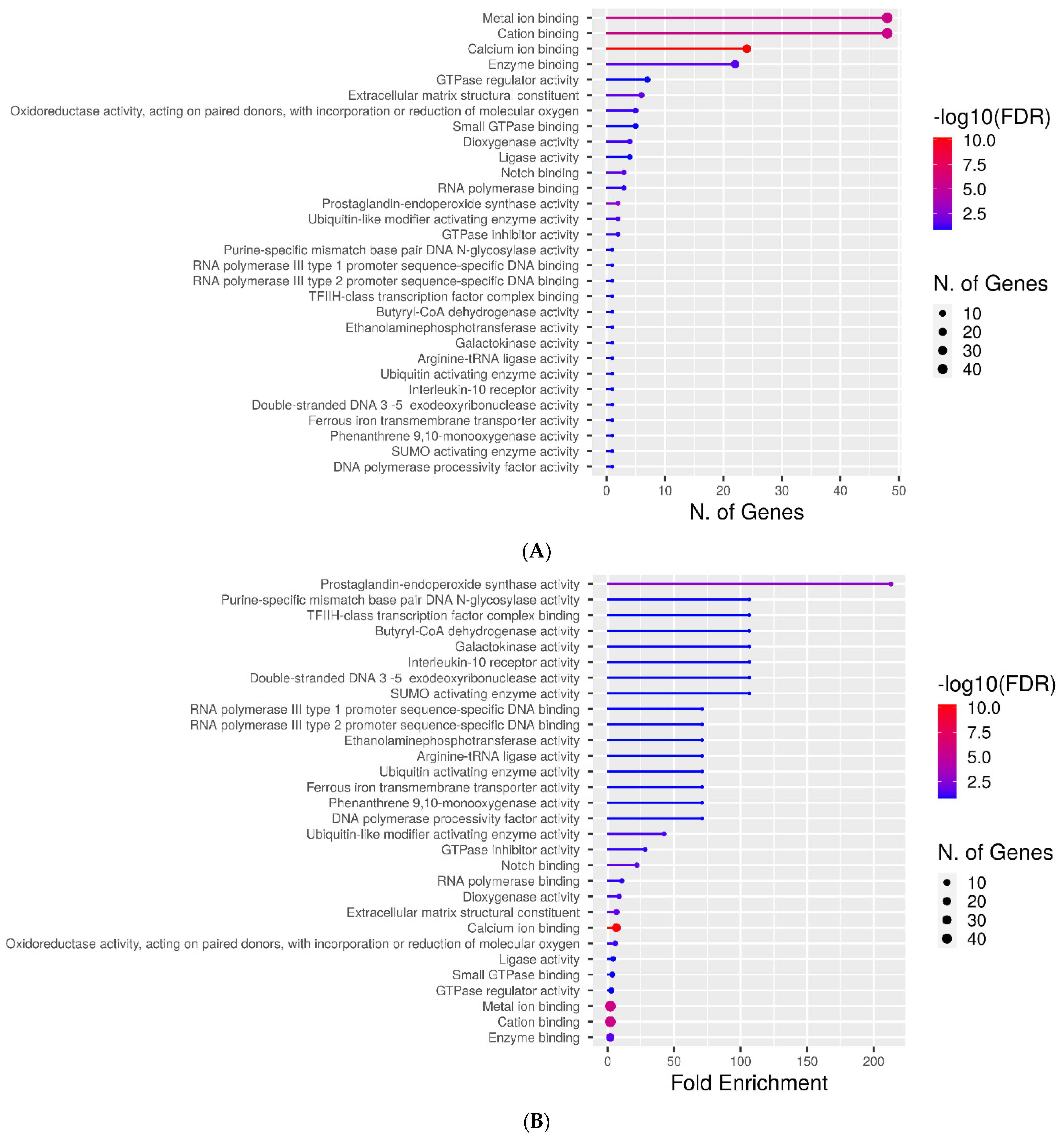
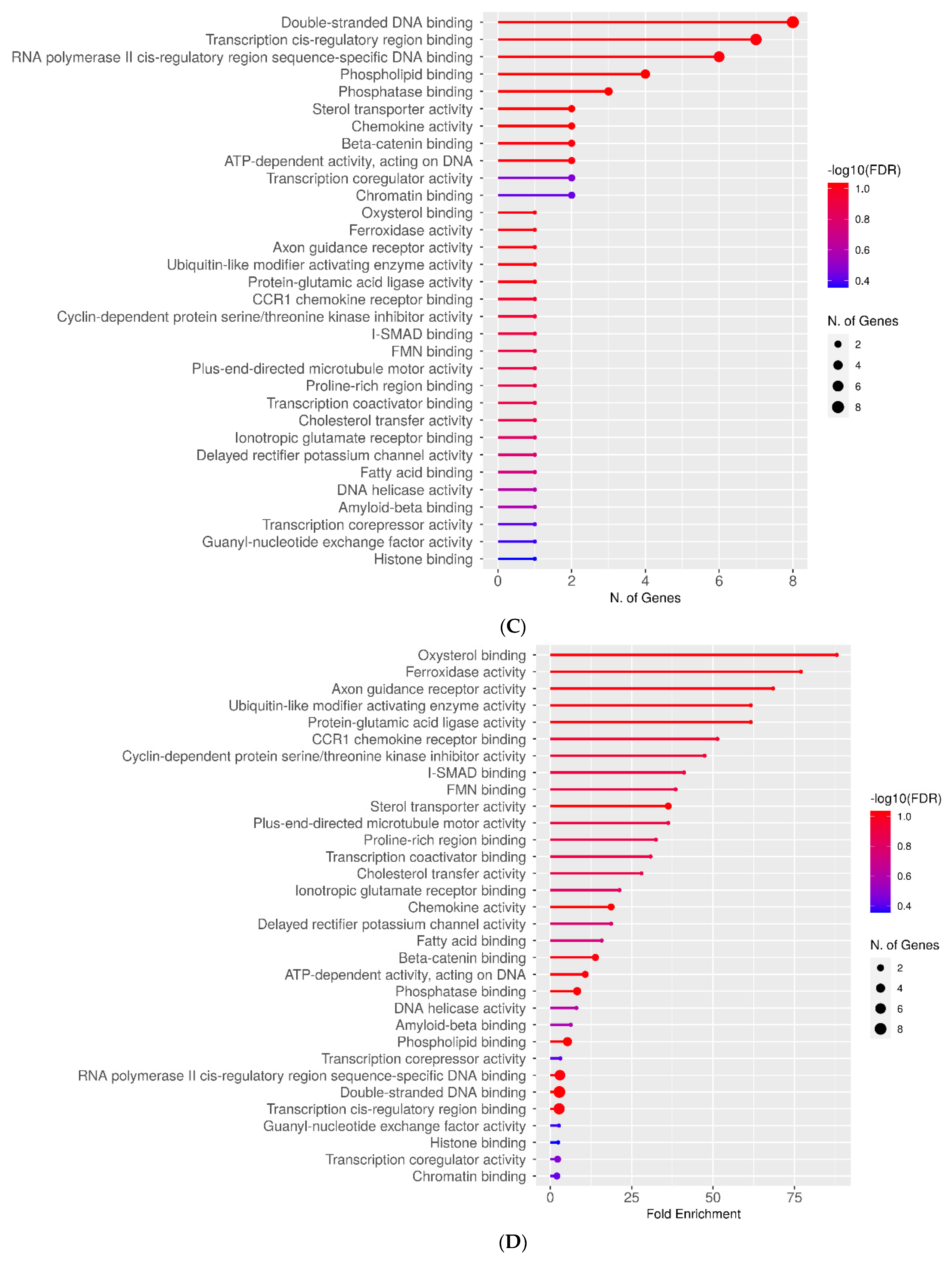
2.3. Molecular Function Categories
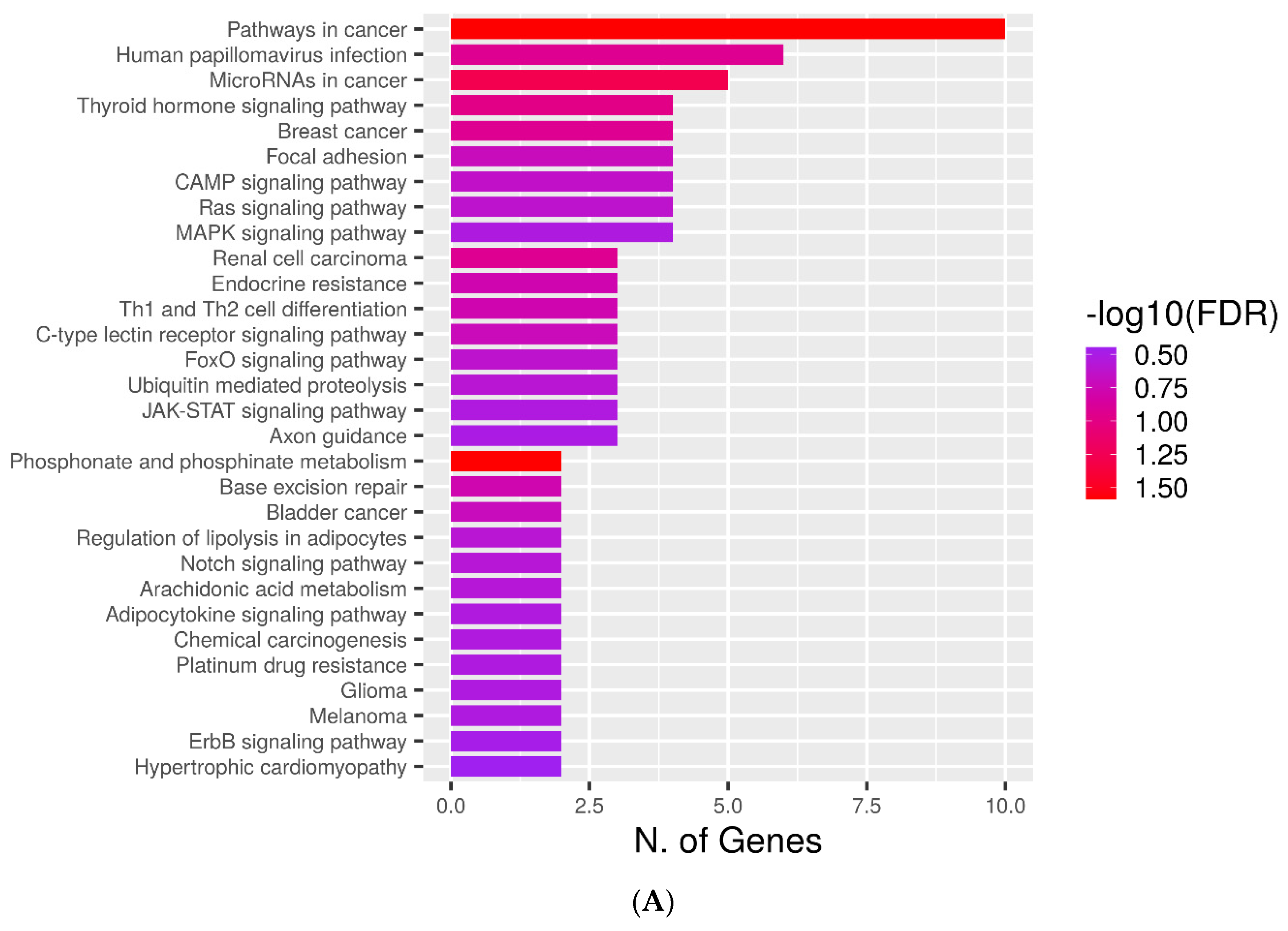
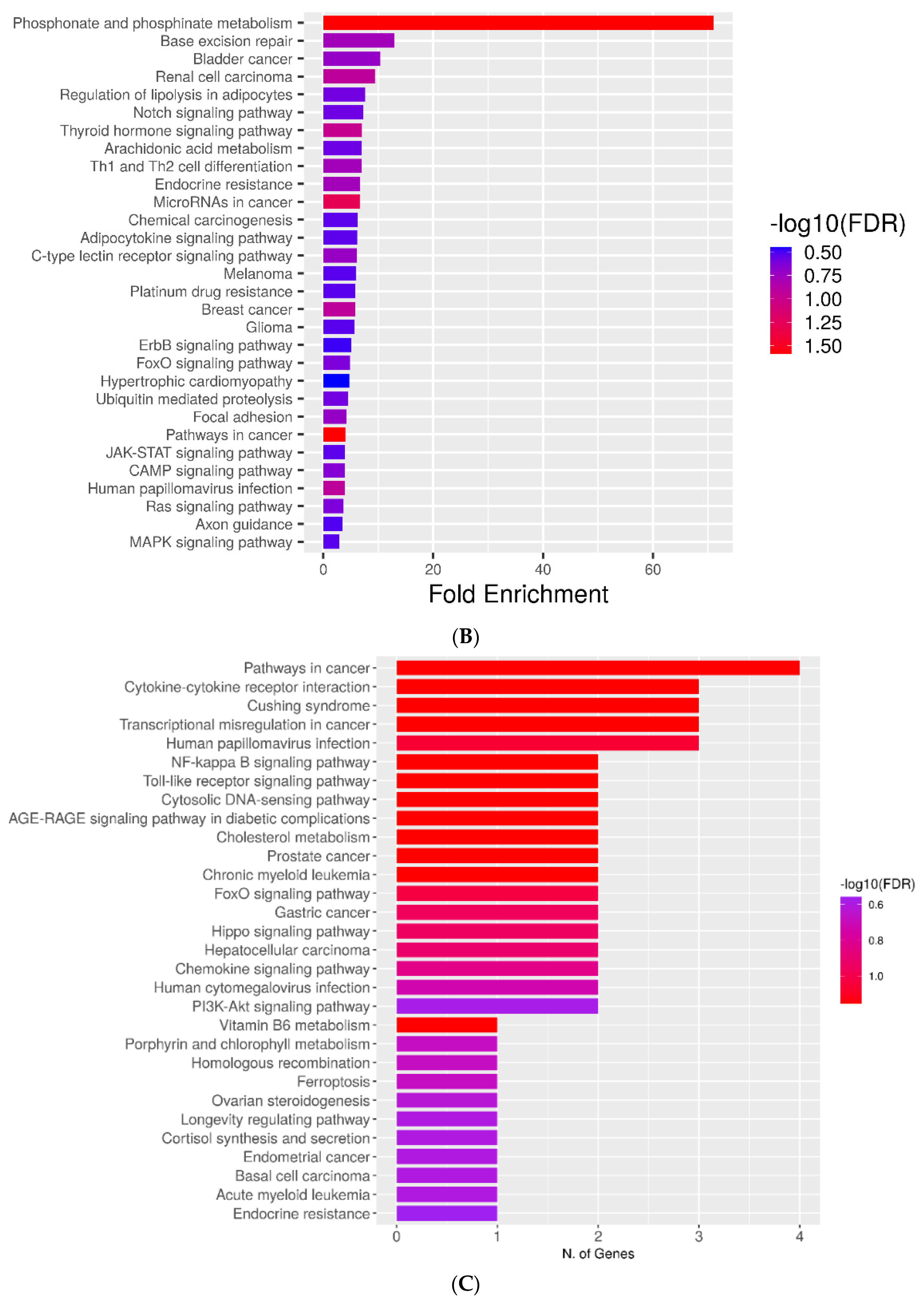
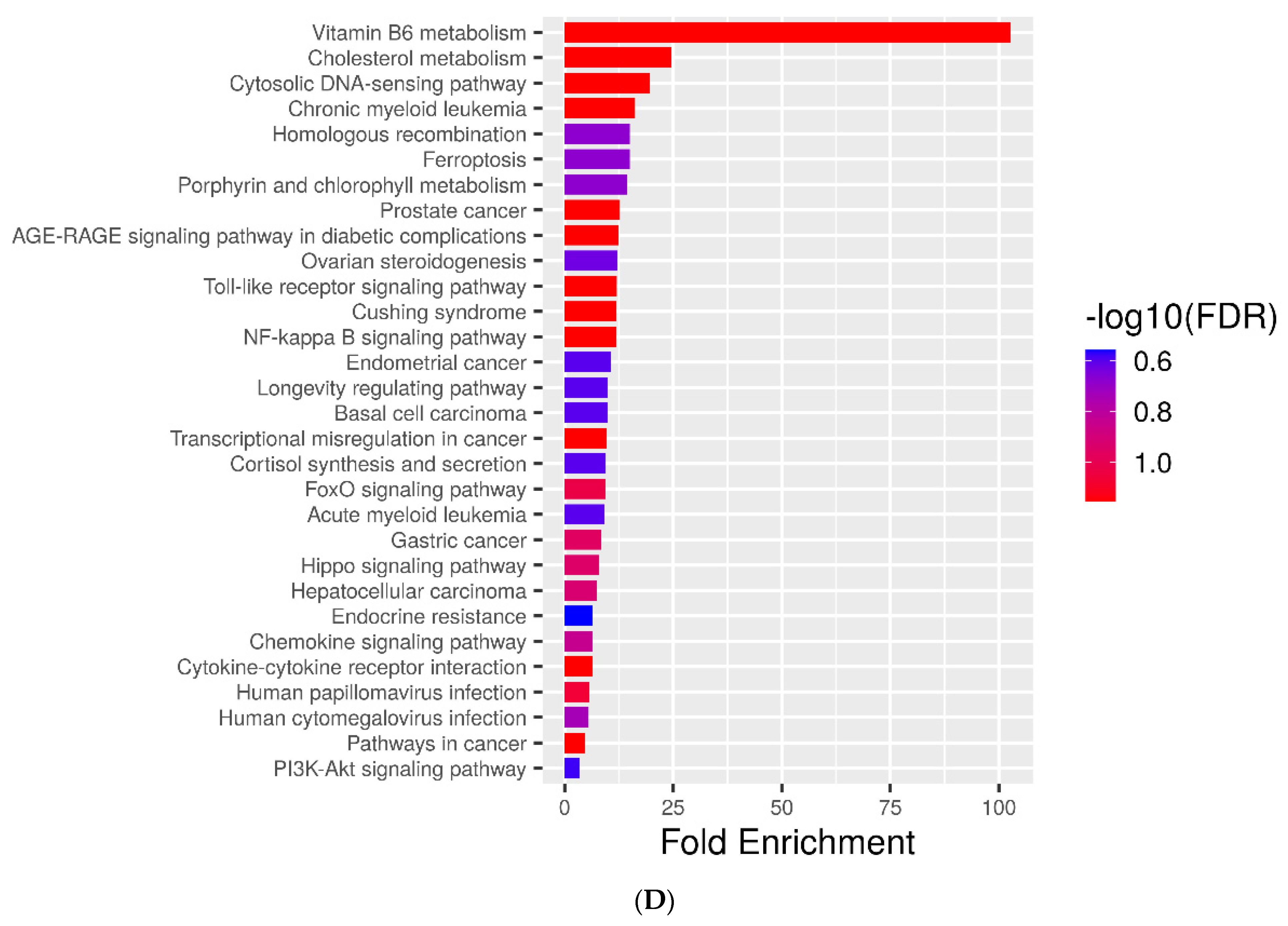
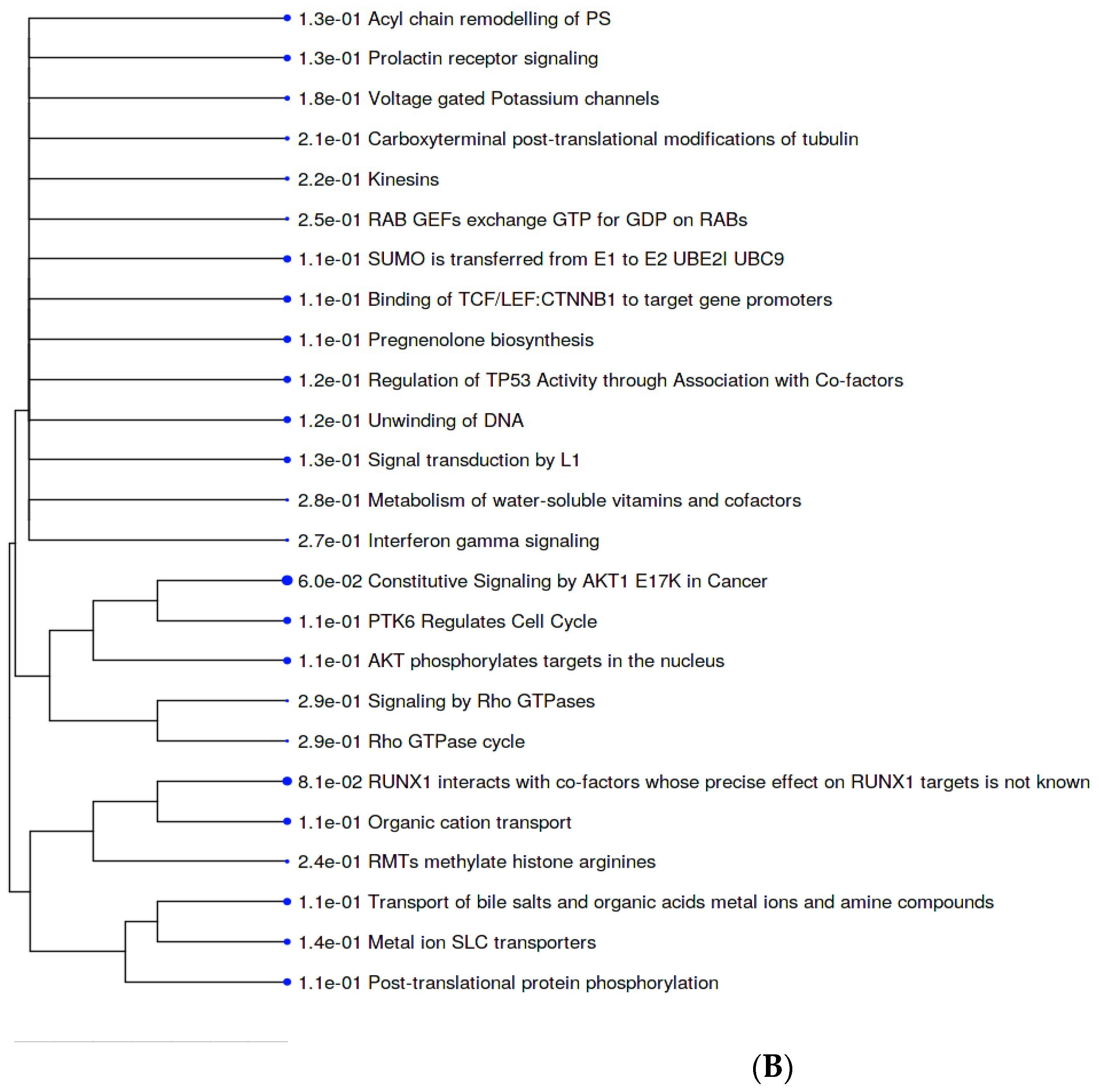
2.4. KEGG Pathways
2.5. Metabolic Pathways
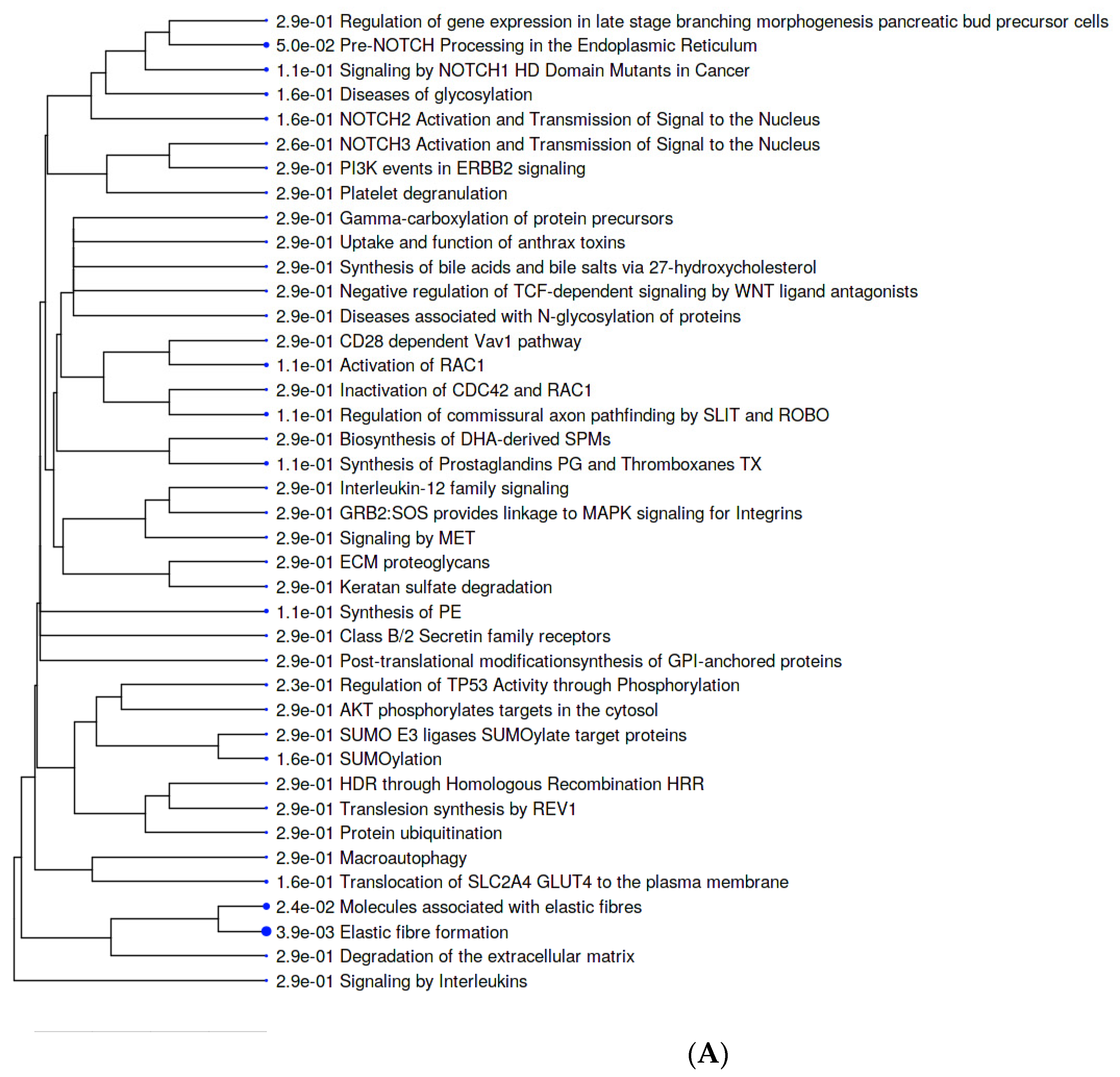
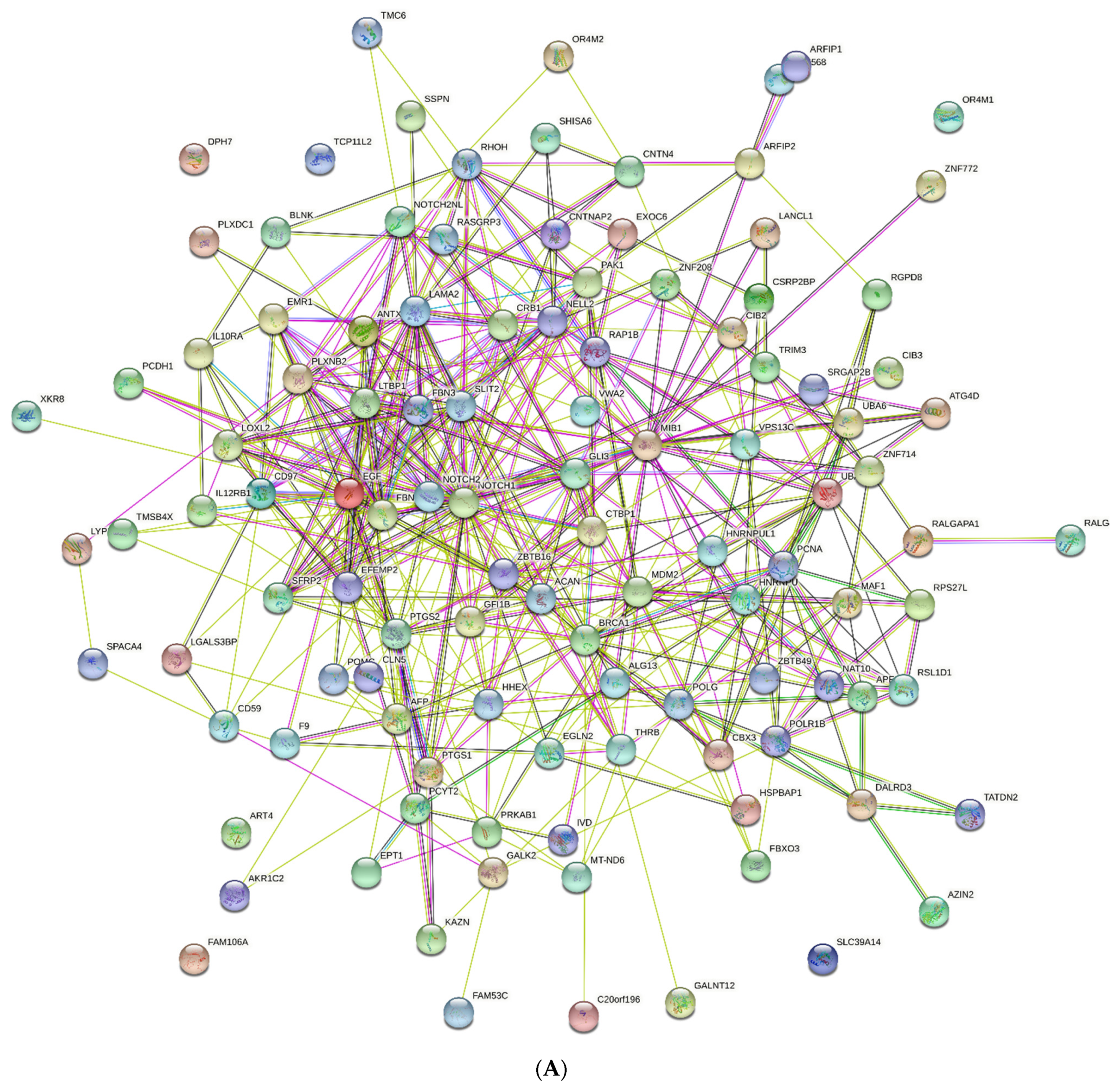
2.6. PPI Networks
3. Discussion
3.1. AFP14–20-like Motif-Containing Proteins
3.2. GIP-9-like Motif-Containing Proteins
4. Materials and Methods
4.1. Search for Short Linear Motifs
4.2. Gene Ontology Analysis
4.3. Gene Set Enrichment Analysis
4.4. KEGG Pathway Enrichment Analysis
4.5. Metabolic Pathway Analysis
4.6. PPI Network Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lesk, A.M. Introduction to Protein Science. In Architecture, Function and Genomics, 3rd ed.; Oxford University Press: Oxford, UK, 2016; p. 466. [Google Scholar]
- Bordin, N.; Sillitoe, I.; Lees, J.G.; Orengo, C. Tracing evolution through protein structures: Nature captured in a few thousand folds. Front. Mol. Biosci. 2021, 8, 668184. [Google Scholar] [CrossRef]
- Narunsky, A.; Ben-Tal, N.; Kolodny, R. Navigating among known structures in protein space. Methods Mol. Biol. 2019, 1851, 233–249. [Google Scholar] [CrossRef]
- Nepomnyachiy, S.; Ben-Tal, N.; Kolodny, R. Complex evolutionary footprints revealed in an analysis of reused protein segments of diverse lengths. Proc. Natl. Acad. Sci. USA 2017, 114, 11703–11708. [Google Scholar] [CrossRef]
- Neduva, V.; Russell, R.B. Linear motifs: Evolutionary interaction switches. FEBS Lett. 2005, 579, 3342–3345. [Google Scholar] [CrossRef]
- Takada, Y.; Ye, X.; Simon, S. The integrins. Genome Biol. 2007, 8, 215. [Google Scholar] [CrossRef]
- Munro, S.; Pelham, H.R. A C-terminal signal prevents secretion of luminal ER proteins. Cell 1987, 48, 899–907. [Google Scholar] [CrossRef]
- Li, S.S. Specificity and versatility of SH3 and other proline-recognition domains: Structural basis and implications for cellular signal transduction. Biochem. J. 2005, 390, 641–653. [Google Scholar] [CrossRef]
- Verschueren, E.; Vanhee, P.; Van der Sloot, A.M.; Serrano, L.; Rousseau, F.; Schymkowitz, J. Protein design with fragment databases. Curr. Opin. Struct. Biol. 2011, 21, 452–459. [Google Scholar] [CrossRef]
- Mackenzie, C.O.; Grigoryan, G. Protein structural motifs in prediction and design. Curr. Opin. Struct. Biol. 2017, 44, 161–167. [Google Scholar] [CrossRef]
- Akiva, E.; Friedlander, G.; Itzhaki, Z.; Margalit, H. A dynamic view of domain-motif interactions. PLoS Comput. Biol. 2012, 8, e1002341. [Google Scholar] [CrossRef]
- Kliche, J.; Ivarsson, Y. Orchestrating serine/threonine phosphorylation and elucidating downstream effects by short linear motifs. Biochem. J. 2022, 479, 1–22. [Google Scholar] [CrossRef]
- Hartooni, N.; Sung, J.; Jain, A.; Morgan, D.O. Single-molecule analysis of specificity and multivalency in binding of short linear substrate motifs to the APC/C. Nat. Commun. 2022, 13, 341. [Google Scholar] [CrossRef]
- Kelil, A.; Levy, E.D.; Michnick, S.W. Evolution of domain-peptide interactions to coadapt specificity and affinity to functional diversity. Proc. Natl. Acad. Sci. USA 2016, 113, E3862–E3871. [Google Scholar] [CrossRef]
- Dionne, U.; Bourgault, É.; Dubé, A.K.; Bradley, D.; Chartier, F.J.M.; Dandage, R.; Dibyachintan, S.; Després, P.C.; Gish, G.D.; Pham, N.T.H.; et al. Protein context shapes the specificity of SH3 domain-mediated interactions in vivo. Nat. Commun. 2021, 12, 1597. [Google Scholar] [CrossRef]
- Reys, V.; Labesse, G. SLiMAn: An integrative web server for exploring short linear motif-mediated interactions in interactome. BioRxiv 2022. Available online: https://www.biorxiv.org/content/10.1101/2022.01.14.476361v1 (accessed on 21 April 2022).
- Moldogazieva, N.T.; Terent’ev, A.A.; Shaĭtan, K.V. Relationship between structure and function of alpha-fetoprotein: Conformational status and biological activity. Biomeditsinskaia Khimiia 2005, 51, 127–151. [Google Scholar]
- Moldogazieva, N.T.; Shaitan, K.V.; Antonov, M.Y.; Mokhosoev, I.M.; Levtsova, O.V.; Terentiev, A.A. Human EGF-derived direct and reverse short linear motifs: Conformational dynamics insight into the receptor-binding residues. J. Biomol. Struct. Dyn. 2018, 36, 1286–1305. [Google Scholar] [CrossRef]
- Zhu, Z.; West, G.R.; Wang, D.C.; Collins, A.B.; Xiao, H.; Bai, Q.; Mesfin, F.B.; Wakefield, M.R.; Fang, Y. AFP peptide (AFPep) as a potential growth factor for prostate cancer. Med. Oncol. 2021, 39, 2. [Google Scholar] [CrossRef]
- Mizejewski, G.J.; Eisele, L.; Maccoll, R. Anticancer versus antigrowth activities of three analogs of the growth-inhibitory peptide: Relevance to physicochemical properties. Anticancer Res. 2006, 26, 3071–3076. [Google Scholar]
- Moldogazieva, N.T.; Terentiev, A.A.; Antonov, M.Y.; Kazimirsky, A.N.; Shaitan, K.V. Correlation between biological activity and conformational dynamics properties of tetra- and pentapeptides derived from fetoplacental proteins. Biochemistry 2012, 77, 469–484. [Google Scholar] [CrossRef]
- Isernia, C.; Malgieri, G.; Russo, L.; D’Abrosca, G.; Baglivo, I.; Pedone, P.V.; Fattorusso, R. Zinc fingers. Met. Ions Life Sci. 2020, 20, 415–435. [Google Scholar] [CrossRef]
- Tachmatzidi, E.C.; Galanopoulou, O.; Talianidis, I. Transcription control of liver development. Cells 2021, 10, 2026. [Google Scholar] [CrossRef] [PubMed]
- Zhou, B.; Lin, W.; Long, Y.; Yang, Y.; Zhang, H.; Wu, K.; Chu, Q. Notch signaling pathway: Architecture, disease, and therapeutics. Signal Transduct. Target. Ther. 2022, 7, 95. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.; Dong, X.; Baldauf, C.; Chen, H.; Zhou, Y.; Springer, T.A.; Luo, X.; Zhong, C.; Gräter, F.; Ding, J. A novel calcium-binding site of von Willebrand factor A2 domain regulates its cleavage by ADAMTS13. Blood 2011, 117, 4623–4631. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ibrahim, A.M.; Sabet, S.; El-Ghor, A.A.; Kamel, N.; Anis, S.E.; Morris, J.S.; Stein, T. Fibulin-2 is required for basement membrane integrity of mammary epithelium. Sci. Rep. 2018, 8, 14139. [Google Scholar] [CrossRef]
- Zhu, W.; Jarman, K.E.; Lokman, N.A.; Neubauer, H.A.; Davies, L.T.; Gliddon, B.L.; Taing, H.; Moretti, P.A.B.; Oehler, M.K.; Pitman, M.R.; et al. CIB2 negatively regulates oncogenic signaling in ovarian cancer via sphingosine kinase 1. Cancer Res. 2017, 77, 4823–4834. [Google Scholar] [CrossRef]
- Fossati, M.; Pizzarelli, R.; Schmidt, E.R.; Kupferman, J.V.; Stroebel, D.; Polleux, F.; Charrier, C. SRGAP2 and its human-specific paralog co-regulate the development of excitatory and inhibitory synapses. Neuron 2016, 91, 356–369. [Google Scholar] [CrossRef]
- Spike, C.A.; Tsukamoto, T.; Greenstein, D. Ubiquitin ligases and a processive proteasome facilitate protein clearance during the oocyte-to-embryo transition in Caenorhabditis elegans. Genetics 2022, 221, iyac051. [Google Scholar] [CrossRef]
- Groettrup, M.; Pelzer, C.; Schmidtke, G.; Hofmann, K. Activating the ubiquitin family: UBA6 challenges the field. Trends Biochem. Sci. 2008, 33, 230–237. [Google Scholar] [CrossRef]
- Arshad, M.; Abdul Hamid, N.; Chan, M.C.; Ismail, F.; Tan, G.C.; Pezzella, F.; Tan, K.L. NUB1 and FAT10 proteins as potential novel biomarkers in cancer: A translational perspective. Cells 2021, 10, 2176. [Google Scholar] [CrossRef]
- Bodnar, R.J. Epidermal growth factor and epidermal growth factor r4eceptor: The yin and yang in the treatment of cutaneous wounds and cancer. Adv. Wound Care 2013, 2, 24–29. [Google Scholar] [CrossRef] [PubMed]
- González-Magaña, A.; Blanco, F.J. Human PCNA structure, function and interactions. Biomolecules 2020, 10, 570. [Google Scholar] [CrossRef] [PubMed]
- Vítová, M.; Lanta, V.; Čížková, M.; Jakubec, M.; Rise, F.; Halskau, Ø.; Bišová, K.; Furse, S. The biosynthesis of phospholipids is linked to the cell cycle in a model eukaryote. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2021, 1866, 158965. [Google Scholar] [CrossRef]
- Agrotis, A.; Pengo, N.; Burden, J.J.; Ketteler, R. Redundancy of human ATG4 protease isoforms in autophagy and LC3/GABARAP processing revealed in cells. Autophagy 2019, 15, 976–997. [Google Scholar] [CrossRef] [PubMed]
- Bakovic, M.; Fullerton, M.D.; Michel, V. Metabolic and molecular aspects of ethanolamine phospholipid biosynthesis: The role of CTP:phosphoethanolamine cytidylyltransferase (Pcyt2). Biochem. Cell Biol. 2007, 85, 283–300. [Google Scholar] [CrossRef] [PubMed]
- Kurata, M.; Onishi, I.; Takahara, T.; Yamazaki, Y.; Ishibashi, S.; Goitsuka, R.; Kitamura, D.; Takita, J.; Hayashi, Y.; Largaesapda, D.A.; et al. C/EBPβ induces B-cell acute lymphoblastic leukemia and cooperates with BLNK mutations. Cancer Sci. 2021, 112, 4920–4930. [Google Scholar] [CrossRef]
- Sherbet, D.P.; Papari-Zareei, M.; Khan, N.; Sharma, K.K.A.; Rambally, S.; Chattopadhyay, A.; Andersson, S.; Agarwal, A.K.; Auchus, R.J. Cofactors, redox state, and directional preferences of hydroxysteroid dehydrogenases. Mol. Cell Endocrinol. 2007, 265, 83–88. [Google Scholar] [CrossRef]
- Goltsov, A.; Swat, M.; Peskov, K.; Kosinsky, Y. Cycle network model of prostaglandin H synthase-1. Pharmaceuticals 2020, 13, 265. [Google Scholar] [CrossRef]
- Tang, R.; Wu, Z.; Lu, F.; Wang, C.; Wu, B.; Wang, J.; Zhu, Y. Identification of critical pathways and hub genes in LanCL1-overexpressed prostate cancer cells. OncoTargets Ther. 2020, 13, 7653–7664. [Google Scholar] [CrossRef]
- Saeed, K.; Östling, P.; Björkman, M.; Mirtti, T.; Alanen, K.; Vesterinen, T.; Sankila, A.; Lundin, J.; Lundin, M.; Rannikko, A.; et al. Androgen receptor-interacting protein HSPBAP1 facilitates growth of prostate cancer cells in androgen-deficient conditions. Int. J. Cancer 2015, 136, 2535–2545. [Google Scholar] [CrossRef]
- Marxsen, J.H.; Stengel, P.; Doege, K.; Heikkinen, P.; Jokilehto, T.; Wagner, T.; Jelkmann, W.; Jaakkola, P.; Metzen, E. Hypoxia-inducible factor-1 (HIF-1) promotes its degradation by induction of HIF-alpha-prolyl-4-hydroxylases. Biochem. J. 2004, 381, 761–767. [Google Scholar] [CrossRef] [PubMed]
- Hannan, N.J.; Jones, R.L.; White, C.A.; Salamonsen, L.A. The chemokines, CX3CL1, CCL14, and CCL4, promote human trophoblast migration at the feto-maternal interface. Biol. Reprod. 2006, 74, 896–904. [Google Scholar] [CrossRef] [PubMed]
- Hlouskova, A.; Bielik, P.; Bonczek, O.; Balcar, V.J.; Šerý, O. Mutations in AXIN2 gene as a risk factor for tooth agenesis and cancer: A review. Neuro. Endocrinol. Lett. 2017, 38, 131–137. [Google Scholar] [PubMed]
- Samatov, T.R.; Wicklein, D.; Tonevitsky, A.G. L1CAM: Cell adhesion and more. Prog. Histochem. Cytochem. 2016, 51, 25–32. [Google Scholar] [CrossRef]
- Jouhilahti, E.M.; Madissoon, E.; Vesterlund, L.; Töhönen, V.; Krjutškov, K.; Plaza Reyes, A.; Petropoulos, S.; Månsson, R.; Linnarsson, S.; Bürglin, T.; et al. The human PRD-like homeobox gene LEUTX has a central role in embryo genome activation. Development 2016, 143, 3459–3469. [Google Scholar] [CrossRef] [PubMed]
- Yan, T.; Ooi, W.F.; Qamra, A.; Cheung, A.; Ma, D.; Sundaram, G.M.; Xu, C.; Xing, M.; Poon, L.; Wang, J.; et al. HoxC5 and miR-615-3p target newly evolved genomic regions to repress hTERT and inhibit tumorigenesis. Nat. Commun. 2018, 9, 100. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Yang, J.; Liao, W.; Liu, X.; Zhang, H.; Wang, S.; Wang, D.; Feng, J.; Yu, L.; Zhu, W.G. Cytosolic FoxO1 is essential for the induction of autophagy and tumour suppressor activity. Nat. Cell Biol. 2010, 12, 665–675. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Zheng, B.; Xue, S.; Chen, C. Knockdown of miR-183 enhances the cisplatin-induced apoptosis in esophageal cancer through increase of FOXO1 expression. OncoTargets Ther. 2020, 13, 8463–8474. [Google Scholar] [CrossRef]
- Zhao, Y.; Zhang, T.; Zhao, Y.; Zhou, J. Distinct association of RUNX family expression with genetic alterations and clinical outcome in acute myeloid leukemia. Cancer Biomark. 2020, 29, 387–397. [Google Scholar] [CrossRef]
- McGrath, D.A.; Fifield, B.A.; Marceau, A.H.; Tripathi, S.; Porter, L.A.; Rubin, S.M. Structural basis of divergent cyclin-dependent kinase activation by Spy1/RINGO proteins. EMBO J. 2017, 36, 2251–2262. [Google Scholar] [CrossRef]
- Jones, M.L.; Baris, Y.; Taylor, M.R.G.; Yeeles, J.T.P. Structure of a human replisome shows the organisation and interactions of a DNA replication machine. EMBO J. 2021, 40, e108819. [Google Scholar] [CrossRef] [PubMed]
- Lobito, A.A.; Ramani, S.R.; Tom, I.; Bazan, J.F.; Luis, E.; Fairbrother, W.J.; Ouyang, W.; Gonzalez, L.C. Murine insulin growth factor-like (IGFL) and human IGFL1 proteins are induced in inflammatory skin conditions and bind to a novel tumor necrosis factor receptor family member, IGFLR1. J. Biol. Chem. 2011, 286, 18969–18981. [Google Scholar] [CrossRef] [PubMed]
- Song, W.; Shao, Y.; He, X.; Gong, P.; Yang, Y.; Huang, S.; Zeng, Y.; Wei, L.; Zhang, J. IGFLR1 as a novel prognostic biomarker in clear cell renal cell cancer correlating with immune infiltrates. Front. Mol. Biosci. 2020, 7, 565173. [Google Scholar] [CrossRef] [PubMed]
- Chao, A.; Tsai, C.L.; Jung, S.M.; Chuang, W.C.; Kao, C.; Hsu, A.; Chen, S.H.; Lin, C.Y.; Lee, Y.C.; Lee, Y.S.; et al. BAI1-associated protein 2-Like 1 (BAIAP2L1) is a potential biomarker in ovarian cancer. PLoS ONE 2015, 10, e0133081. [Google Scholar] [CrossRef] [PubMed]
- Mathema, V.B.; Na-Bangchang, K. Regulatory roles of brain-specific angiogenesis inhibitor 1(BAI1) protein in inflammation, tumorigenesis and phagocytosis: A brief review. Crit. Rev. Oncol. Hematol. 2017, 111, 81–86. [Google Scholar] [CrossRef]
- Sun, N.; Jiang, L.; Ye, M.; Wang, Y.; Wang, G.; Wan, X.; Zhao, Y.; Wen, X.; Liang, L.; Ma, S.; et al. TRIM35 mediates protection against influenza infection by activating TRAF3 and degrading viral PB2. Protein Cell 2020, 11, 894–914. [Google Scholar] [CrossRef]
- Vlasova, I.I.; Sokolov, A.V.; Kostevich, V.A.; Mikhalchik, E.V.; Vasilyev, V.B. Myeloperoxidase-induced oxidation of albumin and ceruloplasmin: Role of tyrosines. Biochemistry 2019, 84, 652–662. [Google Scholar] [CrossRef]
- Zhang, L.; Zhou, D.W.; Guan, W.; Ren, W.; Sun, W.; Shi, J.; Lin, Q.; Zhang, J.; Qiao, T.; Ye, Y.; et al. Pyridoxine 5′-phosphate oxidase is a novel therapeutic target and regulated by the TGF-β signalling pathway in epithelial ovarian cancer. Cell Death Dis. 2017, 8, 3214. [Google Scholar] [CrossRef]
- Riedel, E.O.; Hinrichs, A.; Kemter, E.; Dahlhoff, M.; Backman, M.; Rathkolb, B.; Prehn, C.; Adamski, J.; Renner, S.; Blutke, A.; et al. Functional changes of the liver in the absence of growth hormone (GH) action—Proteomic and metabolomic insights from a GH receptor deficient pig model. Mol. Metab. 2020, 36, 100978. [Google Scholar] [CrossRef]
- Moldogazieva, N.T.; Ostroverkhova, D.S.; Kuzmich, N.N.; Kadochnikov, V.V.; Terentiev, A.A.; Porozov, Y.B. Elucidating binding sites and affinities of ERα agonists and antagonists to human alpha-fetoprotein by in silico modeling and point mutagenesis. Int. J. Mol. Sci. 2020, 21, 893. [Google Scholar] [CrossRef]
- Terentiev, A.A.; Moldogazieva, N.T. Structural and functional mapping of alpha-fetoprotein. Biochemistry 2006, 71, 120–132. [Google Scholar] [CrossRef] [PubMed]
- Lu, C.C.; Chen, Y.T.; Chen, S.Y.; Hsu, Y.M.; Lin, C.C.; Tsao, J.W.; Juan, Y.N.; Yang, J.S.; Tsai, F.J. Hematopoietically expressed homeobox gene is associated with type 2 diabetes in KK Cg-Ay/J mice and a Taiwanese Han Chinese population. Exp. Ther. Med. 2018, 16, 185–191. [Google Scholar] [CrossRef] [PubMed]
- Ye, Q.; Liu, J.; Xie, K. Zinc finger proteins and regulation of the hallmarks of cancer. Histol. Histopathol. 2019, 34, 1097–1109. [Google Scholar] [CrossRef] [PubMed]
- Jen, J.; Wang, Y.C. Zinc finger proteins in cancer progression. J. Biomed. Sci. 2016, 23, 53. [Google Scholar] [CrossRef]
- Di, D.; Chen, L.; Guo, Y.; Wang, L.; Zhao, C.; Ju, J. BCSC-1 suppresses human breast cancer metastasis by inhibiting NF-κB signaling. Int. J. Oncol. 2018, 52, 1674–1684. [Google Scholar] [CrossRef]
- Fasoulakis, Z.; Daskalakis, G.; Theodora, M.; Antsaklis, P.; Sindos, M.; Diakosavvas, M.; Angelou, K.; Loutradis, D.; Kontomanolis, E.N. The relevance of Notch signaling in cancer progression. Adv. Exp. Med. Biol. 2021, 1287, 169–181. [Google Scholar] [CrossRef]
- Tekcham, D.S.; Chen, D.; Liu, Y.; Ling, T.; Zhang, Y.; Chen, H.; Wang, W.; Otkur, W.; Qi, H.; Xia, T.; et al. F-box proteins and cancer: An update from functional and regulatory mechanism to therapeutic clinical prospects. Theranostics 2020, 10, 4150–4167. [Google Scholar] [CrossRef]
- Ma, C.; Hoffmann, F.W.; Marciel, M.P.; Page, K.E.; Williams-Aduja, M.A.; Akana, E.N.L.; Gojanovich, G.S.; Gerschenson, M.; Urschitz, J.; Moisyadi, S.; et al. Upregulated ethanolamine phospholipid synthesis via selenoprotein I is required for effective metabolic reprogramming during T cell activation. Mol. Metab. 2021, 47, 101170. [Google Scholar] [CrossRef]
- Moldogazieva, N.T.; Mokhosoev, I.M.; Mel’nikova, T.I.; Zavadskiy, S.P.; Kuz’menko, A.N.; Terentiev, A.A. Dual character of reactive oxygen, nitrogen, and halogen species: Endogenous sources, interconversions and neutralization. Biochemistry 2020, 85, S56–S78. [Google Scholar] [CrossRef]
- Spinelli, S.; Begani, G.; Guida, L.; Magnone, M.; Galante, D.; D’Arrigo, C.; Scotti, C.; Iamele, L.; De Jonge, H.; Zocchi, E.; et al. LANCL1 binds abscisic acid and stimulates glucose transport and mitochondrial respiration in muscle cells via the AMPK/PGC-1α/Sirt1 pathway. Mol. Metab. 2021, 53, 101263. [Google Scholar] [CrossRef]
- Kim, L.C.; Simon, M.C. Hypoxia-inducible factors in cancer. Cancer Res. 2022, 82, 195–196. [Google Scholar] [CrossRef] [PubMed]
- Pearson, W.R. Rapid and sensitive sequence comparison with FASTP and FASTA. Methods Enzymol. 1990, 183, 63–98. [Google Scholar] [CrossRef] [PubMed]
- UniProt Consortium. UniProt: The universal protein knowledgebase in 2021. Nucleic Acids Res. 2021, 49, D480–D489. [Google Scholar] [CrossRef] [PubMed]
- Mi, H.; Ebert, D.; Muruganujan, A.; Mills, C.; Albou, L.P.; Mushayamaha, T.; Thomas, P.D. PANTHER version 16: A revised family classification, tree-based classification tool, enhancer regions and extensive API. Nucleic Acids Res. 2021, 49, D394–D403. [Google Scholar] [CrossRef]
- Safran, M.; Rosen, N.; Twik, M.; BarShir, R.; Iny Stein, T.; Dahary, D.; Fishilevich, S.; Lancet, D. The GeneCards Suite. In Practical Guide to Life Science Databases; Dahary, D., Ed.; LifeMap Sciences Inc.: Marshfield, MA, USA, 2022. [Google Scholar]
- Cunningham, F.; Allen, J.E.; Allen, J.; Alvarez-Jarreta, J.; Amode, M.R.; Armean, I.M.; Austine-Orimoloye, O.; Azov, A.G.; Barnes, I.; Bennett, R.; et al. Ensembl 2022. Nucleic Acids Res. 2022, 50, D988–D995. [Google Scholar] [CrossRef]
- Ge, S.X.; Jung, D.; Yao, R. ShinyGO: A graphical gene-set enrichment tool for animals and plants. Bioinformatics 2020, 36, 2628–2629. [Google Scholar] [CrossRef]
- Gillespie, M.; Jassal, B.; Stephan, R.; Milacic, M.; Rothfels, K.; Senff-Ribeiro, A.; Griss, J.; Sevilla, C.; Matthews, L.; Gong, C.; et al. The reactome pathway knowledgebase 2022. Nucleic Acids Res. 2022, 50, D687–D692. [Google Scholar] [CrossRef]
- Kanehisa, M.; Sato, Y.; Kawashima, M. KEGG mapping tools for uncovering hidden features in biological data. Protein Sci. 2022, 31, 47–53. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Nastou, K.C.; Lyon, D.; Kirsch, R.; Pyysalo, S.; Doncheva, N.T.; Legeay, M.; Fang, T.; Bork, P.; et al. The STRING database in 2021: Customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021, 49, D605–D612. [Google Scholar] [CrossRef]

| Protein Name | Entry Code | Gene Symbol | Alignment | Aa Positions | Identity | E-Value | GO Molecular Functions | GO Biological Processes | Reference |
|---|---|---|---|---|---|---|---|---|---|
| Tripartite motif-containing protein 3 (RING finger protein HAC1) | TR:Q1KXY7 | TRIM3 | LDSYQCT : : . : : : LDRYQCP | 26–32 | 71.4% | 2.2 × 10−4 | Metal ion binding, ubiquitin-protein ligase/transferase activity | Transcriptional regulation, UPS-mediated protein degradation | [22] |
| Zinc finger protein 714 | TR:A0A087WV13 | ZNF714 | LDSYQCT . : : : : . ENSYQCE | 15–21 | 57.1% | 3.0 × 10−2 | Transcription factor | Transcriptional regulation | [22] |
| Hematopoietically-expressed homeobox protein HHEX | TR:F8VU08 | HHEX | LDSYQCT : : : : : . LDSSQCS | 59–65 | 71.4% | 9.4 × 10−4 | DNA binding, transcription activator activity | Transcriptional regulation, anterior–posterior pattern specification, B- cell differentiation | [23] |
| Neurogenic locus notchhomolog protein 2 | TR:A0A494C1F1 | NOTCH2 | LDSYQCT : . : . : : RDTYECT | 87–93 | 57.1% | 7.9 × 10−2 | Calcium ion binding, signaling receptor activity | Tissue morphogenesis, cell fate determination | [24] |
| von Willebrand factor A domain-containing protein 2 | SP: Q5GFL6-2 | VWA2 | LDSYQCT : : : : : LDGYQCL | 315–321 | 71.4% | 0.36 | Calcium binding activity | Cell–matrix adhesin, insulin-receptor signaling | [25] |
| EGF-containing fibulin-like extracellular matrix protein 2 | TR: E9PRQ8 | EFEMP2 | LDSYQCT . : : : : : PGSYQCT | 144–150 | 71.4% | 4.0 | Calcium ion binding | Extracellular matrix assembly, developmental processes | [26] |
| Slit-Robo RhoGTPase-activating protein 2B | TR:A0A087 × 1G6 | SRGAP2B | LDSYQCT : : . : : . LYSHQCS | 22–28 | 57.1% | 0.17 | GTPase activator activity | Neuronal morphogenesis developmental process | [27] |
| Calcium and integrin-binding family member 2 | TR:H0YND4 | CIB2 | LDSYQ-CT : : . : : : : LDNYQDCT | 13–20 | 75.0% | 3.3 × 10−2 | Calcium ion binding, integrin binding | Calcium ion homeostasis, response to ATP | [28] |
| F-box protein Fbx3 | TR: Q9UKC5 | FBX3 | LDSYQCT : : . : . : . LDDYRCS | 137–143 | 57.1% | 4.0 × 10−2 | Ubiquitin-protein transferase activity | Protein ubiquitination and degradation | [29] |
| Ubiquitin-like modifier-activating enzyme 6 | TR:Q2MD40_ | UBA6 | LDSYQCT : : . : : : LDKYQCV | 151–157 | 71.4% | 5.0 × 10−3 | ATP binding, ubiquitin-activating enzyme activity | Response to DNA damage, protein ubiquitination, embryonic development | [30,31] |
| Epidermal growth factor | TR:Q6QBS2 | EGF | LDSYQCT : : . : : . LDKYACN | 26–32 | 57.1% | 3.8 × 10−2 | Growth factor activity | Cell proliferation and survival | [32] |
| Proliferating cell nuclear antigen | TR:Q7Z6A3 | PCNA | LDSYQCT . : . : . : FDTYRCD | 57–63 | 42.9% | 0.42 | DNA binding | Cell cycle regulation, DNA replication and repair | [33] |
| Ethanolamine-phosphate cytidylyltransferase | TR:I3L1F9 | PCYT2 | LDSYQCT : : . : . : . LDKYNCD | 24–30 | 57.1% | 5.7 × 10−3 | Catalytic activity | Biosynthetic process, cell division, cell fusion, and apoptosis | [34] |
| Cysteine protease ATG4D | SP: Q86TL0-2 | ATG4D | LDSYQCT : . : . . : : LESFHCT | 40–46 | 57.1% | 0.18 | Peptidase activity | Apoptosis/autophagy/mitophagy/proteolysis | [35] |
| CTP:phosphoethanolamine cytidylyltransferase | TR:I3L1C4 | PCYT2 | LDSYQCT : : . : . : LDKYNCD | 24–30 | 57.1% | 0.21 | Transferase activity | Biosynthetic process | [36] |
| B-cell linker protein | TR: Q2MD40 | BLNK | LDSYQCT . : : : . : MDSYSCL | 1–7 | 57.1% | 9.7 × 10−4 | SH2-domain binding, signaling adaptor activity | B-cell differentiation, immune and inflammatory response | [37] |
| 3-alpha hydroxysteroid dehydrogenase III | TR:Q1KXY7 | AKR1C2 | LDS – YQCT . : : : : : MDSKYQCV | 1–7 | 62.5% | 4.4 × 10−5 | Oxidoreductase, metabolic activity | Steroid hormone metabolism | [38] |
| Prostaglandin G/H synthase 1 | SP: P23219-3 | PTGS1 | LDSYQCT : : . : : : LDRYQCD | 26–32 | 71.4% | 0.35 | Cyclooxygenase/peroxidase activity, heme binding, metal ion binding | Response to oxidative stress, inflammatory process | [39] |
| Glutathione S-transferase LANCL1 | TR:H7C2E3 | LANCL1 | LDSYQCT : : : : CDAYQCA | 59–65 | 57.1% | 0.22 | Glutathione binding, zinc ion binding | Oxidative stress response | [40] |
| HSPB1-associated protein 1 | SP: Q96EW2-2 | HSPBAP1 | LDSYQCT : : : : : : LDSYGCN | 176–182 | 71.4% | 0.12 | Oxidoreductase, dioxygenase activity | Brain development | [41] |
| Prolyl hydroxylase EGLN2 | TR:M0R2X9 | EGLN2 | LDSYQCT : : : . : LPSYHCP | 45–51 | 57.1% | 2.5 | Dioxygenase activity, oxygen sensor activity | Cell redox homeostasis, response to hypoxia | [42] |
| Protein Name | Entry Code | Gene Symbol | Alignment | Aa Positions | Identity | E-Value | Go Molecular Functions | Go Biological Processes | Reference |
|---|---|---|---|---|---|---|---|---|---|
| Zinc finger protein 547 | TR: M0QYW2 | ZNF547 | EMTPVNPGV : : . . : : : EEAPLEPGV | 60–68 | 55.6% | 3.4 | DNA binding, metal ion binding, transcription factor activity | Transcriptional regulation | [22] |
| C-C motif chemokine 4-like | SP: Q8NHW4-7 | CCL4L1 | EMTPVNPGV . : : : . : : ALTPVSPGS | 31–39 | 66.7% | 1.7 × 10−2 | Chemokine activity | Response to INF-γ, IL-1, and TNF-α; cell signaling | [43] |
| Axin 2 | TR: A0A024R8M3 | AXIN2 | EMTPVNPGV : : : : : . : . EMTPVEPAT | 361–369 | 66.7% | 1.7 | Beta-catenin binding, ubiquitin protein ligase binding | Regulation of Wnt signaling, cell death, bone mineralization | [44] |
| L1 cell adhesion molecule | TR: Q7Z2J6 | L1CAM | EMTPVNPGV . : . : : . : ATSP I NPAV | 54–62 | 44.4%1 | 1.0 | Cell adhesion molecule activity | Nervous system development | [45] |
| Paired-like homeodomain transcription factor LEUTX | SP: A8MZ59-1 | LEUTX | EMTPVNPGV . . . : : . : : . N I RPVSPG I | 69–77 | 44.4% | 2.3 | DNA binding activity | Transcriptional regulation, embryogenesis | [46] |
| Homeobox protein Hox-C5 | SP: Q00444 | HOXC5 | EMTPVNPGV : . : . : : : . EAAPLNPGM | 90–98 | 55.6% | 0.48 | DNA-binding activity, transcription factor | Anterior/posterior specification, embryonic development | [47] |
| Forkhead box protein O1 | SP: Q12778 | FOXO1 | EMTPVNPGV : : : : . : : : IMTPVDPGV | 476–484 | 77.8% | 0.12 | DNA-binding activity, transcription factor | Transcriptional regulation, metabolic response to oxidative stress | [48,49] |
| RUNX1/CBFA2T2 fusion protein type 1 | TR:D1LYX4 | RUNX1/CBFA2T2 | EMTPVNPGV . : . : : : . PLP P I NPGG | 50–58 | 44.4% | 8.5 | Transcription corepressor activity | Transcriptional regulation | [50] |
| Cyclin-dependent kinase inhibitor 1B | TR: H7C2T1 | CDKN1B | EMTPVNPGV : : : . : : . EQTPKKPGL | 91–99 | 55.6% | 2.3 | Cyclin binding, chaperone binding | Cell-cycle regulation, autophagy, response to chemicals | [51] |
| DNA replication complex GINS protein PSF2 | SP: Q9Y248 | GINS2 | EMTPVNPGV . . : . : : : . DLGPFNPGL | 32–40 | 44.4% | 7.2 | DNA binding | DNA replication, DNA repair | [52] |
| IGF-like family receptor 1 | TR: K7ESC2 | IGFLR1 | EMTPVNPGV . : : . : : : . PLTPGNPGA | 124–132 | 55.6% | 2.7 | Receptor activity | IGF-mediated signaling, inflammation process | [53,54] |
| Brain-specific angiogenesis inhibitor 1-associated protein 2-like protein 2 | TR: B0QYF0 | BAIAP2L2 | EMTPVNPGV : : : . : : : PMTPMNPGN | 98–106 | 66.7% | 4.5 × 10−2 | Cadherin-binding and cytoskeletal-binding activities | Actin cytoskeleton organization, brain development | [55,56] |
| E3 ubiquitin-protein ligase TRIM35 | TR: H0YBF3 | TRIM35 | EMTPVNPGV : . . : : . : : . EPEPVQPGM | 33–41 | 55.6% | 4.6 | Zinc ion binding, ubiquitin-protein ligase activity | Protein ubiquitination, innate immune response, apoptotic process | [57] |
| Ceruloplasmin | TR: H7C5N5 | CP | EMTPVNPGV : : : . : . EMFPRTGG I | 162–170 | 55.6% | 2.8 | Oxidoreductase activity, copper binding | Redox homeostasis | [58] |
| Pyridoxine-5’-phosphate oxidase | TR: A0A286YF38 | PNPO | EMTPVNPGV : . : . : : EVPPLGPGL | 46–54 | 44.4% | 1.5 | Oxidoreductase activity | Biosynthetic process | [59] |
| Growth hormone receptor | TR: Q9NRZ8 | GHR | EMTPVNPGV . . . : : : : . SLQSVNPGL | 11–19 | 44.4% | 1.3 | Cytokine receptor activity | Response to stimulus, cell signaling | [60] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sologova, S.S.; Zavadskiy, S.P.; Mokhosoev, I.M.; Moldogazieva, N.T. Short Linear Motifs Orchestrate Functioning of Human Proteins during Embryonic Development, Redox Regulation, and Cancer. Metabolites 2022, 12, 464. https://doi.org/10.3390/metabo12050464
Sologova SS, Zavadskiy SP, Mokhosoev IM, Moldogazieva NT. Short Linear Motifs Orchestrate Functioning of Human Proteins during Embryonic Development, Redox Regulation, and Cancer. Metabolites. 2022; 12(5):464. https://doi.org/10.3390/metabo12050464
Chicago/Turabian StyleSologova, Susanna S., Sergey P. Zavadskiy, Innokenty M. Mokhosoev, and Nurbubu T. Moldogazieva. 2022. "Short Linear Motifs Orchestrate Functioning of Human Proteins during Embryonic Development, Redox Regulation, and Cancer" Metabolites 12, no. 5: 464. https://doi.org/10.3390/metabo12050464
APA StyleSologova, S. S., Zavadskiy, S. P., Mokhosoev, I. M., & Moldogazieva, N. T. (2022). Short Linear Motifs Orchestrate Functioning of Human Proteins during Embryonic Development, Redox Regulation, and Cancer. Metabolites, 12(5), 464. https://doi.org/10.3390/metabo12050464








