Biosynthesis, Molecular Regulation, and Application of Bacilysin Produced by Bacillus Species
Abstract
1. Introduction
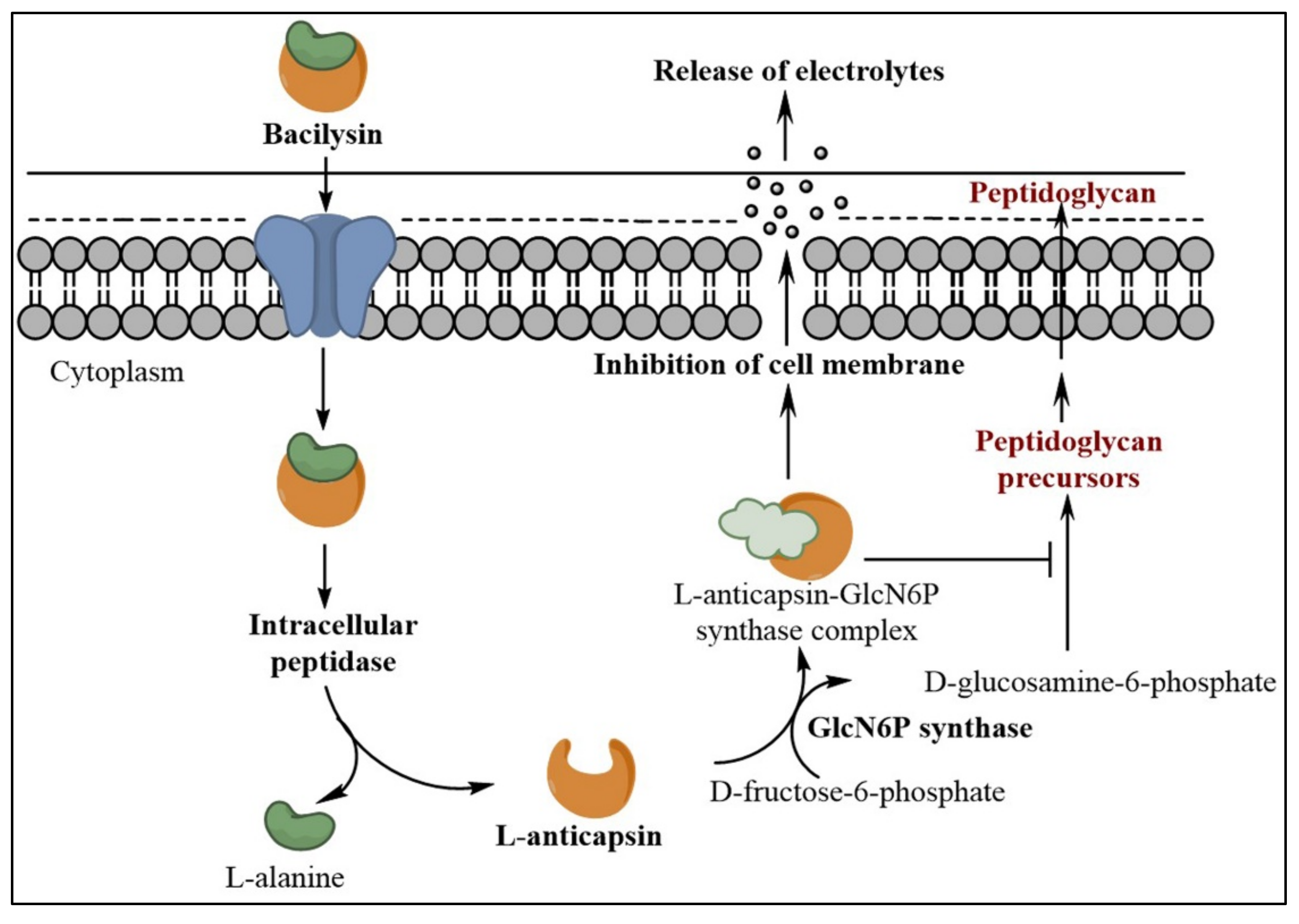
2. Mode of Action of Bacilysin and Inhibitory Effect on Pathogenic Microbes
3. Strain Specificity of Bacillus Species in Bacilysin Production
4. Biosynthesis of Bacilysin by the bac Operon
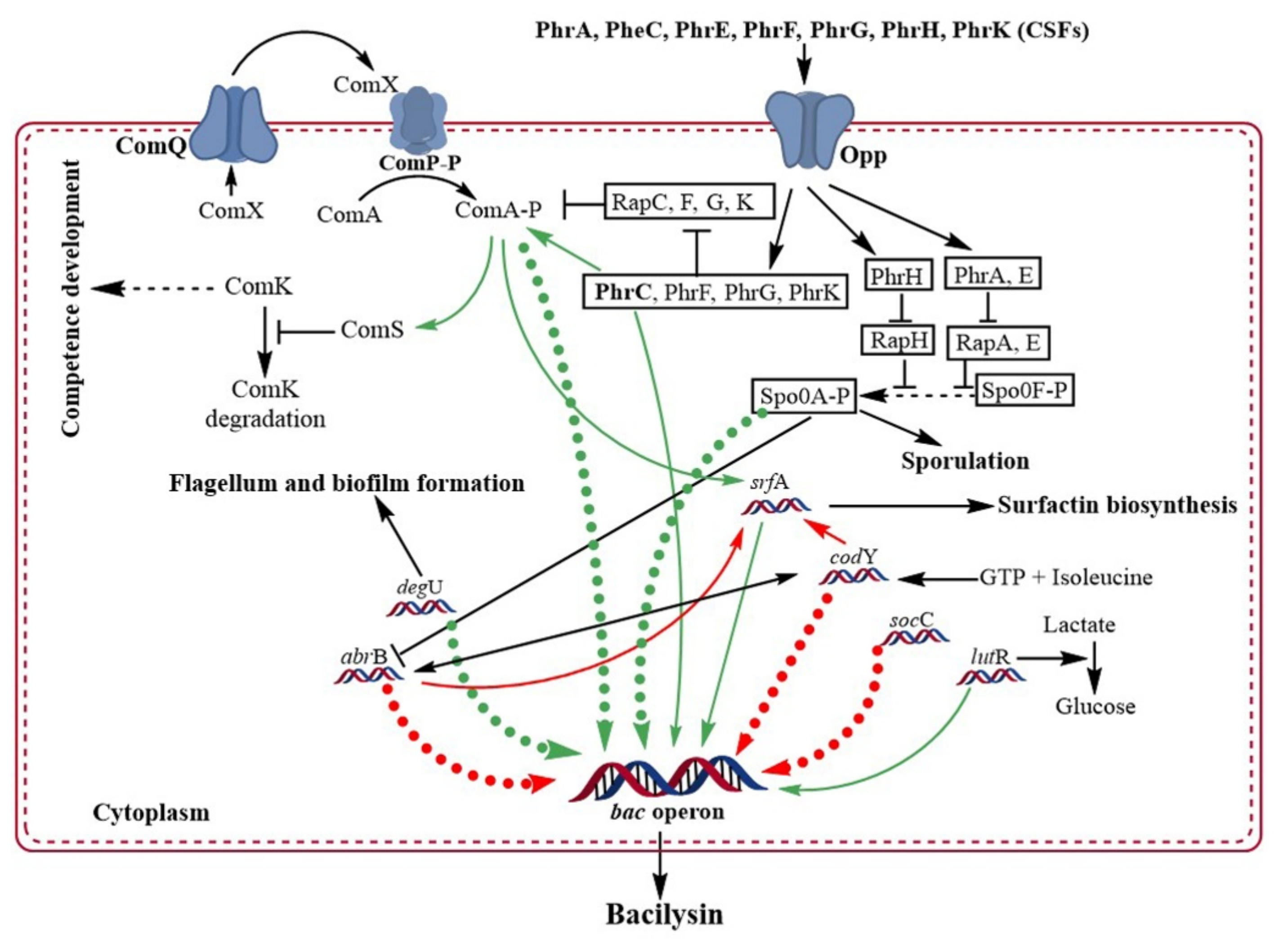
5. Regulatory Role of Signaling Molecules in Bacilysin Biosynthesis
6. Conclusions and Future Prospects
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Clark, S.; Jowitt, T.A.; Harris, L.K.; Knight, C.G.; Dobson, C.B. The lexicon of antimicrobial peptides: A complete set of arginine and tryptophan sequences. Commun. Biol. 2021, 4, 605. [Google Scholar] [CrossRef] [PubMed]
- Kleinkauf, H.; Von Döhren, H. Peptide Antibiotics. In Biotechnology, 2nd ed.; Wiley: Berlin, Germany, 2008; Volume 7, pp. 277–322. [Google Scholar]
- Li, J.; Koh, J.J.; Liu, S.; Lakshminarayanan, R.; Verma, C.S.; Beuerman, R.W. Membrane active antimicrobial peptides: Translating mechanistic insights to design. Front. Neurosci. 2017, 11, 73. [Google Scholar] [CrossRef] [PubMed]
- Benfield, A.H.; Henriques, S.T. Mode-of-action of antimicrobial peptides: Membrane disruption vs. intracellular mechanisms. Front. Med. Technol. 2020, 2, 610997. [Google Scholar] [CrossRef] [PubMed]
- Palmer, N.; Maasch, J.R.M.A.; Torres, M.D.T.; De La Fuente-Nunez, C. Molecular dynamics for antimicrobial peptide discovery. Infect. Immun. 2021, 89, e00703-20. [Google Scholar] [CrossRef] [PubMed]
- Fjell, C.D.; Hiss, J.A.; Hancock, R.E.W.; Schneider, G. Designing antimicrobial peptides: Form follows function. Nat. Rev. DrugDiscov. 2012, 11, 37–51. [Google Scholar] [CrossRef]
- Brogden, K.A. Antimicrobial peptides: Pore formers or metabolic inhibitors in bacteria? Nat. Rev. Microbiol. 2005, 3, 238–250. [Google Scholar] [CrossRef]
- Sumi, C.D.; Yang, B.W.; Yeo, I.C.; Hahm, Y.T. Antimicrobial peptides of the genus Bacillus: A new era for antibiotics. Can. J. Microbiol. 2015, 61, 93–103. [Google Scholar] [CrossRef]
- Sarkar, N.; Paulus, H. Function of peptide antibiotics in sporulation. Nat. New Biol. 1972, 239, 228–230. [Google Scholar] [CrossRef]
- Frangou-Lazaridis, M.; Seddon, B. Effect of gramicidin s on the transcription system of the producer Bacillus brevis Nagano. Micro. 1985, 131, 437–449. [Google Scholar] [CrossRef]
- Ristow, H.; Pschorn, W.; Hansen, J.; Winkel, U. Induction of sporulation in Bacillus brevis by peptide antibiotics. Nature 1979, 280, 165–166. [Google Scholar] [CrossRef]
- Özcengiz, G.; Öğülür, I. Biochemistry, genetics and regulation of bacilysin biosynthesis and its significance more than an antibiotic. New Biotechnol. 2015, 32, 612–619. [Google Scholar] [CrossRef] [PubMed]
- Hilton, M.D.; Alaeddinoglu, N.G.; Demain, A.L. Bacillus subtilis mutant deficient in the ability to produce the dipeptide antibiotic bacilysin: Isolation and mapping of the mutation. J. Bacteriol. 1988, 170, 1018–1020. [Google Scholar] [CrossRef] [PubMed]
- Özcengiz, G.; Alaeddinoglu, N.G. Bacilysin production by Bacillus subtilis: Effects of bacilysin, pH and temperature. Folia Microbiol. 1991, 36, 522–526. [Google Scholar] [CrossRef] [PubMed]
- Özcengiz, G.; Alaeddinoglu, N.G. Bacilysin production and sporulation in Bacillus subtilis. Curr. Microbiol. 1991, 23, 61–64. [Google Scholar] [CrossRef]
- Kenig, M.; Abraham, E.P. Antimicrobial activities and antagonists of bacilysin and anticapsin. J. Gen. Microbiol. 1976, 94, 37–45. [Google Scholar] [CrossRef]
- Wang, T.; Liu, X.H.; Wu, M.B.; Ge, S. Molecular insights into the antifungal mechanism of bacilysin. J. Mol. Model. 2018, 24, 118. [Google Scholar] [CrossRef]
- Foster, J.W.; Woodruff, H.B. Bacillin, a New Antibiotic Substance from a Soil Isolate of Bacillus subtilis. J. bacteriol. 1946, 51, 363–369. [Google Scholar] [CrossRef]
- Newton, G.G. Antibiotics from a Strain of Bacillus subtilis: Bacilipin A and B and Bacilysin. Br. J. Exp. Pathol. 1949, 30, 306. [Google Scholar]
- Rogers, H.; Newton, G.; Abraham, E. Production and purification of bacilysin. Biochem. J. 1965, 97, 573–578. [Google Scholar] [CrossRef]
- Walker, J.E.; Abraham, E.P. The structure of bacilysin and other products of Bacillus subtilis. Biochem. J. 1970, 118, 563–570. [Google Scholar] [CrossRef]
- Kamiński, K.; Sokolowska, T. The probable identity of bagilysin and tetaine. J. Antibiot. 1973, 26, 184–185. [Google Scholar] [CrossRef] [PubMed]
- Atsumi, K.; Ōiwa, R.; Ōmura, S. Production of bacillin by Bacillus sp. strain no. KM-208 and its identity with tetaine (bacilysin). J. Antibiot. 1975, 28, 77–78. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Parker, J.B.; Walsh, C.T. Action and Timing of BacC and BacD in the Late Stages of Biosynthesis of the Dipeptide Antibiotic Bacilysin. Biochemistry 2013, 52, 889–901. [Google Scholar] [CrossRef]
- Wu, L.; Wu, H.; Chen, L.; Lin, L.; Borriss, R.; Gao, X. Bacilysin overproduction in Bacillus amyloliquefaciens FZB42 markerless derivative strains FZBREP and FZBSPA enhances antibacterial activity. Appl. Microbiol. Biotechnol. 2015, 99, 4255–4263. [Google Scholar] [CrossRef] [PubMed]
- Roscoe, J.; Abraham, E.P. Experiments relating to the biosynthesis of bacilysin. Biochem. J. 1966, 99, 793–800. [Google Scholar] [CrossRef]
- Walker, J.E. Antibiotic production and sporulation in Bacillus subtilis. Biochem. J. 1971, 121, 571–573. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.A.; Göpel, Y.; Milewski, S.; Görke, B. Two small RNAs conserved in enterobacteriaceae provide intrinsic resistance to antibiotics targeting the cell wall biosynthesis enzyme glucosamine-6-phosphate synthase. Front. Microbiol. 2016, 7, 908. [Google Scholar] [CrossRef] [PubMed]
- Kenig, M.; Vandamme, E.; Abraham, E.P. The mode of action of bacilysin and anticapsin and biochemical properties of bacilysin resistant mutants. J. Gen. Microbiol. 1976, 94, 46–54. [Google Scholar] [CrossRef]
- Wojciechowski, M.; Milewski, S.; Mazerski, J.; Borowski, E. Glucosamine-6-phosphate synthase, a novel target for antifungal agents. Molecular modelling studies in drug design. Acta Biochim. Pol. 2005, 52, 647–653. [Google Scholar] [CrossRef]
- Wu, L.; Wu, H.; Chen, L.; Xie, S.; Zang, H.; Borriss, R.; Gao, X. Bacilysin from Bacillus amyloliquefaciens FZB42 has specific bactericidal activity against harmful algal bloom species. Appl. Environ. Microbiol. 2014, 80, 7512–7520. [Google Scholar] [CrossRef]
- Perry, D. Peptide transport in Staphylococcus aureus. J. Gen. Microbiol. 1981, 124, 425–428. [Google Scholar] [CrossRef] [PubMed]
- Perry, D.; Abraham, E.P. Transport and metabolism of bacilysin and other peptides by suspensions of Staphylococcus aureus. J. Gen. Microbiol. 1979, 115, 213–221. [Google Scholar] [CrossRef] [PubMed]
- Steinborn, G.; Hajirezaei, M.R.; Hofemeister, J. bac genes for recombinant bacilysin and anticapsin production in Bacillus host strains. Arch. Microbiol. 2005, 183, 71–79. [Google Scholar] [CrossRef] [PubMed]
- Yazgan, A.; Özcengiz, G.; Marahiel, M.A. Tn10 insertional mutations of Bacillus subtilis that block the biosynthesis of bacilysin. Biochim. Biophys. Acta 2001, 1518, 87–94. [Google Scholar] [CrossRef]
- Inaoka, T.; Takahashi, K.; Ohnishi-Kameyama, M.; Yoshida, M.; Ochi, K. Guanine nucleotides guanosine 5′-diphosphate 3′-diphosphate and GTP co-operatively regulate the production of an antibiotic bacilysin in Bacillus subtilis. J. Biol. Chem. 2003, 278, 2169–2176. [Google Scholar] [CrossRef] [PubMed]
- Karata, A.Y.; Çetin, S.; Özcengiz, G. The effects of insertional mutations in comQ, comP, srfA, spo0H, spo0A and abrB genes on bacilysin biosynthesis in Bacillus subtilis. Biochim. Biophys. Acta 2003, 1626, 51–56. [Google Scholar] [CrossRef]
- Rabbee, M.F.; Ali, M.; Choi, J.; Hwang, B.; Jeong, S.; Baek, K. Bacillus velezensis: A Valuable Member of Bioactive Molecules within Plant Microbiomes. Molecules 2019, 24, 1046. [Google Scholar] [CrossRef]
- Chen, X.H.; Scholz, R.; Borriss, M.; Junge, H.; Mögel, G.; Kunz, S.; Borriss, R. Difficidin and bacilysin produced by plant-associated Bacillus amyloliquefaciens are efficient in controlling fire blight disease. J. Biotechnol. 2009, 140, 38–44. [Google Scholar] [CrossRef]
- Rabbee, M.F.; Baek, K.H. Antimicrobial Activities of Lipopeptides and Polyketides of Bacillus velezensis for Agricultural Applications. Molecules 2020, 25, 4973. [Google Scholar] [CrossRef]
- Han, X.; Shen, D.; Xiong, Q.; Bao, B.; Zhang, W.; Dai, T.; Zhao, Y.; Borriss, R.; Fan, B. The plant-beneficial rhizobacterium Bacillus velezensis FZB42 controls the soybean pathogen phytophthora sojae due to bacilysin production. Appl. Environ. Microbiol. 2021, 87, e0160121. [Google Scholar] [CrossRef]
- Wu, L.; Wu, H.; Chen, L.; Yu, X.; Borriss, R.; Gao, X. Difficidin and bacilysin from Bacillus amyloliquefaciens FZB42 have antibacterial activity against Xanthomonas oryzae rice pathogens. Sci. Rep. 2015, 5, 12975. [Google Scholar] [CrossRef] [PubMed]
- Ba§alp, A.; Özcengiz, G.; Alaeddinoĝlu, N.G. Changes in patterns of alkaline serine protease and bacilysin formation caused by common effectors of sporulation in Bacillus subtilis 168. Curr. Microbiol. 1992, 24, 129–135. [Google Scholar] [CrossRef]
- Nannan, C.; Vu, H.Q.; Gillis, A.; Caulier, S.; Nguyen, T.T.T.; Mahillon, J. Bacilysin within the Bacillus subtilis group: Gene prevalence versus antagonistic activity against Gram-negative foodborne pathogens. J. Biotechnol. 2021, 327, 28–35. [Google Scholar] [CrossRef] [PubMed]
- Zaid, D.S.; Cai, S.; Hu, C.; Li, Z.; Li, Y.; Gralnick, J.A. Comparative Genome Analysis Reveals Phylogenetic Identity of Bacillus velezensis HNA3 and Genomic Insights into Its Plant Growth Promotion and Biocontrol Effects. Microbiol. Spectr. 2022, 10, e02169-21. [Google Scholar] [CrossRef]
- Han, L.-L.; Liu, Y.-C.; Miao, C.-C.; Feng, H. Disruption of the pleiotropic gene scoC causes transcriptomic and phenotypical changes in Bacillus pumilus BA06. BMC Genom. 2019, 20, 327. [Google Scholar] [CrossRef]
- Caldwell, R.; Sapolsky, R.; Weyler, W.; Maile, R.R.; Causey, S.C.; Ferrari, E. Correlation between Bacillus subtilis scoC phenotype and gene expression determined using microarrays for transcriptome analysis. J. Bacteriol. 2001, 183, 7329–7340. [Google Scholar] [CrossRef]
- Barbieri, G.; Albertini, A.M.; Ferrari, E.; Sonenshein, A.L.; Belitsky, B.R. Interplay of CodY and ScoC in the regulation of major extracellular protease genes of Bacillus subtilis. J. Bacteriol. 2016, 198, 907–920. [Google Scholar] [CrossRef] [PubMed]
- Shank, E.A.; Kolter, R. Extracellular signaling and multicellularity in Bacillus subtilis. Curr. Opin. Microbiol. 2011, 14, 741–747. [Google Scholar] [CrossRef] [PubMed]
- Mahlstedt, S.; Fielding, E.N.; Moore, B.S.; Walsh, C.T. Prephenate Decarboxylases: A New Prephenate-Utilizing Enzyme Family That Performs Nonaromatizing Decarboxylation en Route to Diverse Secondary Metabolites. Biochemistry 2010, 49, 9021–9023. [Google Scholar] [CrossRef]
- Parker, J.B.; Walsh, C.T. Olefin Isomerization Regiochemistries during Tandem Action of BacA and BacB on Prephenate in Bacilysin Biosynthesis. Biochemistry 2012, 51, 3241–3251. [Google Scholar] [CrossRef]
- Mahlstedt, S.A.; Walsh, C.T. Investigation of anticapsin biosynthesis reveals a four-enzyme pathway to tetrahydrotyrosine in Bacillus subtilis. Biochemistry 2010, 49, 912–923. [Google Scholar] [CrossRef] [PubMed]
- Parker, J.B.; Walsh, C.T. Stereochemical Outcome at Four Stereogenic Centers during Conversion of Prephenate to Tetrahydrotyrosine by bacABGF in the Bacilysin Pathway. Biochemistry 2012, 51, 5622–5632. [Google Scholar] [CrossRef] [PubMed]
- Rajavel, M.; Mitra, A.; Gopal, B. Role of Bacillus subtilis bacB in the synthesis of bacilysin. J. Biol. Chem. 2009, 284, 31882–31892. [Google Scholar] [CrossRef] [PubMed]
- Grossman, A.D. Genetic Networks Controlling the Initiation of Sporulation and the Development of Genetic Competence in Bacillus subtilis. Annu. Rev. Genet. 1995, 29, 477–508. [Google Scholar] [CrossRef]
- Comella, N.; Grossman, A.D. Conservation of genes and processes controlled by the quorum response in bacteria: Characterization of genes controlled by the quorum-sensing transcription factor ComA in Bacillus subtilis. Mol. Microbiol. 2005, 57, 1159–1174. [Google Scholar] [CrossRef]
- Deshmukh, A.; Gopal, B. Structural insights into the catalytic mechanism of Bacillus subtilis bacF. Acta Crystallogr. 2020, 76, 145–151. [Google Scholar] [CrossRef]
- Perego, M.; Glaser, P.; Hoch, J.A. Aspartyl-phosphate phosphatases deactivate the response regulator components of the sporulation signal transduction system in Bacillus subtilis. Mol. Microbiol. 1996, 19, 1151–1157. [Google Scholar] [CrossRef]
- Weinrauch, Y.; Penchev, R.; Dubnau, E.; Smith, E.; Dubnau, D. A Bacillus subtilis regulatory gene product for genetic competence and sporulation resembles sensor protein members of the bacterial two-component signal-transduction systems. Genes Dev. 1990, 4, 860–872. [Google Scholar] [CrossRef]
- Solomon, J.M.; Magnuson, R.; Srivastava, A.; Grossman, A.D. Convergent sensing pathways mediate response to two extracellular competence factors in Bacillus subtilis. Genes Dev. 1995, 9, 547–558. [Google Scholar] [CrossRef]
- Magnuson, R.; Solomon, J.; Grossman, A.D. Biochemical and genetic characterization of a competence pheromone from Bacillus subtilis. Cell 1994, 77, 207–216. [Google Scholar] [CrossRef]
- Core, L.; Perego, M. TPR-mediated interaction of RapC with ComA inhibits response regulator-DNA binding for competence development in Bacillus subtilis. Mol. Microbiol. 2003, 49, 1509–1522. [Google Scholar] [CrossRef] [PubMed]
- Lazazzera, B.A.; Solomon, J.M.; Grossman, A.D. An exported peptide functions intracellularly to contribute to cell density signaling in Bacillus subtilis. Cell 1997, 89, 917–925. [Google Scholar] [CrossRef]
- Lazazzera, B.A.; Kurtser, I.G.; Mcquade, R.S.; Grossman, A.D. An autoregulatory circuit affecting peptide signaling in Bacillus subtilis. J. Bacteriol. 1999, 181, 5193–5200. [Google Scholar] [CrossRef] [PubMed]
- Wolf, D.; Rippa, V.; Mobarec, J.C.; Sauer, P.; Adlung, L.; Kolb, P.; Bischofs, I.B. The quorum-sensing regulator ComA from Bacillus subtilis activates transcription using topologically distinct DNA motifs. Nucleic Acids Res. 2016, 44, 2160–2172. [Google Scholar] [CrossRef] [PubMed]
- Horinouchi, S.; Ueda, K.; Nakayama, J.; Ikeda, T. Cell-to-cell communications among microorganisms. Chem. Biol. 2010, 4, 283–337. [Google Scholar]
- Bongiorni, C.; Ishikawa, S.; Stephenson, S.; Ogasawara, N.; Perego, M. Synergistic regulation of competence development in Bacillus subtilis by two Rap-Phr systems. J. Bacteriol. 2005, 187, 4353–4361. [Google Scholar] [CrossRef]
- Auchtung, J.M.; Lee, C.A.; Grossman, A.D. Modulation of the ComA-dependent quorum response in Bacillus subtilis by multiple Rap proteins and Phr peptides. J. Bacteriol. 2006, 188, 5273–5285. [Google Scholar] [CrossRef]
- Smits, W.K.; Bongiorni, C.; Veening, J.-W.; Hamoen, L.W.; Kuipers, O.P.; Perego, M. Temporal separation of distinct differentiation pathways by a dual specificity Rap-Phr system in Bacillus subtilis. Mol. Microbiol. 2007, 65, 103–120. [Google Scholar] [CrossRef]
- Köroğlu, T.E.; Öğülür, İ.; Mutlu, S.; Yazgan-Karataş, A.; Özcengiz, G. Global Regulatory Systems Operating in Bacilysin Biosynthesis in Bacillus subtilis. J. Mol. Microbiol. Biotechnol. 2011, 20, 144–155. [Google Scholar] [CrossRef]
- Pottathil, M.; Jung, A.; Lazazzera, B.A. CSF, a species-specific extracellular signaling peptide for communication among strains of Bacillus subtilis and Bacillus mojavensis. J. Bacteriol. 2008, 190, 4095–4099. [Google Scholar] [CrossRef]
- Fernandes, P.A.V.; Arruda, I.R.D.; Santos, A.F.A.B.D.; Araújo, A.A.D.; Maior, A.M.S.; Ximenes, E.A. Antimicrobial activity of surfactants produced by Bacillus subtilis R14 against multidrug-resistant bacteria. Braz. J. Microbiol. 2007, 38, 704–709. [Google Scholar] [CrossRef]
- Sarwar, A.; Hassan, M.N.; Imran, M.; Iqbal, M.; Majeed, S.; Brader, G.; Sessitsch, A.; Hafeez, F.Y. Biocontrol activity of surfactin A purified from Bacillus NH-100 and NH-217 against rice bakanae disease. Microbiol. Res. 2018, 209, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Sabaté, D.C.; Audisio, M.C. Inhibitory activity of surfactin, produced by different Bacillus subtilis subsp. subtilis strains, against Listeria monocytogenes sensitive and bacteriocin-resistant strains. Microbiol. Res. 2013, 168, 125–129. [Google Scholar] [CrossRef] [PubMed]
- Robertson, J.B.; Gocht, M.; Marahiel, M.A.; Zuber, P. AbrB, a regulator of gene expression in Bacillus, interacts with the transcription initiation regions of a sporulation gene and an antibiotic biosynthesis gene. Proc. Natl. Acad. Sci. USA 1989, 86, 8457–8461. [Google Scholar] [CrossRef] [PubMed]
- Barbieri, G.; Voigt, B.; Albrecht, D.; Hecker, M.; Albertini, A.M.; Sonenshein, A.L.; Ferrari, E.; Belitsky, B.R. CodY regulates expression of the Bacillus subtilis extracellular proteases Vpr and Mpr. J. Bacteriol. 2015, 197, 1423–1432. [Google Scholar] [CrossRef] [PubMed]
- Sonenshein, A.L. CodY, a global regulator of stationary phase and virulence in Gram-positive bacteria. Curr. Opin. Microbiol. 2005, 8, 203–207. [Google Scholar] [CrossRef]
- Kallios, P.T.; Fagelson, J.E.; Hoch, J.A.; Straucht, M.A. The Transition State Regulator Hpr of Bacillus subtilis Is a DNA-binding Protein. J. Biol. Chem. 1991, 266, 13411–13417. [Google Scholar] [CrossRef]
- Inaoka, T.; Wang, G.; Ochi, K. ScoC regulates bacilysin production at the transcription level in Bacillus subtilis. J. Bacteriol. 2009, 191, 7367–7371. [Google Scholar] [CrossRef]
- Kobayashi, K. Gradual activation of the response regulator DegU controls serial expression of genes for flagellum formation and biofilm formation in Bacillus subtilis. Mol. Microbiol. 2007, 66, 395–409. [Google Scholar] [CrossRef]
- Dahl, M.K.; Msadek, T.; Kunst, F.; Rapoport, G. Mutational analysis of the Bacillus subtilis DegU regulator and its phosphorylation by the DegS protein kinase. J. Bacteriol. 1991, 173, 2539–2547. [Google Scholar] [CrossRef]
- Mariappan, A.; Makarewicz, O.; Chen, X.-H.; Borriss, R. Two-Component Response Regulator DegU Controls the Expression of Bacilysin in Plant-Growth-Promoting Bacterium Bacillus amyloliquefaciens FZB42. J. Mol. Microbiol. 2012, 22, 114–125. [Google Scholar]
- Köroǧlu, T.E.; Kurt-Gür, G.; Ünlü, E.C.; Yazgan-Karataş, A. The novel gene yvfI in Bacillus subtilis is essential for bacilysin biosynthesis. Antonie Van Leeuwenhoek. 2008, 94, 471–479. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.; Gao, L.; Bie, X.; Lu, Z.; Liu, H.; Zhang, C.; Lu, F.; Zhao, H. Identification of novel surfactin derivatives from NRPS modification of Bacillus subtilis and its antifungal activity against Fusarium moniliforme. BMC Microbiol. 2016, 16, 31. [Google Scholar] [CrossRef] [PubMed]

| Genes | Gene Product Sizes (aa) | Gene Products | Functions of Gene Products | References |
|---|---|---|---|---|
| Genes directly involved in bacilysin biosynthesis | ||||
| bacA | 204 | Decarboxylase | Acts on prephenate | [24,50,51] |
| bacB | 235 | 3E-ex-H2HPP isomerase | Synthesizes epoxy-3E-H2HPP | [50,51,52] |
| bacC | 255 | Dehydrogenase | Synthesizes L-anticapsin | [24,52] |
| bacD | 472 | Ligase | Ligases L-anticapsin and L-alanine | [24,34] |
| bacE | 394 | Bacilysin exporter | Provides host resistance to bacilysin and effluxes it from cell | [34] |
| bacF | 399 | Aminotransferase | Synthesizes L-dihydroanticapsin from L-phenylalanine | [24,51,79] |
| bacG | 259 | Reductase | Synthesizes epoxy-4S-H4HPP precursor of L-anticapsin | [24,51] |
| Genes positively regulate bacilysin biosynthesis | ||||
| srfA | 3588 | Surfactin synthase subunit 1 | Regulates bacilysin biosynthesis positively. | [24,84] |
| degU | 229 | Transcriptional regulatory protein DegU | Binds bacA operon and bacG genes | [82] |
| comX | 55 | Competence pheromone ComX | Activates comA which positively regulates bac operon (Quorum sensing). | [56] |
| phrC | 40 | Phosphatase | Controls comA activity by blocking RapC | [56,67] |
| lutR | 219 | HTH-type transcriptional regulator LutR | Controls lactate utilization, regulates bac operon positively | [80,83] |
| Genes negatively regulate bacilysin biosynthesis | ||||
| socC | 203 | Deoxyfructose oxidoreductase | Negatively control the expression of bacA gene | [79] |
| abrB | 96 | Transition state regulatory protein AbrB | Binds to the bac operon and regulates bacilysin biosynthesis negatively. Acts mutually with CodY | [37,75] |
| codY | 259 | Transcriptional regulatory protein CodY | Binds to the bac operon and regulates bacilysin biosynthesis negatively. Acts mutually with abrB | [36,70] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Islam, T.; Rabbee, M.F.; Choi, J.; Baek, K.-H. Biosynthesis, Molecular Regulation, and Application of Bacilysin Produced by Bacillus Species. Metabolites 2022, 12, 397. https://doi.org/10.3390/metabo12050397
Islam T, Rabbee MF, Choi J, Baek K-H. Biosynthesis, Molecular Regulation, and Application of Bacilysin Produced by Bacillus Species. Metabolites. 2022; 12(5):397. https://doi.org/10.3390/metabo12050397
Chicago/Turabian StyleIslam, Tarequl, Muhammad Fazle Rabbee, Jinhee Choi, and Kwang-Hyun Baek. 2022. "Biosynthesis, Molecular Regulation, and Application of Bacilysin Produced by Bacillus Species" Metabolites 12, no. 5: 397. https://doi.org/10.3390/metabo12050397
APA StyleIslam, T., Rabbee, M. F., Choi, J., & Baek, K.-H. (2022). Biosynthesis, Molecular Regulation, and Application of Bacilysin Produced by Bacillus Species. Metabolites, 12(5), 397. https://doi.org/10.3390/metabo12050397







