Histamine: A Mediator of Intestinal Disorders—A Review
Abstract
1. Histamine
1.1. Synthesis and Degradation of Histamine
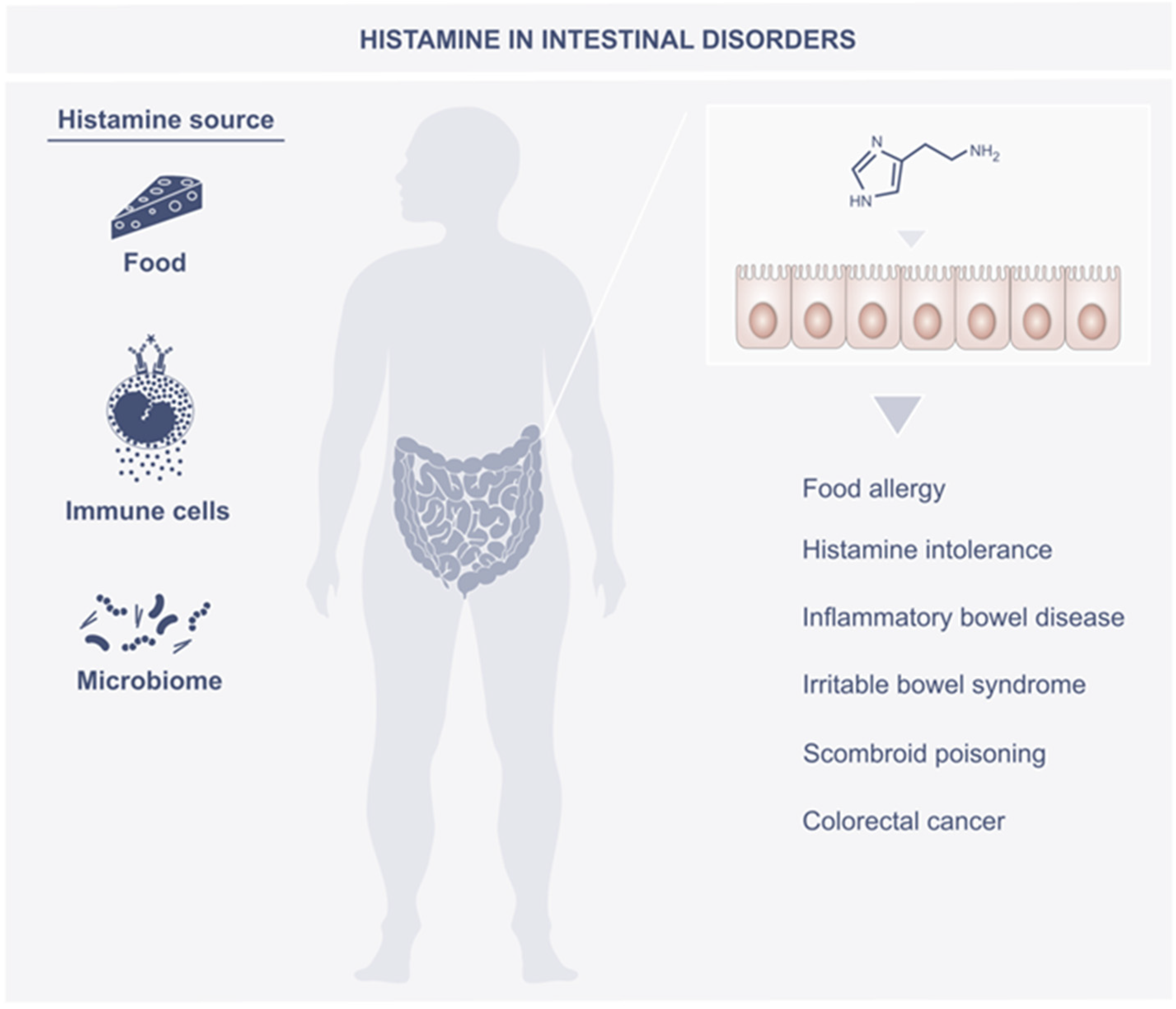
1.2. Sources of Histamine in the Body
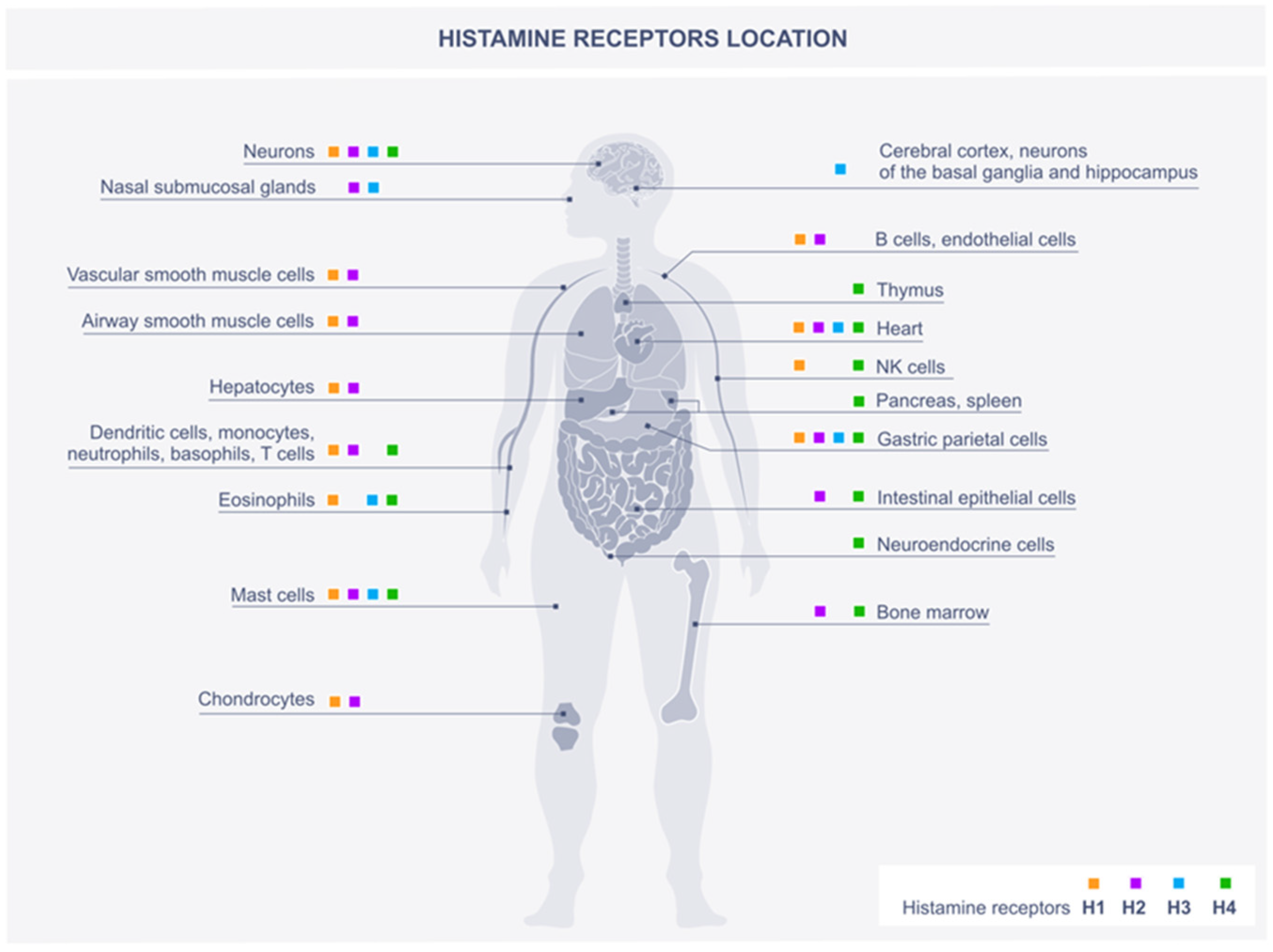
2. Histamine Scheme of Action through Histamine Receptors
2.1. H1R
2.2. H2R
2.3. H3R
2.4. H4R
3. Histamine in the Intestines
4. Role of Histamine in Intestine Disorders
4.1. Food Allergy
4.2. Histamine Intolerance
4.3. Inflammatory Bowel Disease
4.4. Irritable Bowel Syndrome
4.5. Scombroid Poisoning
4.6. Colorectal Cancer
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Barger, G.; Dale, H.H. Chemical structure and sympathomimetic action of amines. J. Physiol. 1910, 41, 19–59. [Google Scholar] [CrossRef] [PubMed]
- Dale, H.H.; Laidlaw, P.P. The physiological action of β-iminazolylethylamine. J. Physiol. 1910, 41, 318–344. [Google Scholar] [CrossRef] [PubMed]
- Lindskog, M. Histamine Receptors in the Cross-Talk between Periphery and Brain. Int. J. Neuropsychopharmacol. 2017, 20, 400–402. [Google Scholar] [CrossRef][Green Version]
- Hungerford, J.M. Scombroid poisoning: A review. Toxicon 2010, 56, 231–243. [Google Scholar] [CrossRef] [PubMed]
- Obara, I.; Telezhkin, V.; AlRashdi, I.; Chazot, P. Histamine, histamine receptors, and neuropathic pain relief. Br. J. Pharmacol. 2020, 177, 580–599. [Google Scholar] [CrossRef]
- Kovacova-Hanuskova, E.; Buday, T.; Gavliakova, S.; Plevkova, J. Histamine, histamine intoxication and intolerance. Allergol. Immunopathol. 2015, 43, 498–506. [Google Scholar] [CrossRef]
- Maintz, L.; Novak, N. Histamine and histamine intolerance. Am. J. Clin. Nutr. 2007, 85, 1185–1196. [Google Scholar] [CrossRef]
- Comas-Basté, O.; Sánchez-Pérez, S.; Veciana-Nogués, M.T.; Latorre-Moratalla, M.; Vidal-Carou, M.D.C. Histamine Intolerance: The Current State of the Art. Biomolecules 2020, 10, 1181. [Google Scholar] [CrossRef]
- Schwelberger, H.G.; Hittmair, A.; Kohlwein, S.D. Analysis of tissue and subcellular localization of mammalian diamine oxidase by confocal laser scanning fluorescence microscopy. Inflamm. Res. 1998, 47, S60–S61. [Google Scholar] [CrossRef]
- Panula, P.; Chazot, P.L.; Cowart, M.; Gutzmer, R.; Leurs, R.; Liu, W.L.S.; Stark, H.; Thurmond, R.L.; Haas, H.L. International Union of Basic and Clinical Pharmacology. XCVIII. Histamine Receptors. Pharmacol. Rev. 2015, 67, 601–655. [Google Scholar] [CrossRef]
- Zhang, Z.; Kurashima, Y. Two Sides of the Coin: Mast Cells as a Key Regulator of Allergy and Acute/Chronic Inflammation. Cells 2021, 10, 1615. [Google Scholar] [CrossRef] [PubMed]
- Borriello, F.; Iannone, R.; Marone, G. Histamine Release from Mast Cells and Basophils. Histamine Histamine Recept. Health Dis. 2017, 241, 121–139. [Google Scholar] [CrossRef]
- Landete, J.M.; Pardo, I.; Ferrer, S. Histamine, Histidine, and Growth-Phase Mediated Regulation of the Histidine Decarboxylase Gene in Lactic Acid Bacteria Isolated from Wine. FEMS Microbiol. Lett. 2006, 260, 84–90. [Google Scholar] [CrossRef] [PubMed]
- Visciano, P.; Schirone, M.; Tofalo, R.; Suzzi, G. Histamine poisoning and control measures in fish and fishery products. Front. Microbiol. 2014, 5, 500. [Google Scholar] [CrossRef]
- Zhao, Y.; Zhang, X.; Jin, H.; Chen, L.; Ji, J.; Zhang, Z. Histamine Intolerance-A Kind of Pseudoallergic Reaction. Biomolecules 2022, 12, 454. [Google Scholar] [CrossRef]
- Barcik, W.; Pugin, B.; Westermann, P.; Perez, N.R.; Ferstl, R.; Wawrzyniak, M.; Smolinska, S.; Jutel, M.; Hessel, E.M.; Michalovich, D.; et al. Histamine-secreting microbes are increased in the gut of adult asthma patients. J. Allergy Clin. Immunol. 2016, 138, 1491–1494.e7. [Google Scholar] [CrossRef]
- Smolinska, S.; O’Mahony, L. Microbiome—Host Immune System Interactions. Semin. Liver Dis. 2016, 36, 317–326. [Google Scholar] [CrossRef]
- Krell, T.; Gavira, J.; Velando, F.; Fernández, M.; Roca, A.; Monteagudo-Cascales, E.; Matilla, M. Histamine: A Bacterial Signal Molecule. Int. J. Mol. Sci. 2021, 22, 6312. [Google Scholar] [CrossRef]
- Landete, J.M.; De las Rivas, B.; Marcobal, A.; Muñoz, R. Updated molecular knowledge about histamine biosynthesis by bacteria. Crit. Rev. Food Sci. Nutr. 2008, 48, 697–714. [Google Scholar] [CrossRef]
- Smolinska, S.; Jutel, M.; Crameri, R.; O’Mahony, L. Histamine and gut mucosal immune regulation. Allergy 2014, 69, 273–281. [Google Scholar] [CrossRef]
- Barcik, W.; Wawrzyniak, M.; Akdis, C.A.; O’Mahony, L. Immune regulation by histamine and histamine-secreting bacteria. Curr. Opin. Immunol. 2017, 48, 108–113. [Google Scholar] [CrossRef] [PubMed]
- De la Torre, M.; Gomez-Botran, J.L.; Olivera, E.R.; Bermejo, F.; Rodríguez-Morán, J.; Luengo, J.M. Histamine catabolism in Pseudomonas putida U: Identification of the genes, catabolic enzymes and regulators. Environ. Microbiol. 2018, 20, 1828–1841. [Google Scholar] [CrossRef] [PubMed]
- Thomas, C.M.; Hong, T.; Van Pijkeren, J.P.; Hemarajata, P.; Trinh, D.V.; Hu, W.; Britton, R.A.; Kalkum, M.; Versalovic, J. Histamine derived from probiotic lactobacillus reuteri suppresses TNF via modulation of PKA and ERK signaling. PLoS ONE 2012, 7, e31951. [Google Scholar] [CrossRef] [PubMed]
- Frei, R.; Ferstl, R.; Konieczna, P.; Ziegler, M.; Simon, T.; Rugeles, T.M.; Mailand, S.; Watanabe, T.; Lauener, R.; Akdis, C.A.; et al. Histamine receptor 2 modifies dendritic cell responses to microbial ligands. J. Allergy Clin. Immunol. 2013, 132, 194–204.e12. [Google Scholar] [CrossRef] [PubMed]
- Ferstl, R.; Frei, R.; Schiavi, E.; Konieczna, P.; Barcik, W.; Ziegler, M.; Lauener, R.P.; Chassard, C.; Lacroix, C.; Akdis, C.A.; et al. Histamine receptor 2 is a key influence in immune responses to intestinal histamine-secreting microbes. J. Allergy Clin. Immunol. 2014, 134, 744–746.e3. [Google Scholar] [CrossRef] [PubMed]
- Weiss, G.A.; Hennet, T. Mechanisms and consequences of intestinal dysbiosis. Cell. Mol. Life Sci. 2017, 74, 2959–2977. [Google Scholar] [CrossRef]
- Carthy, E.; Ellender, T. Histamine, Neuroinflammation and Neurodevelopment: A Review. Front. Neurosci. 2021, 15, 680214. [Google Scholar] [CrossRef]
- Shulpekova, Y.O.; Nechaev, V.M.; Popova, I.R.; Deeva, T.A.; Kopylov, A.T.; Malsagova, K.A.; Kaysheva, A.L.; Ivashkin, V.T. Food Intolerance: The Role of Histamine. Nutrients 2021, 13, 3207. [Google Scholar] [CrossRef]
- Smolinska, S.; Groeger, D. Biology of the Microbiome 1: Interactions with the Host Immune Response. Gastroenterol. Clin. N. Am. 2017, 46, 19–35. [Google Scholar] [CrossRef]
- Xia, R.; Na Wang, N.; Xu, Z.; Lu, Y.; Song, J.; Zhang, A.; Guo, C.; He, Y. Cryo-EM structure of the human histamine H1 receptor/Gq complex. Nat. Commun. 2021, 12, 2086. [Google Scholar] [CrossRef]
- Simons, F.E.R. Advances in H1-Antihistamines. New Engl. J. Med. 2004, 351, 2203–2217. [Google Scholar] [CrossRef] [PubMed]
- Togias, A. H1-receptors: Localization and role in airway physiology and in immune functions. J. Allergy Clin. Immunol. 2003, 112 (Suppl. 4), S60–S68. [Google Scholar] [CrossRef]
- Jutel, M.; Klunker, S.; Akdis, M.; Małolepszy, J.; Thomet, O.A.; Żak-Nejmark, T.; Blaser, K.; Akdis, C. Histamine Upregulates Th1 and Downregulates Th2 Responses due to Different Patterns of Surface Histamine 1 and 2 Receptor Expression. Int. Arch. Allergy Immunol. 2001, 124, 190–192. [Google Scholar] [CrossRef] [PubMed]
- Ohsawa, Y.; Hirasawa, N. The Role of Histamine H1 and H4 Receptors in Atopic Dermatitis: From Basic Research to Clinical Study. Allergol. Int. 2014, 63, 533–542. [Google Scholar] [CrossRef] [PubMed]
- Park, P.; Sanderson, T.M.; Amici, M.; Choi, S.-L.; Bortolotto, Z.A.; Zhuo, M.; Kaang, B.-K.; Collingridge, G.L. Calcium-Permeable AMPA Receptors Mediate the Induction of the Protein Kinase A-Dependent Component of Long-Term Potentiation in the Hippocampus. J. Neurosci. 2016, 36, 622–631. [Google Scholar] [CrossRef]
- Shi, Z.; Fultz, R.; Engevik, M.A.; Gao, C.; Hall, A.; Major, A.; Mori-Akiyama, Y.; Versalovic, J. Distinct roles of histamine H1- and H2-receptor signaling pathways in inflammation-associated colonic tumorigenesis. Am. J. Physiol. Gastrointest. Liver Physiol. 2019, 316, 205–216. [Google Scholar] [CrossRef] [PubMed]
- Mazzoni, A.; Young, H.A.; Spitzer, J.H.; Visintin, A.; Segal, D.M. Histamine regulates cytokine production in maturing dendritic cells, resulting in altered T cell polarization. J. Clin. Investig. 2001, 108, 1865–1873. [Google Scholar] [CrossRef]
- Teuscher, C.; Poynter, M.; Offner, H.; Zamora, A.; Watanabe, T.; Fillmore, P.D.; Zachary, J.F.; Blankenhorn, E.P. Attenuation of Th1 Effector Cell Responses and Susceptibility to Experimental Allergic Encephalomyelitis in Histamine H2 Receptor Knockout Mice Is Due to Dysregulation of Cytokine Production by Antigen-Presenting Cells. Am. J. Pathol. 2004, 164, 883–892. [Google Scholar] [CrossRef]
- Dai, H.; Kaneko, K.; Kato, H.; Fujii, S.; Jing, Y.; Xu, A.; Sakurai, E.; Kato, M.; Okamura, N.; Kuramasu, A.; et al. Selective cognitive dysfunction in mice lacking histamine H1 and H2 receptors. Neurosci. Res. 2007, 57, 306–313. [Google Scholar] [CrossRef]
- Mobarakeh, J.I.; Takahashi, K.; Sakurada, S.; Kuramasu, A.; Yanai, K. Enhanced antinociceptive effects of morphine in histamine H2 receptor gene knockout mice. Neuropharmacology 2006, 51, 612–622. [Google Scholar] [CrossRef]
- Nieto-Alamilla, G.; Márquez-Gómez, R.; García-Gálvez, A.-M.; Morales-Figueroa, G.-E.; Arias-Montaño, J.-A. The Histamine H3 Receptor: Structure, Pharmacology, and Function. Mol. Pharmacol. 2016, 90, 649–673. [Google Scholar] [CrossRef] [PubMed]
- Singh, M.; Jadhav, H.R. Histamine H3 Receptor Function and Ligands: Recent Developments. Mini-Rev. Med. Chem. 2013, 13, 47–57. [Google Scholar] [CrossRef]
- Kotańska, M.; Kuder, K.J.; Szczepańska, K.; Sapa, J.; Kieć-Kononowicz, K. The histamine H3 receptor inverse agonist pitolisant reduces body weight in obese mice. Naunyn-Schmiedeberg’s Arch. Pharmacol. 2018, 391, 875–881. [Google Scholar] [CrossRef] [PubMed]
- Abdulrazzaq, Y.M.; Bastaki, S.M.; Adeghate, E. Histamine H3 receptor antagonists—Roles in neurological and endocrine diseases and diabetes mellitus. Biomed. Pharmacother. 2022, 19, 112947. [Google Scholar] [CrossRef] [PubMed]
- Sander, L.E.; Lorentz, A.; Sellge, G.; Coëffier, M.; Neipp, M.; Veres, T.; Frieling, T.; Meier, P.; Manns, M.; Bischoff, S. Selective expression of histamine receptors H1R, H2R, and H4R, but not H3R, in the human intestinal tract. Gut 2006, 55, 498–504. [Google Scholar] [CrossRef] [PubMed]
- Schneider, E.H.; Seifert, R. The histamine H4-receptor and the central and peripheral nervous system: A critical analysis of the literature. Neuropharmacology 2016, 106, 116–128. [Google Scholar] [CrossRef]
- Thangam, E.B.; Jemima, E.A.; Singh, H.; Baig, M.S.; Khan, M.; Mathias, C.B.; Church, M.K.; Saluja, R. The Role of Histamine and Histamine Receptors in Mast Cell-Mediated Allergy and Inflammation: The Hunt for New Therapeutic Targets. Front. Immunol. 2018, 9, 1873. [Google Scholar] [CrossRef]
- Cataldi, M.; Borriello, F.; Granata, F.; Annunziato, L.; Marone, G. Histamine receptors and antihistamines: From discovery to clinical applications. Chem. Immunol. Allergy 2014, 100, 214–226. [Google Scholar] [CrossRef]
- Deiteren, A.; De Man, J.G.; Pelckmans, P.A.; De Winter, B.Y. Histamine H4 Receptors in the Gastrointestinal Tract: H4 Receptors in the Gastrointestinal Tract. Br. J. Pharmacol. 2015, 172, 1165–1178. [Google Scholar] [CrossRef]
- Pfanzagl, B.; Mechtcheriakova, D.; Meshcheryakova, A.; Aberle, S.W.; Pfragner, R.; Jensen-Jarolim, E. Activation of the ileal neuroendocrine tumor cell line P-STS by acetylcholine is amplified by histamine: Role of H3R and H4R. Sci. Rep. 2017, 7, 1313. [Google Scholar] [CrossRef]
- Beghdadi, W.; Porcherie, A.; Schneider, B.S.; Dubayle, D.; Peronet, R.; Huerre, M.; Watanabe, T.; Ohtsu, H.; Louis, J.; Mécheri, S. Inhibition of histamine-mediated signaling confers significant protection against severe malaria in mouse models of disease. J. Exp. Med. 2008, 205, 395–408. [Google Scholar] [CrossRef] [PubMed]
- Metz, M.; Doyle, E.; Bindslev-Jensen, C.; Watanabe, T.; Zuberbier, T.; Maurer, M. Effects of Antihistamines on Innate Immune Responses to Severe Bacterial Infection in Mice. Int. Arch. Allergy Immunol. 2011, 155, 355–360. [Google Scholar] [CrossRef] [PubMed]
- Sullivant, A.; Mackin, A.; Pharr, T.; Cooley, J.; Wills, R.; Archer, T. Identification of histamine receptors in the canine gastrointestinal tract. Vet. Immunol. Immunopathol. 2016, 182, 29–36. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Dwyer, L.; Song, J.H.; Martin-Cano, F.E.; Bahney, J.; Peri, L.; Britton, F.C.; Sanders, K.M.; Koh, S.D. Identification of histamine receptors and effects of histamine on murine and simian colonic excitability. Neurogastroenterol. Motil. 2011, 23, 949-e409. [Google Scholar] [CrossRef] [PubMed]
- Schirmer, B.; Neumann, D. The Function of the Histamine H4 Receptor in Inflammatory and Inflammation-Associated Diseases of the Gut. Int. J. Mol. Sci. 2021, 22, 6116. [Google Scholar] [CrossRef]
- Scholl, I.; Untersmayr, E.; Bakos, N.; Roth-Walter, F.; Gleiss, A.; Boltz-Nitulescu, G.; Scheiner, O.; Jensen-Jarolim, E. Antiulcer drugs promote oral sensitization and hypersensitivity to hazelnut allergens in BALB/c mice and humans. Am. J. Clin. Nutr. 2005, 81, 154–160. [Google Scholar] [CrossRef]
- Anvari, S.; Miller, J.; Yeh, C.-Y.; Davis, C.M. IgE-Mediated Food Allergy. Clin. Rev. Allergy Immunol. 2019, 57, 244–260. [Google Scholar] [CrossRef]
- Raithel, M.; Weidenhiller, M.; Abel, R.; Baenkler, H.; Hahn, E. Colorectal mucosal histamine release by mucosa oxygenation in comparison with other established clinical tests in patients with gastrointestinally mediated allergy. World J. Gastroenterol. 2006, 12, 4699–4705. [Google Scholar] [CrossRef][Green Version]
- Raithel, M.; Matek, M.; Baenkler, H.W.; Jorde, W.; Hahn, E.G. Mucosal histamine content and histamine secretion in Crohn’s disease, ulcerative colitis and allergic enteropathy. Int. Arch. Allergy Immunol. 1995, 108, 127–133. [Google Scholar] [CrossRef]
- Bijlsma, P.B.; Backhaus, B.; Weidenhiller, M.; Donhauser, N.; Hahn, E.G.; Raithel, M. Food llergy diagnosis by detection of antigeninduced electrophysiological changes and histamine release in human intestinal biopsies during mucosa-oxygenation. Inflamm. Res. 2004, 53 (Suppl. 1), S29–S30. [Google Scholar] [CrossRef]
- Nolte, H.; Schiotz, P.O.; Kruse, A.; Skov, P.S. Comparison of intestinal mast-cell and basophil histamine-release in children with food allergic reactions. Allergy 1989, 44, 554–565. [Google Scholar] [CrossRef] [PubMed]
- Raithel, M.; Hagel, A.F.; Zopf, Y.; Bijlsma, P.B.; De Rossi, T.M.; Gabriel, S.; Weidenhiller, M.; Kressel, J.; Hahn, E.G.; Konturek, P.C. Analysis of immediate ex vivo release of nitric oxide from human colonic mucosa in gastrointestinally mediated allergy, inflammatory bowel disease and controls. J. Physiol. Pharmacol. 2012, 63, 317–325. [Google Scholar] [PubMed]
- Nowak-Wegrzyn, A.; Assa’Ad, A.H.; Bahna, S.L.; Bock, S.A.; Sicherer, S.H.; Teuber, S.S. Work Group report: Oral food challenge testing. J. Allergy Clin. Immunol. 2009, 123 (Suppl. 6), S365–S383. [Google Scholar] [CrossRef]
- Ramsey, N.; Berin, M.C. Pathogenesis of IgE-mediated food allergy and implications for future immunotherapeutics. Pediatr. Allergy Immunol. 2021, 32, 1416–1425. [Google Scholar] [CrossRef]
- Chinthrajah, R.S.; Purington, N.; Andorf, S.; Long, A.; O’Laughlin, K.L.; Lyu, S.C.; Manohar, M.; Boyd, S.D.; Tibshirani, R.; Maecker, H.; et al. Sustained outcomes in oral immunotherapy for peanut allergy (POISED study): A large, randomised, double-blind, placebo-controlled, phase 2 study. Lancet 2019, 394, 1437–1449. [Google Scholar] [CrossRef]
- Anagnostou, K.; Islam, S.; King, Y.; Foley, L.; Pasea, L.; Bond, S.; Palmer, C.; Deighton, J.; Ewan, P.; Clark, A. Assessing the efficacy of oral immunotherapy for the desensitisation of peanut allergy in children (STOP II): A phase 2 randomised controlled trial. Lancet 2014, 383, 1297–1304. [Google Scholar] [CrossRef]
- Peanut Allergen Powder (Palforzia). JAMA 2020, 324, 192–193. [CrossRef]
- Kacik, J.; Wróblewska, B.; Lewicki, S.; Zdanowski, R.; Kalicki, B. Serum Diamine Oxidase in Pseudoallergy in the Pediatric Population. Adv. Exp. Med. Biol. 2017, 1039, 35–44. [Google Scholar] [CrossRef]
- Schwelberger, H.G. Histamine intolerance: Overestimated or underestimated? Inflamm. Res. 2009, 58 (Suppl. 1), 51–52. [Google Scholar] [CrossRef]
- Schnedl, W.J.; Enko, D. Histamine Intolerance Originates in the Gut. Nutrients 2021, 13, 1262. [Google Scholar] [CrossRef]
- Schink, M.; Konturek, P.C.; Tietz, E.; Dieterich, W.; Pinzer, T.C.; Wirtz, S.; Neurath, M.F.; Zopf, Y. Microbial patterns in patients with histamine intolerance. J. Physiol. Pharmacol. 2018, 69, 579–593. [Google Scholar] [CrossRef]
- Sanchez-Perez, S.; Comas-Baste, O.; Duelo, A.; Veciana-Nogués, M.T.; Berlanga, M.; Latorre-Moratalla, M.L.; Vidal-Carou, M.C. Intestinal Dysbiosis in Patients with Histamine Intolerance. Nutrients 2022, 14, 1774. [Google Scholar] [CrossRef]
- Schwelberger, H.G. Histamine intolerance: A metabolic disease? Inflamm. Res. 2010, 59 (Suppl. 2), 219–221. [Google Scholar] [CrossRef] [PubMed]
- Dyer, J.; Warren, K.; Merlin, S.; Metcalfe, D.D.; Kaliner, M. Measurement of plasma histamine: Description of an improved method and normal values. J. Allergy Clin. Immunol. 1982, 70, 82–87. [Google Scholar] [CrossRef]
- Hrubisko, M.; Danis, R.; Huorka, M.; Wawruch, M. Histamine Intolerance—The More We Know the Less We Know. A Review. Nutrients 2021, 13, 2228. [Google Scholar] [CrossRef]
- Schnedl, W.J.; Schenk, M.; Lackner, S.; Enko, D.; Mangge, H.; Forster, F. Diamine oxidase supplementation improves symptoms in patients with histamine intolerance. Food Sci. Biotechnol. 2019, 28, 1779–1784. [Google Scholar] [CrossRef]
- Martin, I.S.M.; Brachero, S.; Vilar, E.G. Histamine intolerance and dietary management: A complete review. Allergol. Immunopathol. 2016, 44, 475–483. [Google Scholar] [CrossRef]
- Mokhtar, S.; Mostafa, G.; Taha, R.; Eldeep, G.S.S. Effect of different starter cultures on the biogenic amines production as a critical control point in fresh fermented sausages. Eur. Food Res. Technol. 2012, 235, 527–535. [Google Scholar] [CrossRef]
- Winterkamp, S.; Weidenhiller, M.; Otte, P.; Stolper, J.; Schwab, D.; Hahn, E.G.; Raithel, M. Urinary Excretion of N-Methylhistamine as a Marker of Disease Activity in Inflammatory Bowel Disease. Am. J. Gastroenterol. 2002, 97, 3071–3077. [Google Scholar] [CrossRef]
- Hagel, A.F.; de Rossi, T.; Konturek, P.C.; Lbrecht, H.; Lker, S.; Hahn, E.G.; Raithel, M. Plasma histamine and tumour necrosis factor-alpha levels in Crohn’s disease and ulcerative colitis at various stages of disease. J. Physiol. Pharmacol. 2015, 66, 549–556. [Google Scholar]
- Smolinska, S.; Groeger, D.; Perez, N.R.; Schiavi, E.; Ferstl, R.; Frei, R.; Konieczna, P.; Akdis, C.A.; Jutel, M.; O’mahony, L. Histamine Receptor 2 is Required to Suppress Innate Immune Responses to Bacterial Ligands in Patients with Inflammatory Bowel Disease. Inflamm. Bowel Dis. 2016, 22, 1575–1586. [Google Scholar] [CrossRef] [PubMed]
- Petersen, J.; Raithel, M.; Schwelberger, H.G. Histamine N-methyltransferase and diamine oxidase gene polymorphisms in patients with inflammatory and neoplastic intestinal diseases. Inflamm. Res. 2002, 51, S91–S92. [Google Scholar] [CrossRef]
- Schulze, H.A.; Hasler, R.; Mah, N.; Lu, T.; Nikolaus, S.; Costello, C.M.; Schreiber, S. From model cell line to in vivo gene expression: Disease-related intestinal gene expression in IBD. Genes Immun. 2008, 9, 240–248. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Borghini, R.; Donato, G.; Alvaro, D.; Picarelli, A. New insights in IBS-like disorders: Pandora’s box has been opened; a review. Gastroenterol. Hepatol. Bed Bench 2017, 10, 79–89. [Google Scholar]
- Mac Sharry, J.; O’Mahony, L.; Fanning, A.; Bairead, E.; Sherlock, G.; Tiesman, J.; Fulmer, A.; Kiely, B.; Dinan, T.G.; Shanahan, F.; et al. Mucosal cytokine imbalance in irritable bowel syndrome. Scand. J. Gastroenterol. 2008, 43, 1467–1476. [Google Scholar] [CrossRef]
- Dinan, T.G.; Quigley, E.M.M.; Ahmed, S.M.M.; Scully, P.; O’Brien, S.; O’Mahony, L.; O’Mahony, S.; Shanahan, F.; Keeling, P.W.N. Hypothalamic-Pituitary-Gut Axis Dysregulation in Irritable Bowel Syndrome: Plasma Cytokines as a Potential Biomarker? Gastroenterology 2006, 130, 304–311. [Google Scholar] [CrossRef]
- Bohn, L.; Störsrud, S.; Törnblom, H.; Bengtsson, U.; Simrén, M. Self-Reported Food-Related Gastrointestinal Symptoms in IBS Are Common and Associated With More Severe Symptoms and Reduced Quality of Life. Am. J. Gastroenterol. 2013, 108, 634–641. [Google Scholar] [CrossRef]
- Barbara, G.; Stanghellini, V.; De Giorgio, R.; Cremon, C.; Cottrell, G.S.; Santini, D.; Pasquinelli, G.; Morselli/labate, A.M.; Grady, E.F.; Bunnett, N.W.; et al. Activated mast cells in proximity to colonic nerves correlate with abdominal pain in irritable bowel syndrome. Gastroenterology 2004, 126, 693–702. [Google Scholar] [CrossRef]
- Klooker, T.K.; Braak, B.; Koopman, K.; Welting, O.; Wouters, M.M.; Van Der Heide, S.; Schemann, M.; Bischoff, S.C.; Wijngaard, R.M.V.D.; Boeckxstaens, G.E. The mast cell stabiliser ketotifen decreases visceral hypersensitivity and improves intestinal symptoms in patients with irritable bowel syndrome. Gut 2010, 59, 1213–1221. [Google Scholar] [CrossRef]
- Buhner, S.; Li, Q.; Vignali, S.; Barbara, G.; De Giorgio, R.; Stanghellini, V.; Cremon, C.; Zeller, F.; Langer, R.; Daniel, H.; et al. Activation of human enteric neurons by supernatants of colonic biopsy specimens from patients with irritable bowel syndrome. Gastroenterology 2009, 137, 1425–1434. [Google Scholar] [CrossRef]
- Chen, H.; Nwe, P.-K.; Yang, Y.; Rosen, C.E.; Bielecka, A.A.; Kuchroo, M.; Cline, G.W.; Kruse, A.C.; Ring, A.M.; Crawford, J.M.; et al. A Forward Chemical Genetic Screen Reveals Gut Microbiota Metabolites That Modulate Host Physiology. Cell 2019, 177, 1217–1231.e18. [Google Scholar] [CrossRef] [PubMed]
- Barbara, G.; Wang, B.; Stanghellini, V.; de Giorgio, R.; Cremon, C.; Di Nardo, G.; Trevisani, M.; Campi, B.; Geppetti, P.; Tonini, M.; et al. Mast cell-dependent excitation of visceral-nociceptive sensory neurons in irritable bowel syndrome. Gastroenterology 2007, 132, 26–37. [Google Scholar] [CrossRef] [PubMed]
- Mishima, Y.; Ishihara, S. Molecular Mechanisms of Microbiota-Mediated Pathology in Irritable Bowel Syndrome. Int. J. Mol. Sci. 2020, 21, 8664. [Google Scholar] [CrossRef] [PubMed]
- Codling, C.; O’Mahony, L.; Shanahan, F.; Quigley, E.M.M.; Marchesi, J.R. A Molecular Analysis of Fecal and Mucosal Bacterial Communities in Irritable Bowel Syndrome. Am. J. Dig. Dis. 2009, 55, 392–397. [Google Scholar] [CrossRef] [PubMed]
- O’Mahony, L.; McCarthy, J.; Kelly, P.; Hurley, G.; Luo, F.; Chen, K.; O’Sullivan, G.C.; Kiely, B.; Collins, J.K.; Shanahan, F.; et al. Lactobacillus and bifidobacterium in irritable bowel syndrome: Symptom responses and relationship to cytokine profiles. Gastroenterology 2005, 128, 541–551. [Google Scholar] [CrossRef]
- Whorwell, P.J.; Altringer, L.; Morel, J.; Bond, Y.; Charbonneau, D.; O’Mahony, L.; Kiely, B.; Shanahan, F.; Quigley, E.M.M. Efficacy of an encapsulated probiotic Bifidobacterium infantis 35624 in women with irritable bowel syndrome. Am. J. Gastroenterol. 2006, 101, 1581–1590. [Google Scholar] [CrossRef]
- Hungerford, J.M. Histamine and Scombrotoxins. Toxicon 2021, 201, 115–126. [Google Scholar] [CrossRef]
- Hui, J.Y.; Taylor, S.L. Inhibition of in vivo histamine metabolism in rats by foodborne and pharmacologic inhibitors of diamine oxidase, histamine N-methyltransferase, and monoamine oxidase. Toxicol. Appl. Pharmacol. 1985, 81, 241–249. [Google Scholar] [CrossRef]
- Ijomah, P.; Clifford, M.N.; Walker, R.; Wright, J.; Hardy, R.; Murray, C.K. The importance of endogenous histamine relative to dietary histamine in the aetiology of scombrotoxicosis. Food Addit. Contam. 1991, 8, 531–542. [Google Scholar] [CrossRef]
- Siegel, R.; DeSantis, C.; Jemal, A. Colorectal cancer statistics. CA Cancer J. Clin. 2014, 64, 104–117. [Google Scholar] [CrossRef]
- Jess, T.; Simonsen, J.; Jørgensen, K.T.; Pedersen, B.V.; Nielsen, N.M.; Frisch, M. Decreasing risk of colorectal cancer in patients with inflammatory bowel disease over 30 years. Gastroenterology 2012, 143, 375–381.e1. [Google Scholar] [CrossRef] [PubMed]
- Herrinton, L.J.; Liu, L.; Levin, T.R.; Allison, J.E.; Lewis, J.D.; Velayos, F. Incidence and mortality of colorectal adenocarcinoma in persons with inflammatory bowel disease from 1998 to 2010. Gastroenterology 2012, 143, 382–389. [Google Scholar] [CrossRef] [PubMed]
- Beaugerie, L.; Svrcek, M.; Seksik, P.; Bouvier, A.; Simon, T.; Allez, M.; Brixi, H.; Gornet, J.; Altwegg, R.; Beau, P.; et al. CESAME Study Group: Risk of colorectal high-grade dysplasia and cancer in a prospective observational cohort of patients with inflammatory bowel disease. Gastroenterology 2013, 145, 166–175.e8. [Google Scholar] [CrossRef] [PubMed]
- Louis, P.; Hold, G.L.; Flint, H.J. The gut microbiota, bacterial metabolites and colorectal cancer. Nat. Rev. Microbiol. 2014, 12, 661–672. [Google Scholar] [CrossRef] [PubMed]
- Sears, C.L.; Garrett, W.S. Microbes, Microbiota, and Colon Cancer. Cell Host Microbe 2014, 15, 317–328. [Google Scholar] [CrossRef] [PubMed]
- Schwabe, R.F.; Jobin, C. The microbiome and cancer. Nat. Rev. Cancer 2013, 13, 800–812. [Google Scholar] [CrossRef] [PubMed]
- Tjalsma, H.; Boleij, A.; Marchesi, J.R.; Dutilh, B.E. A bacterial Driver–passenger model for colorectal cancer: Beyond the usual suspects. Nat. Rev. Microbiol. 2012, 10, 575–582. [Google Scholar] [CrossRef]
- Arthur, J.C.; Perez-Chanona, E.; Mühlbauer, M.; Tomkovich, S.; Uronis, J.M.; Fan, T.-J.; Campbell, B.J.; Abujamel, T.; Dogan, B.; Rogers, A.B.; et al. Intestinal Inflammation Targets Cancer-Inducing Activity of the Microbiota. Science 2012, 338, 120–123. [Google Scholar] [CrossRef]
- Arthur, J.C.; Gharaibeh, R.Z.; Mühlbauer, M.; Perez-Chanona, E.; Uronis, J.M.; McCafferty, J.; Fodor, A.A.; Jobin, C. Microbial genomic analysis reveals the essential role of inflammation in bacteria-induced colorectal cancer. Nat. Commun. 2014, 5, 4724. [Google Scholar] [CrossRef]
- Goodwin, A.C.; Destefano Shields, C.E.; Wu, S.; Huso, D.L.; Wu, X.; Murray-Stewart, T.R.; Hacker-Prietz, A.; Rabizadeh, S.; Woster, P.M.; Sears, C.L.; et al. Polyamine catabolism contributes to enterotoxigenic Bacteroides fragilis-induced colon tumorigenesis. Proc. Natl. Acad. Sci. USA 2011, 108, 15354–15359. [Google Scholar] [CrossRef]
- Rowland, I.R.; Weisz, J.; Fritz-Wolz, G.; Clawson, G.; Benedict, C.M.; Abendroth, C.; Creveling, C.R. Effect of Bifidobacterium longum and inulin on gut bacterial metabolism and carcinogen-induced aberrant crypt foci in rats. Carcinogenesis 1998, 19, 281–285. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.-C.; Lin, W.-C.; Kong, M.-S.; Shi, H.N.; Walker, W.A.; Lin, C.-Y.; Huang, C.-T.; Lin, Y.-C.; Jung, S.-M.; Lin, T.-Y. Oral inoculation of probiotics Lactobacillus acidophilus NCFM suppresses tumour growth both in segmental orthotopic colon cancer and extra-intestinal tissue. Br. J. Nutr. 2012, 107, 1623–1634. [Google Scholar] [CrossRef] [PubMed]
- Verma, A.; Shukla, G. Probiotics Lactobacillus rhamnosus GG, Lactobacillus acidophilus suppresses DMH-induced procarcinogenic fecal enzymes and preneoplastic aberrant crypt foci in early colon carcinogenesis in Sprague Dawley rats. Nutr. Cancer 2013, 65, 84–91. [Google Scholar] [CrossRef]
- Cianchi, F.; Cortesini, C.; Schiavone, N.; Perna, F.; Magnelli, L.; Fanti, E.; Bani, D.; Messerini, L.; Fabbroni, V.; Perigli, G.; et al. The role of cyclooxygenase-2 in mediating the effects of histamine on cell proliferation and vascular endothelial growth factor production in colorectal cancer. Clin. Cancer Res. 2005, 11, 6807–6815. [Google Scholar] [CrossRef] [PubMed]
- Masini, E.; Fabbroni, V.; Giannini, L.; Vannacci, A.; Messerini, L.; Perna, F.; Cortesini, C.; Cianchi, F. Histamine and histidine decarboxylase up-regulation in colorectal cancer: Correlation with tumor stage. Agents Actions 2005, 54, S80–S81. [Google Scholar] [CrossRef]
- Gao, C.; Ganesh, B.P.; Shi, Z.; Shah, R.R.; Fultz, R.; Major, A.; Venable, S.; Lugo, M.; Hoch, K.; Chen, X.; et al. Gut Microbe–Mediated Suppression of Inflammation-Associated Colon Carcinogenesis by Luminal Histamine Production. Am. J. Pathol. 2017, 187, 232–236. [Google Scholar] [CrossRef]
- Yang, X.D.; Ai, W.; Asfaha, S.; Bhagat, G.; Friedman, R.A.; Jin, G.; Park, H.; Shykind, B.; Diacovo, T.G.; Falus, A.; et al. Histamine deficiency promotes inflammation-associated carcinogenesis through reduced myeloid maturation and accumulation of CD11b+Ly6G+ immature myeloid cells. Nat. Med. 2011, 17, 87–95. [Google Scholar] [CrossRef]
- Chen, X.; Churchill, M.; Nagar, K.K.; Tailor, Y.H.; Chu, T.; Rush, B.S.; Jiang, Z.; Wang, E.B.; Renz, B.W.; Wang, H.; et al. IL-17 producing mast cells promote the expansion of myeloid-derived suppressor cells in a mouse allergy model of colorectal cancer. Oncotarget 2015, 6, 32966–32979. [Google Scholar] [CrossRef]
- He, G.-H.; Ding, J.-Q.; Zhang, X.; Xu, W.-M.; Lin, X.-Q.; Huang, M.-J.; Feng, J.; Wang, P.; Cai, W.-K. Activation of histamine H4 receptor suppresses the proliferation and invasion of esophageal squamous cell carcinoma via both metabolism and non-metabolism signaling pathways. Klin. Wochenschr. 2018, 96, 951–964. [Google Scholar] [CrossRef]
- Tanaka, T.; Ishikawa, H. Mast cells and inflammation-associated colorectal carcinogenesis. Semin. Immunopathol. 2013, 35, 245–254. [Google Scholar] [CrossRef]
- Malfettone, A.; Silvestris, N.; Saponaro, C.; Ranieri, G.; Russo, A.; Caruso, S.; Popescu, O.; Simone, G.; Paradiso, A.; Mangia, A. High density of tryptase-positive mast cells in human colorectal cancer: A poor prognostic factor related to protease-activated receptor 2 expression. J. Cell. Mol. Med. 2013, 17, 1025–1037. [Google Scholar] [CrossRef] [PubMed]
- Schirmer, B.; Rother, T.; Bruesch, I.; Bleich, A.; Werlein, C.; Jonigk, D.; Seifert, R.; Neumann, D. Genetic Deficiency of the Histamine H4-Receptor Reduces Experimental Colorectal Carcinogenesis in Mice. Cancers 2020, 12, 912. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Xiong, Y.; Li, J.; Yang, Y.; Liu, L.; Wang, W.; Wang, L.; Li, M.; Fang, Z. Deletion and down-regulation of HRH4 gene in gastric carcinomas: A potential correlation with tumor progression. PLoS ONE 2012, 7, e31207. [Google Scholar] [CrossRef] [PubMed]
| Symptoms of Histamine Intolerance | |
|---|---|
| Respiratory system | rhinorrhea, rhinitis, nasal congestion, dyspnea, sneezing |
| Cardiovascular system | tachycardia, hypotonia, collapse |
| Gastrointestinal system | bloating, flatulence, postprandial fullness, diarrhea, abdominal pain, constipation, nausea, vomiting |
| Reproductive system | menstrual cramps, dysmenorrhea |
| Skin | pruritus, flushing, urticarial, dermatitis, swelling |
| Nervous system | headache/migraine, dizziness, chronic inappropriate fatigue, nervousness, sleep disturbances (insomnia), anxiety, panic disorder, depression |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Smolinska, S.; Winiarska, E.; Globinska, A.; Jutel, M. Histamine: A Mediator of Intestinal Disorders—A Review. Metabolites 2022, 12, 895. https://doi.org/10.3390/metabo12100895
Smolinska S, Winiarska E, Globinska A, Jutel M. Histamine: A Mediator of Intestinal Disorders—A Review. Metabolites. 2022; 12(10):895. https://doi.org/10.3390/metabo12100895
Chicago/Turabian StyleSmolinska, Sylwia, Ewa Winiarska, Anna Globinska, and Marek Jutel. 2022. "Histamine: A Mediator of Intestinal Disorders—A Review" Metabolites 12, no. 10: 895. https://doi.org/10.3390/metabo12100895
APA StyleSmolinska, S., Winiarska, E., Globinska, A., & Jutel, M. (2022). Histamine: A Mediator of Intestinal Disorders—A Review. Metabolites, 12(10), 895. https://doi.org/10.3390/metabo12100895







